Imaging-Based Staging of Hepatic Fibrosis in Patients with Hepatitis B: A Dynamic Radiomics Model Based on Gd-EOB-DTPA-Enhanced MRI
Abstract
1. Introduction
1.1. Related Studies
1.1.1. Liver Stiffness Measurement
1.1.2. Radiomics Analysis
1.1.3. Deep Learning Model
1.2. Our Contributions
- Time-domain information was fully used based on time-varying curves and discrepancy of imaging features with the contrast enhancement process, which was found to play a critical role in all stages of classification.
- ROI extraction was based on whole-liver region segmentation using three-dimensional (3D) U-net and transfer learning, with the post-processing algorithm for interference information excluded. This method of ROI extraction replaced manual selection and delineation, eliminating the influence of region selection on the results of radiomics analysis while being more automated.
- Feature extraction was performed in liver volume and more valuable information, such as morphological changes in the liver, was considered, which facilitated the classification of cirrhosis.
2. Materials and Methods
2.1. Dataset
2.1.1. Study Population
2.1.2. MRI Protocol
2.1.3. Reference Standard
2.2. Serum Fibrosis Tests
2.3. Normalized Liver Enhancement
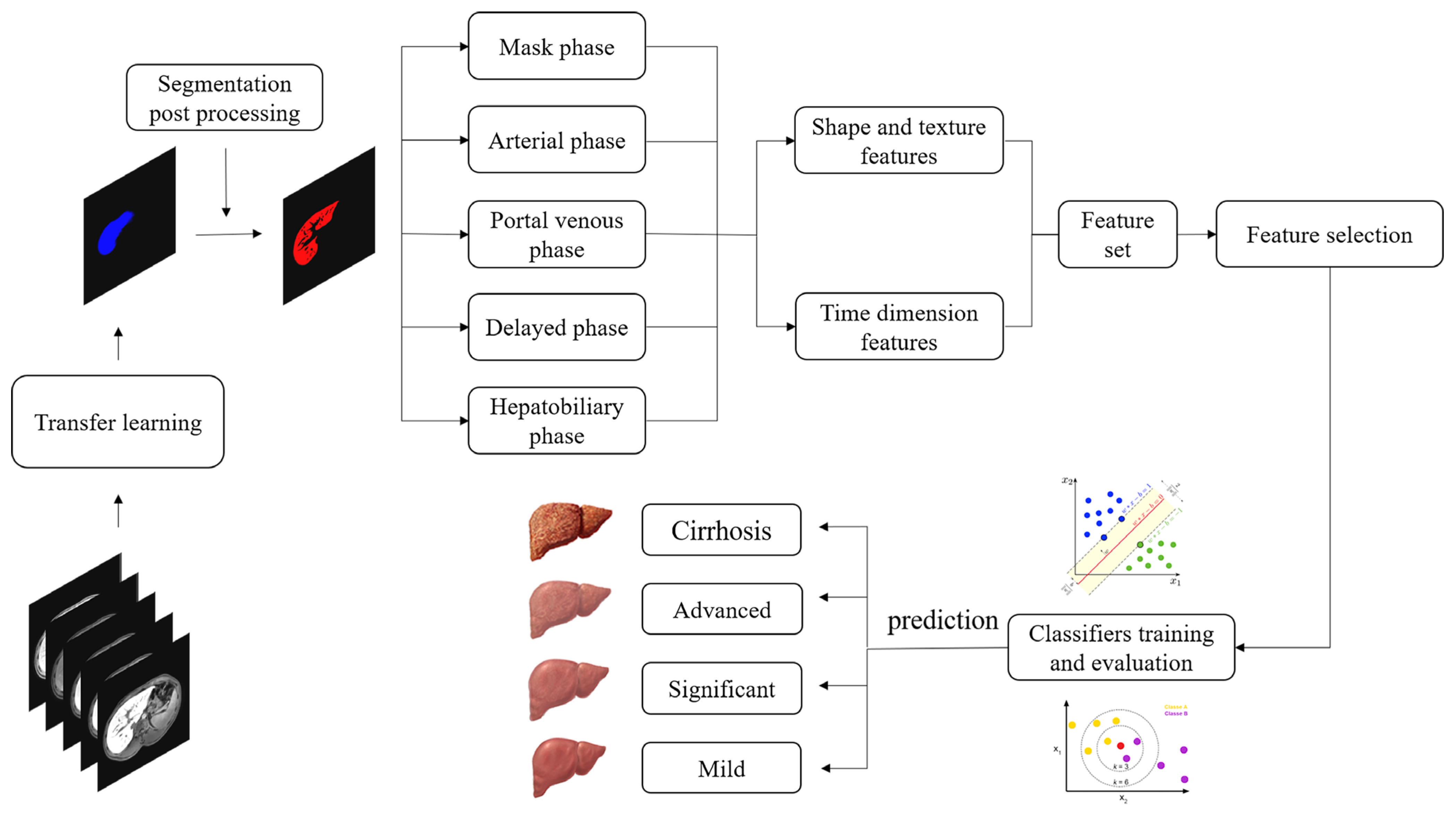
2.4. Overall Framework of the Proposed Dynamic Radiomics Model
2.5. Processing Pipeline
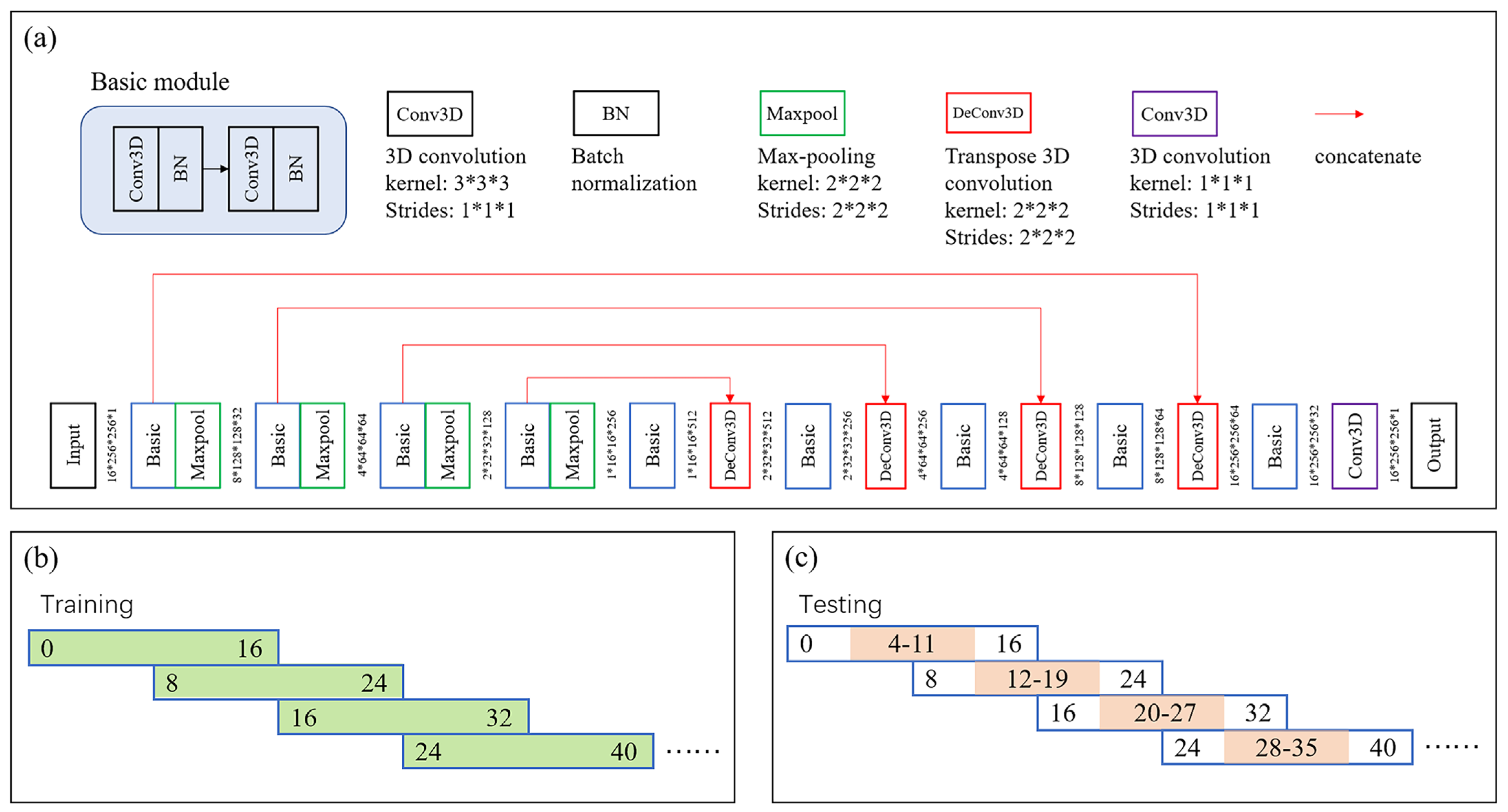
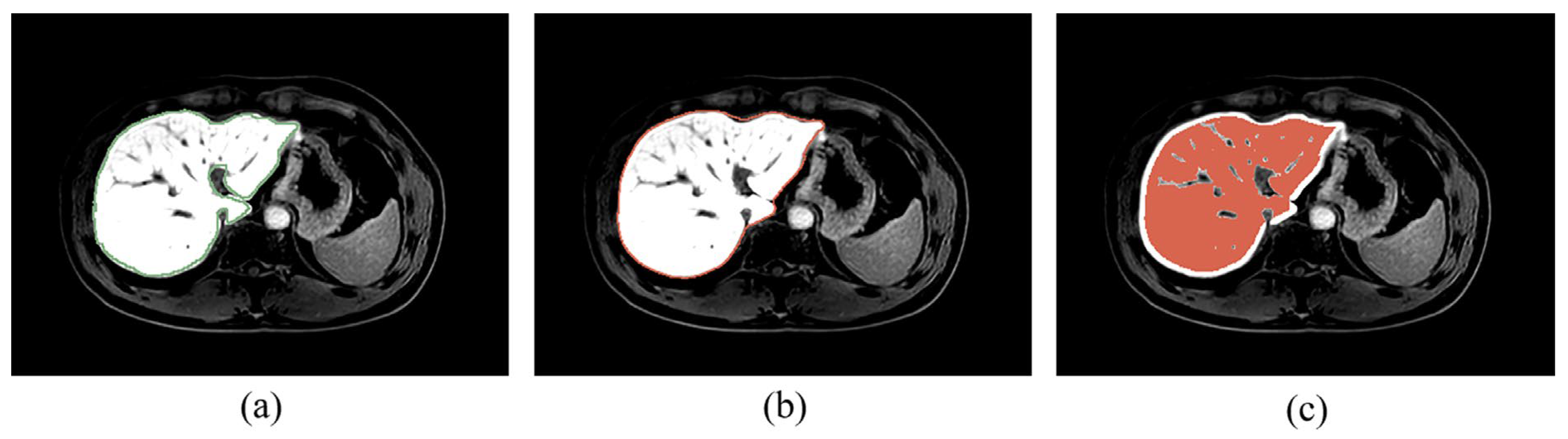
2.5.1. ROI Extraction
2.5.2. Feature Extraction
2.5.3. Feature Selection and Classification
2.6. Data Resampling
2.7. Evaluation Metrics
2.8. Implementation Details
3. Results
3.1. Liver Segmentation
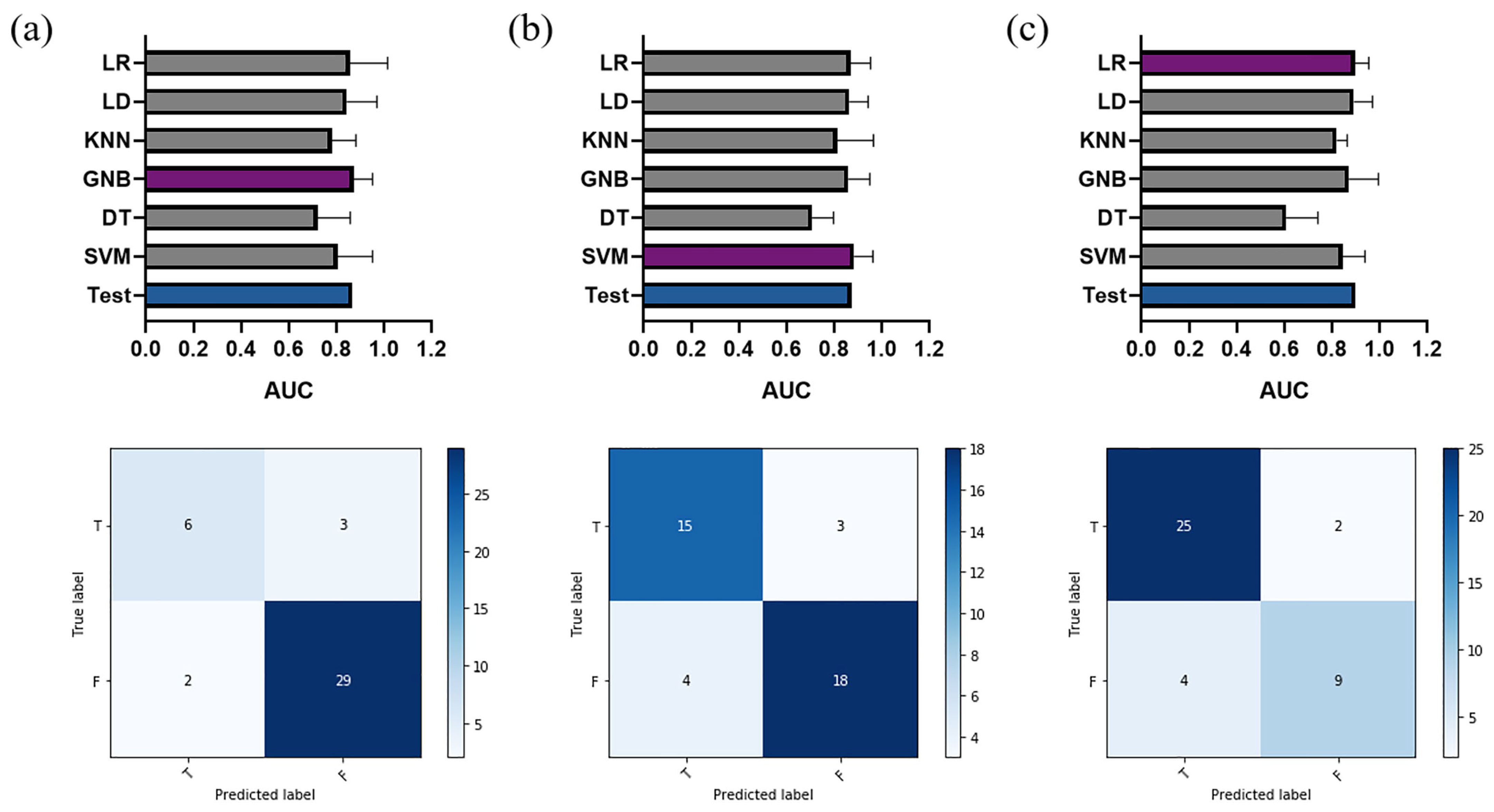
3.2. Proposed Dynamic Radiomics Model
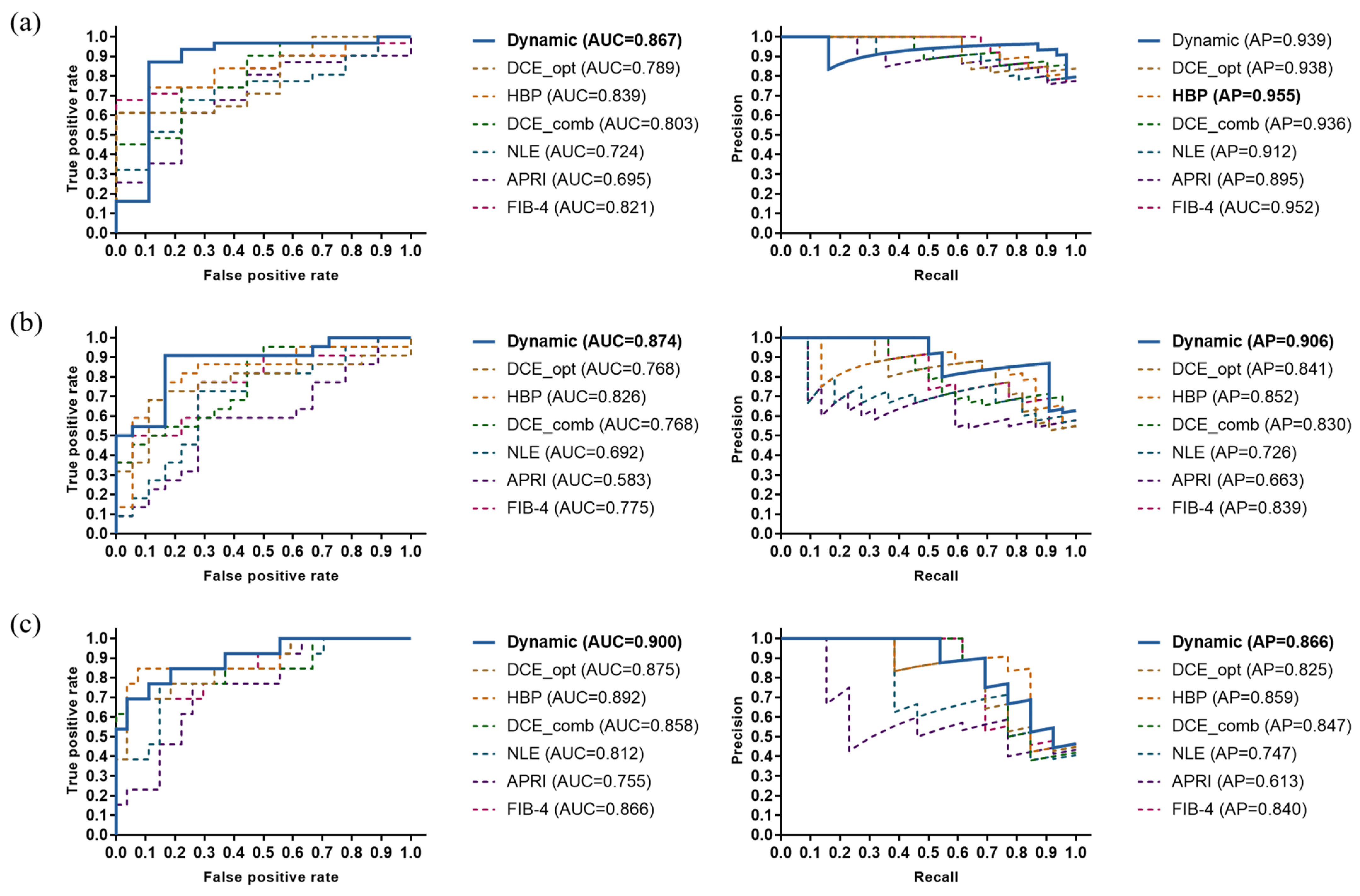
3.3. Performance Comparison
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL-ALEH Clinical Practice Guidelines: Non-invasive tests for evaluation of liver disease severity and prognosis. J. Hepatol. 2015, 63, 237–264. [Google Scholar] [CrossRef]
- Cadranel, J.F.; Rufat, P.; Degos, F. Practices of liver biopsy in France: Results of a prospective nationwide survey. For the Group of Epidemiology of the French Association for the Study of the Liver (AFEF). Hepatology 2000, 32, 477–481. [Google Scholar] [CrossRef]
- Shiha, G.; Ibrahim, A.; Helmy, A.; Sarin, S.K.; Omata, M.; Kumar, A.; Bernstien, D.; Maruyama, H.; Saraswat, V.; Chawla, Y.; et al. Asian-Pacific Association for the Study of the Liver (APASL) consensus guidelines on invasive and non-invasive assessment of hepatic fibrosis: A 2016 update. Hepatol. Int. 2017, 11, 1–30. [Google Scholar] [CrossRef]
- Chang, P.E.; Goh, G.B.; Ngu, J.H.; Tan, H.K.; Tan, C.K. Clinical applications, limitations and future role of transient elastography in the management of liver disease. World J. Gastrointest. Pharmacol. Ther. 2016, 7, 91–106. [Google Scholar] [CrossRef]
- Singh, S.; Venkatesh, S.K.; Loomba, R.; Wang, Z.; Sirlin, C.; Chen, J.; Yin, M.; Miller, F.H.; Low, R.N.; Hassanein, T.; et al. Magnetic resonance elastography for staging liver fibrosis in non-alcoholic fatty liver disease: A diagnostic accuracy systematic review and individual participant data pooled analysis. Eur. Radiol. 2016, 26, 1431–1440. [Google Scholar] [CrossRef]
- Parekh, V.; Jacobs, M.A. Radiomics: A new application from established techniques. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 207–226. [Google Scholar] [CrossRef]
- Tang, M.; Feng, S.-T.; Peng, S.; Kuang, M. IDDF2018-ABS-0098 Preoperative prediction of microvascular invasion in hepatocellular cancer: A radiomics model using GD-EOB-DTPA enhanced MRI. Gut 2018, 67, A96. [Google Scholar]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Dohan, A.; Gallix, B.; Guiu, B.; Le Malicot, K.; Reinhold, C.; Soyer, P.; Bennouna, J.; Ghiringhelli, F.; Barbier, E.; Boige, V. Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut 2020, 69, 531–539. [Google Scholar] [CrossRef]
- Ribeiro, R.T.; Marinho, R.T.; Sanches, J.M. Classification and staging of chronic liver disease from multimodal data. IEEE Trans. Biomed. Eng. 2012, 60, 1336–1344. [Google Scholar] [CrossRef]
- Duan, J.; Hu, C.; Luo, S.; Zhao, X.; Wang, T. Microcomputed tomography with diffraction-enhanced imaging for morphologic characterization and quantitative evaluation of microvessel of hepatic fibrosis in rats. PLoS ONE 2013, 8, e78176. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, X.; Liu, B.J.; Ma, K.; Yan, W.; Liling, L.; Yuhong, H.; Fujita, H. Effective staging of fibrosis by the selected texture features of liver: Which one is better, CT or MR imaging? Comput. Med. Imaging Graph. 2015, 46, 227–236. [Google Scholar] [CrossRef]
- Kato, H.; Kanematsu, M.; Zhang, X.; Saio, M.; Kondo, H.; Goshima, S.; Fujita, H. Computer-aided diagnosis of hepatic fibrosis: Preliminary evaluation of MRI texture analysis using the finite difference method and an artificial neural network. AJR Am. J. Roentgenol. 2007, 189, 117–122. [Google Scholar] [CrossRef]
- House, M.J.; Bangma, S.J.; Thomas, M.; Gan, E.K.; Ayonrinde, O.T.; Adams, L.A.; Olynyk, J.K.; St Pierre, T.G. Texture-based classification of liver fibrosis using MRI. J. Magn. Reson. Imaging 2015, 41, 322–328. [Google Scholar] [CrossRef]
- Cannella, R.; Borhani, A.A.; Tublin, M.; Behari, J.; Furlan, A. Diagnostic value of MR-based texture analysis for the assessment of hepatic fibrosis in patients with nonalcoholic fatty liver disease (NAFLD). Abdom. Radiol. 2019, 44, 1816–1824. [Google Scholar] [CrossRef]
- Wu, Z.; Matsui, O.; Kitao, A.; Kozaka, K.; Koda, W.; Kobayashi, S.; Ryu, Y.; Minami, T.; Sanada, J.; Gabata, T. Hepatitis C related chronic liver cirrhosis: Feasibility of texture analysis of MR images for classification of fibrosis stage and necroinflammatory activity grade. PLoS ONE 2015, 10, e0118297. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.S.; Park, B.; Yun, J.; Sung, Y.S.; Shim, W.H.; Shin, Y.M.; Kim, S.Y.; Lee, S.J.; Lee, M.G. Radiomics analysis of gadoxetic acid–enhanced MRI for staging liver fibrosis. Radiology 2019, 290, 380–387. [Google Scholar] [CrossRef]
- Piechnik, S.K.; Ferreira, V.M.; Dall’Armellina, E.; Cochlin, L.E.; Greiser, A.; Neubauer, S.; Robson, M.D. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J. Cardiovasc. Magn. Reson. 2010, 12, 1–11. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Sahani, D.; Amendola, M.; Balzer, T.; Breuer, J.; Brown, J.J.; Casalino, D.D.; Davis, P.L.; Francis, I.R.; Krinsky, G.; et al. Efficacy and safety of MR imaging with liver-specific contrast agent: U.S. multicenter phase III study. Radiology 2005, 237, 89–98. [Google Scholar] [CrossRef]
- Kobayashi, S.; Matsui, O.; Gabata, T.; Koda, W.; Minami, T.; Ryu, Y.; Kozaka, K.; Kitao, A. Relationship between signal intensity on hepatobiliary phase of gadolinium ethoxybenzyl diethylenetriaminepentaacetic acid (Gd-EOB-DTPA)-enhanced MR imaging and prognosis of borderline lesions of hepatocellular carcinoma. Eur. J. Radiol. 2012, 81, 3002–3009. [Google Scholar] [CrossRef]
- Verloh, N.; Utpatel, K.; Haimerl, M.; Zeman, F.; Fellner, C.; Fichtner-Feigl, S.; Teufel, A.; Stroszczynski, C.; Evert, M.; Wiggermann, P. Liver fibrosis and Gd-EOB-DTPA-enhanced MRI: A histopathologic correlation. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.R.; Lee, J.M.; Yoon, J.H.; Han, J.K.; Choi, B.I. Comparison of magnetic resonance elastography and gadoxetate disodium–enhanced magnetic resonance imaging for the evaluation of hepatic fibrosis. Invest. Radiol. 2013, 48, 607–613. [Google Scholar] [CrossRef]
- Feier, D.; Balassy, C.; Bastati, N.; Stift, J.; Badea, R.; Ba-Ssalamah, A. Liver fibrosis: Histopathologic and biochemical influences on diagnostic efficacy of hepatobiliary contrast-enhanced MR imaging in staging. Radiology 2013, 269, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Verloh, N.; Probst, U.; Utpatel, K.; Zeman, F.; Brennfleck, F.; Werner, J.M.; Fellner, C.; Stroszczynski, C.; Evert, M.; Wiggermann, P.; et al. Influence of hepatic fibrosis and inflammation: Correlation between histopathological changes and Gd-EOB-DTPA-enhanced MR imaging. PLoS ONE 2019, 14, e0215752. [Google Scholar] [CrossRef] [PubMed]
- Haimerl, M.; Verloh, N.; Zeman, F.; Fellner, C.; Nickel, D.; Lang, S.A.; Teufel, A.; Stroszczynski, C.; Wiggermann, P. Gd-EOB-DTPA-enhanced MRI for evaluation of liver function: Comparison between signal-intensity-based indices and T1 relaxometry. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Haimerl, M.; Schlabeck, M.; Verloh, N.; Zeman, F.; Fellner, C.; Nickel, D.; Barreiros, A.P.; Loss, M.; Stroszczynski, C.; Wiggermann, P. Volume-assisted estimation of liver function based on Gd-EOB-DTPA-enhanced MR relaxometry. Eur. Radiol. 2016, 26, 1125–1133. [Google Scholar] [CrossRef] [PubMed]
- Barr, R.G.; Ferraioli, G.; Palmeri, M.L.; Goodman, Z.D.; Garcia-Tsao, G.; Rubin, J.; Garra, B.; Myers, R.P.; Wilson, S.R.; Rubens, D.; et al. Elastography assessment of liver fibrosis: Society of radiologists in ultrasound consensus conference statement. Radiology 2015, 276, 845–861. [Google Scholar] [CrossRef] [PubMed]
- Cassinotto, C.; Lapuyade, B.; Ait-Ali, A.; Vergniol, J.; Gaye, D.; Foucher, J.; Bailacq-Auder, C.; Chermak, F.; Le Bail, B.; de Ledinghen, V. Liver fibrosis: Noninvasive assessment with acoustic radiation force impulse elastography—Comparison with FibroScan M and XL probes and FibroTest in patients with chronic liver disease. Radiology 2013, 269, 283–292. [Google Scholar] [CrossRef]
- Friedrich-Rust, M.; Nierhoff, J.; Lupsor, M.; Sporea, I.; Fierbinteanu-Braticevici, C.; Strobel, D.; Takahashi, H.; Yoneda, M.; Suda, T.; Zeuzem, S.; et al. Performance of Acoustic Radiation Force Impulse imaging for the staging of liver fibrosis: A pooled meta-analysis. J. Viral Hepat. 2012, 19, e212–e219. [Google Scholar] [CrossRef]
- Bota, S.; Sporea, I.; Sirli, R.; Popescu, A.; Danila, M.; Costachescu, D. Intra- and interoperator reproducibility of acoustic radiation force impulse (ARFI) elastography—Preliminary results. Ultrasound Med. Biol. 2012, 38, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.K.; Yin, M.; Ehman, R.L. Magnetic resonance elastography of liver: Technique, analysis, and clinical applications. J. Magn. Reson. Imaging 2013, 37, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Rouviere, O.; Yin, M.; Dresner, M.A.; Rossman, P.J.; Burgart, L.J.; Fidler, J.L.; Ehman, R.L. MR elastography of the liver: Preliminary results. Radiology 2006, 240, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Huwart, L.; Sempoux, C.; Vicaut, E.; Salameh, N.; Annet, L.; Danse, E.; Peeters, F.; ter Beek, L.C.; Rahier, J.; Sinkus, R.; et al. Magnetic resonance elastography for the noninvasive staging of liver fibrosis. Gastroenterology 2008, 135, 32–40. [Google Scholar] [CrossRef]
- Asbach, P.; Klatt, D.; Hamhaber, U.; Braun, J.; Somasundaram, R.; Hamm, B.; Sack, I. Assessment of liver viscoelasticity using multifrequency MR elastography. Magn. Reson. Med. 2008, 60, 373–379. [Google Scholar] [CrossRef]
- Low, G.; Kruse, S.A.; Lomas, D.J. General review of magnetic resonance elastography. World J. Radiol. 2016, 8, 59. [Google Scholar] [CrossRef]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut 2019, 68, 729–741. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Deep learning for staging liver fibrosis on CT: A pilot study. Eur. Radiol. 2018, 28, 4578–4585. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Kunimatsu, A.; Abe, O.; Kiryu, S. Liver fibrosis: Deep convolutional neural network for staging by using gadoxetic acid–enhanced hepatobiliary phase MR images. Radiology 2018, 287, 146–155. [Google Scholar] [CrossRef]
- Choi, K.J.; Jang, J.K.; Lee, S.S.; Sung, Y.S.; Shim, W.H.; Kim, H.S.; Yun, J.; Choi, J.-Y.; Lee, Y.; Kang, B.-K.; et al. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent–enhanced CT Images in the Liver. Radiology 2018, 289, 688–697. [Google Scholar] [CrossRef]
- Scheuer, P.J. Classification of chronic viral hepatitis: A need for reassessment. J. Hepatol. 1991, 13, 372–374. [Google Scholar] [CrossRef]
- Xiao, G.; Yang, J.; Yan, L. Comparison of diagnostic accuracy of aspartate aminotransferase to platelet ratio index and fibrosis-4 index for detecting liver fibrosis in adult patients with chronic hepatitis B virus infection: A systemic review and meta-analysis. Hepatology 2015, 61, 292–302. [Google Scholar] [CrossRef]
- Wai, C.-T.; Greenson, J.K.; Fontana, R.J.; Kalbfleisch, J.D.; Marrero, J.A.; Conjeevaram, H.S.; Lok, A.S.-F. A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 2003, 38, 518–526. [Google Scholar] [CrossRef]
- Sterling, R.K.; Lissen, E.; Clumeck, N.; Sola, R.; Correa, M.C.; Montaner, J.; Sulkowski, M.S.; Torriani, F.J.; Dieterich, D.T.; Thomas, D.L.; et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology 2006, 43, 1317–1325. [Google Scholar] [CrossRef]
- Avants, B.B.; Epstein, C.L.; Grossman, M.; Gee, J.C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008, 12, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Sankur, B.; Sezgin, M. Image thresholding techniques: A survey over categories. Pattern Recognit. 2001, 34, 1573–1583. [Google Scholar]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Bermingham, M.L.; Pong-Wong, R.; Spiliopoulou, A.; Hayward, C.; Rudan, I.; Campbell, H.; Wright, A.F.; Wilson, J.F.; Agakov, F.; Navarro, P.; et al. Application of high-dimensional feature selection: Evaluation for genomic prediction in man. Sci. Rep. 2015, 5, 10312. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Int. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Blagus, R.; Lusa, L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinformatics 2013, 14, 106. [Google Scholar] [CrossRef]
- Avanzo, M.; Pirrone, G.; Vinante, L.; Caroli, A.; Stancanello, J.; Drigo, A.; Massarut, S.; Mileto, M.; Urbani, M.; Trovo, M. Electron density and Biologically Effective Dose (BED) radiomics-based machine learning models to predict late radiation-induced subcutaneous fibrosis. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Guido, M.; Mangia, A.; Faa, G. Chronic viral hepatitis: The histology report. Dig. Liver Dis. 2011, 43, S331–S343. [Google Scholar] [CrossRef]
- Rahn, S.B. Liver biopsy interpretation in chronic hepatitis. J. Insur. Med. 2001, 33, 110–113. [Google Scholar] [PubMed]
- Watanabe, H.; Kanematsu, M.; Goshima, S.; Kondo, H.; Onozuka, M.; Moriyama, N.; Bae, K.T. Staging hepatic fibrosis: Comparison of gadoxetate disodium–enhanced and diffusion-weighted MR imaging—preliminary observations. Radiology 2011, 259, 142–150. [Google Scholar] [CrossRef]
- Wolf, D.C. Evaluation of the Size, Shape, and Consistency of the Liver. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef]
- Yang, F.; Dogan, N.; Stoyanova, R.; Ford, J.C. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys. Med. 2018, 50, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Holloway, L.C.; Brink, C.; Field, M.; Christiansen, R.L.; Sun, Y.; Barton, M.B.; Liney, G.P. Multicenter evaluation of MRI-based radiomic features: A phantom study. Med. Phys. 2020, 47, 3054–3063. [Google Scholar] [CrossRef] [PubMed]
- Ford, J.; Dogan, N.; Young, L.; Yang, F. Quantitative radiomics: Impact of pulse sequence parameter selection on MRI-based textural features of the brain. Contrast Media Mol. Imaging 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Leijenaar, R.T.H.; Carvalho, S.; Velazquez, E.R.; Van Elmpt, W.J.C.; Parmar, C.; Hoekstra, O.S.; Hoekstra, C.J.; Boellaard, R.; Dekker, A.L.A.J.; Gillies, R.J. Stability of FDG-PET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013, 52, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A. Reproducibility and prognosis of quantitative features extracted from CT images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Wang, Y.; Shi, Z.; Yu, J. A universal intensity standardization method based on a many-to-one weak-paired cycle generative adversarial network for magnetic resonance images. IEEE Trans. Med. Imaging 2019, 38, 2059–2069. [Google Scholar] [CrossRef] [PubMed]



| Overall | Significant | Advanced | Cirrhosis | ||||
|---|---|---|---|---|---|---|---|
| Sex | |||||||
| Male | 93 (70.5%) | 79 (77.5%) | 61 (82.4%) | 36 (85.7%) | |||
| Female | 39 (29.6%) | 23 (22.6%) | 13 (17.6%) | 6 (14.3%) | |||
| Age (years) * | 45.8±13.2 | 47.7 ± 13.3 | 50.8 ± 12.6 | 52.6 ± 11.1 | |||
| Fibrosis score | |||||||
| S1 | 30 (22.7%) | 0 (0%) | 0 (0%) | 0 (0%) | |||
| S2 | 28 (21.2%) | 28 (27.45%) | 0 (0%) | 0 (0%) | |||
| S3 | 32 (24.2%) | 32 (31.4%) | 32 (43.2%) | 0 (0%) | |||
| S4 | 42 (31.8%) | 42 (41.2%) | 42 (56.8%) | 42 (100%) | |||
| Group | + | – | + | – | + | – | |
| Training | 71 | 21 | 52 | 40 | 29 | 63 | |
| Test | 31 | 9 | 22 | 18 | 13 | 27 | |
| Feature | Positive | Control | p-Value | Adjusted p-Value |
|---|---|---|---|---|
| Significant fibrosis | ||||
| ∇GLRLM, Run Variance (DYN4-DYN3) | −0.518 (−0.675~−0.257) | 0.181 (−0.643~0.934) | 0.067 | 0.084 |
| ∇GLDM, Dependence Variance (DYN2-DYN1) | 0.208 (−0.088~0.557) | −0.442 (−1.275~0.117) | 0.006 ** | 0.028 * |
| ∇GLDM, the variance of Dependence Variance in time domain | −0.617 (−0.716~−0.110) | −0.247 (−0.410~0.495) | 0.037 * | 0.077 |
| GLDM, Dependence Variance in HBP | −0.479 (−0.953~0.198) | 0.717 (−0.521~1.271) | 0.046 * | 0.077 |
| ∇First Order, Median (DYN4-DYN3) | 0.169 (−0.837~0.868) | 0.527 (−0.192~1.202) | 0.308 | 0.308 |
| Advanced fibrosis | ||||
| GLDM, Large Dependence High Gray Level Emphasis in HBP | −0.310 (−0.920~0.005) | 0.288 (−0.451~0.502) | 0.030 * | 0.037 * |
| ∇First Order, 10th percentile difference (DYN4-DYN3) | 0.048 (−0.548~0.574) | 0.204 (−0.224~1.076) | 0.265 | 0.265 |
| ∇GLCM Informational Measure of Correlation (DYN3-DYN2) | 0.914 (0.214~1.412) | −0.258 (−0.733~0.416) | 0.002 ** | 0.006 ** |
| ∇GLCM Informational Measure of Correlation (HBP-DYN1) | −0.645 (−0.864~−0.048) | 0.648 (−0.279~1.357) | 0.004 ** | 0.006 ** |
| ∇GLCM Correlation difference (DYN3-DYN2) | −0.680 (−0.961~0.039) | 0.400 (0.038~1.064) | 0.002 ** | 0.006 ** |
| Cirrhosis | ||||
| GLDM Large Dependence High Gray Level Emphasis in DYN4 | −0.784 (−1.066~0.256) | 0.601 (−0.403~1.027) | 0.010** | 0.034* |
| Shape-based Maximum 2D diameter | 0.429 (−0.060~0.800) | −0.175 (−0.763~0.481) | 0.022* | 0.034* |
| Shape-based Sphericity | −0.699 (−1.151~0.226) | 0.366 (−0.168~0.817) | 0.027* | 0.034* |
| ∇GLCM Informational Measure of Correlation (HBP-DYN4) | −0.428 (−1.392~0.069) | 0.212 (−0.255~1.274) | 0.020* | 0.034* |
| ∇GLCM Kurtosis of Cluster Prominence in time domain | 0.457 (0.015~1.162) | −0.725 (−1.140~0.344) | 0.055 | 0.055 |
| Feature | Positive Subset (20%) | Control Subset (20%) |
|---|---|---|
| Significant fibrosis | ||
| ∇GLRLM, Run Variance (DYN4-DYN3) | −0.786 | 0.289 |
| ∇GLDM, Dependence Variance (DYN2-DYN1) | 0.090 | −1.684 |
| ∇GLDM, the variance of Dependence Variance in time domain | −0.455 | 0.044 |
| GLDM, Dependence Variance in HBP | −0.775 | 0.397 |
| ∇First Order, Median (DYN4-DYN3) | −1.869 | 0.832 |
| Advanced fibrosis | ||
| GLDM, Large Dependence High Gray Level Emphasis in HBP | −0.883 | 0.149 |
| ∇First Order, 10th percentile difference (DYN4-DYN3) | −0.263 | 0.581 |
| ∇GLCM Informational Measure of Correlation (DYN3-DYN2) | 1.176 | −0.948 |
| ∇GLCM Informational Measure of Correlation (HBP-DYN1) | −1.070 | 1.087 |
| ∇GLCM Correlation difference (DYN3-DYN2) | −0.984 | 0.657 |
| Cirrhosis | ||
| GLDM Large Dependence High Gray Level Emphasis in DYN4 | −0.851 | 1.050 |
| Shape-based Maximum 2D diameter | 0.855 | −0.839 |
| Shape-based Sphericity | −1.809 | 0.531 |
| ∇GLCM Informational Measure of Correlation (HBP-DYN4) | −1.691 | 0.901 |
| ∇GLCM Kurtosis of Cluster Prominence in time domain | 1.161 | −0.562 |
| Model | Accuracy | AUC (95%CI) | AP | F1 |
|---|---|---|---|---|
| Significant fibrosis | ||||
| Dynamic (proposed) | 0.875 | 0.867 (0.723~0.954) | 0.939 | 0.921 |
| DYN1 | 0.750 | 0.778 (0.619~0.894) | 0.934 | 0.839 |
| DYN2 | 0.575 | 0.581 (0.414~0.735) | 0.855 | 0.667 |
| DYN3 | 0.725 | 0.814 (0.659~0.919) | 0.922 | 0.792 |
| DYN4 | 0.775 | 0.789 (0.631~0.901) | 0.938 | 0.857 |
| HBP | 0.750 | 0.839 (0.688~0.936) | 0.955 | 0.828 |
| DYN combined | 0.775 | 0.803 (0.647~0.911) | 0.936 | 0.852 |
| NLE | 0.625 | 0.724 (0.560~0.853) | 0.912 | 0.706 |
| APRI | 0.625 | 0.695 (0.530~0.831) | 0.895 | 0.706 |
| FIB-4 | 0.725 | 0.821 (0.667~0.924) | 0.952 | 0.784 |
| Advanced fibrosis | ||||
| Dynamic (proposed) | 0.825 | 0.874 (0.730~0.957) | 0.906 | 0.837 |
| DYN1 | 0.750 | 0.768 (0.607~0.886) | 0.841 | 0.773 |
| DYN2 | 0.525 | 0.710 (0.545~0.842) | 0.768 | 0.387 |
| DYN3 | 0.575 | 0.692 (0.526~0.828) | 0.716 | 0.485 |
| DYN4 | 0.650 | 0.715 (0.550~0.846) | 0.754 | 0.667 |
| HBP | 0.800 | 0.826 (0.673~0.927) | 0.852 | 0.818 |
| DYN combined | 0.675 | 0.768 (0.607~0.886) | 0.830 | 0.723 |
| NLE | 0.700 | 0.692 (0.526~0.828) | 0.726 | 0.714 |
| APRI | 0.575 | 0.583 (0.417~0.737) | 0.663 | 0.541 |
| FIB-4 | 0.675 | 0.775 (0.616~0.892) | 0.839 | 0.629 |
| Cirrhosis | ||||
| Dynamic (proposed) | 0.850 | 0.900 (0.764~0.972) | 0.866 | 0.750 |
| DYN1 | 0.775 | 0.852 (0.704~0.944) | 0.825 | 0.640 |
| DYN2 | 0.850 | 0.875 (0.732~0.958) | 0.825 | 0.750 |
| DYN3 | 0.825 | 0.855 (0.707~0.946) | 0.833 | 0.741 |
| DYN4 | 0.775 | 0.832 (0.680~0.931) | 0.785 | 0.640 |
| HBP | 0.850 | 0.892 (0.753~0.968) | 0.859 | 0.786 |
| DYN combined | 0.850 | 0.858 (0.711~0.948) | 0.847 | 0.750 |
| NLE | 0.725 | 0.812 (0.657~0.918) | 0.747 | 0.645 |
| APRI | 0.725 | 0.755 (0.593~0.877) | 0.613 | 0.522 |
| FIB-4 | 0.850 | 0.866 (0.721~0.953) | 0.840 | 0.700 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, R.; Shi, C.; Wang, C.; Shi, N.; Qiu, T.; Chen, W.; Shi, Y.; Wang, H. Imaging-Based Staging of Hepatic Fibrosis in Patients with Hepatitis B: A Dynamic Radiomics Model Based on Gd-EOB-DTPA-Enhanced MRI. Biomolecules 2021, 11, 307. https://doi.org/10.3390/biom11020307
Zheng R, Shi C, Wang C, Shi N, Qiu T, Chen W, Shi Y, Wang H. Imaging-Based Staging of Hepatic Fibrosis in Patients with Hepatitis B: A Dynamic Radiomics Model Based on Gd-EOB-DTPA-Enhanced MRI. Biomolecules. 2021; 11(2):307. https://doi.org/10.3390/biom11020307
Chicago/Turabian StyleZheng, Rencheng, Chunzi Shi, Chengyan Wang, Nannan Shi, Tian Qiu, Weibo Chen, Yuxin Shi, and He Wang. 2021. "Imaging-Based Staging of Hepatic Fibrosis in Patients with Hepatitis B: A Dynamic Radiomics Model Based on Gd-EOB-DTPA-Enhanced MRI" Biomolecules 11, no. 2: 307. https://doi.org/10.3390/biom11020307
APA StyleZheng, R., Shi, C., Wang, C., Shi, N., Qiu, T., Chen, W., Shi, Y., & Wang, H. (2021). Imaging-Based Staging of Hepatic Fibrosis in Patients with Hepatitis B: A Dynamic Radiomics Model Based on Gd-EOB-DTPA-Enhanced MRI. Biomolecules, 11(2), 307. https://doi.org/10.3390/biom11020307








