Tissue-Specific Landscape of Metabolic Dysregulation during Ageing
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. NMR Sample Preparation, Data Acquisition and Analysis
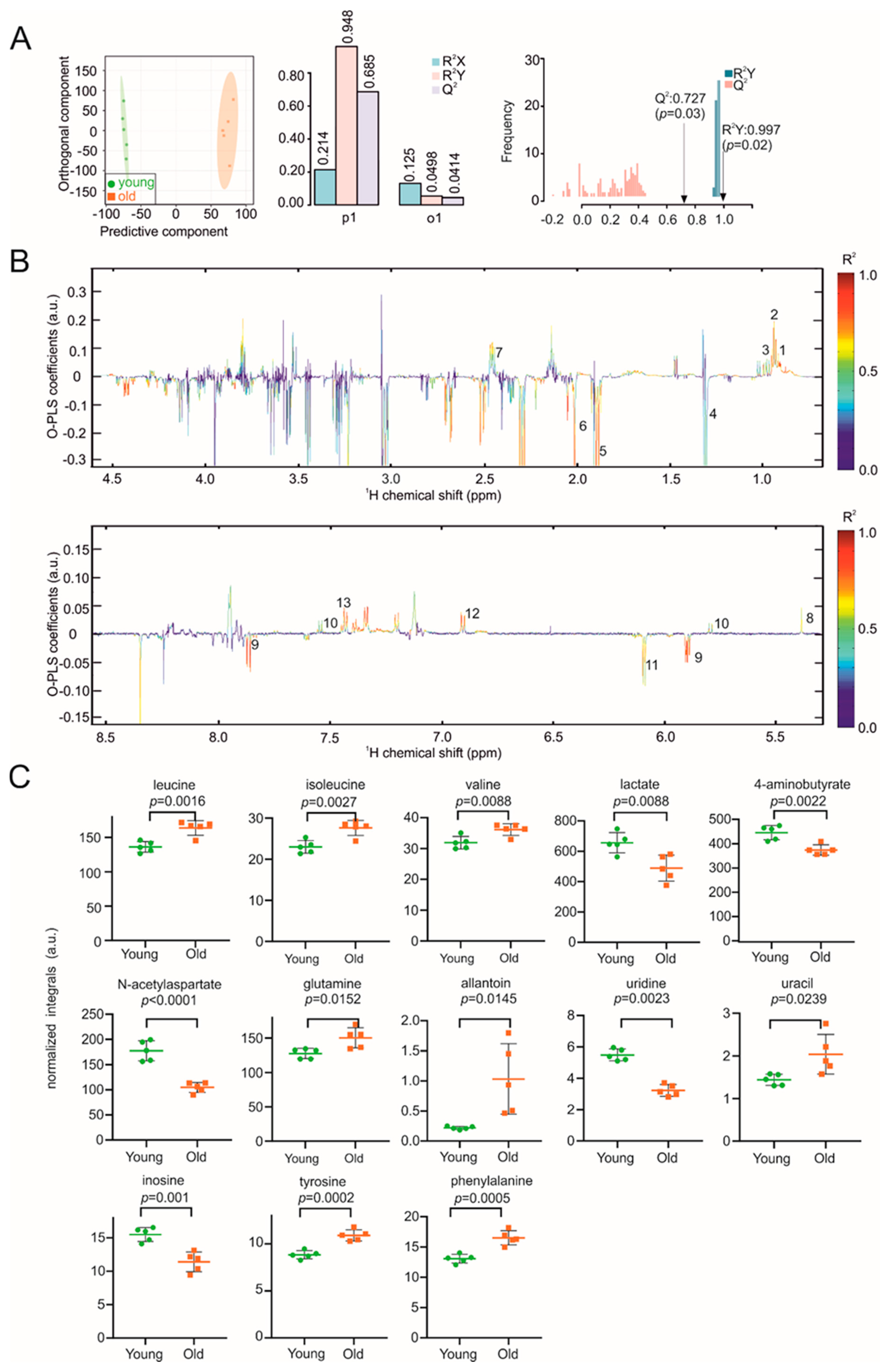
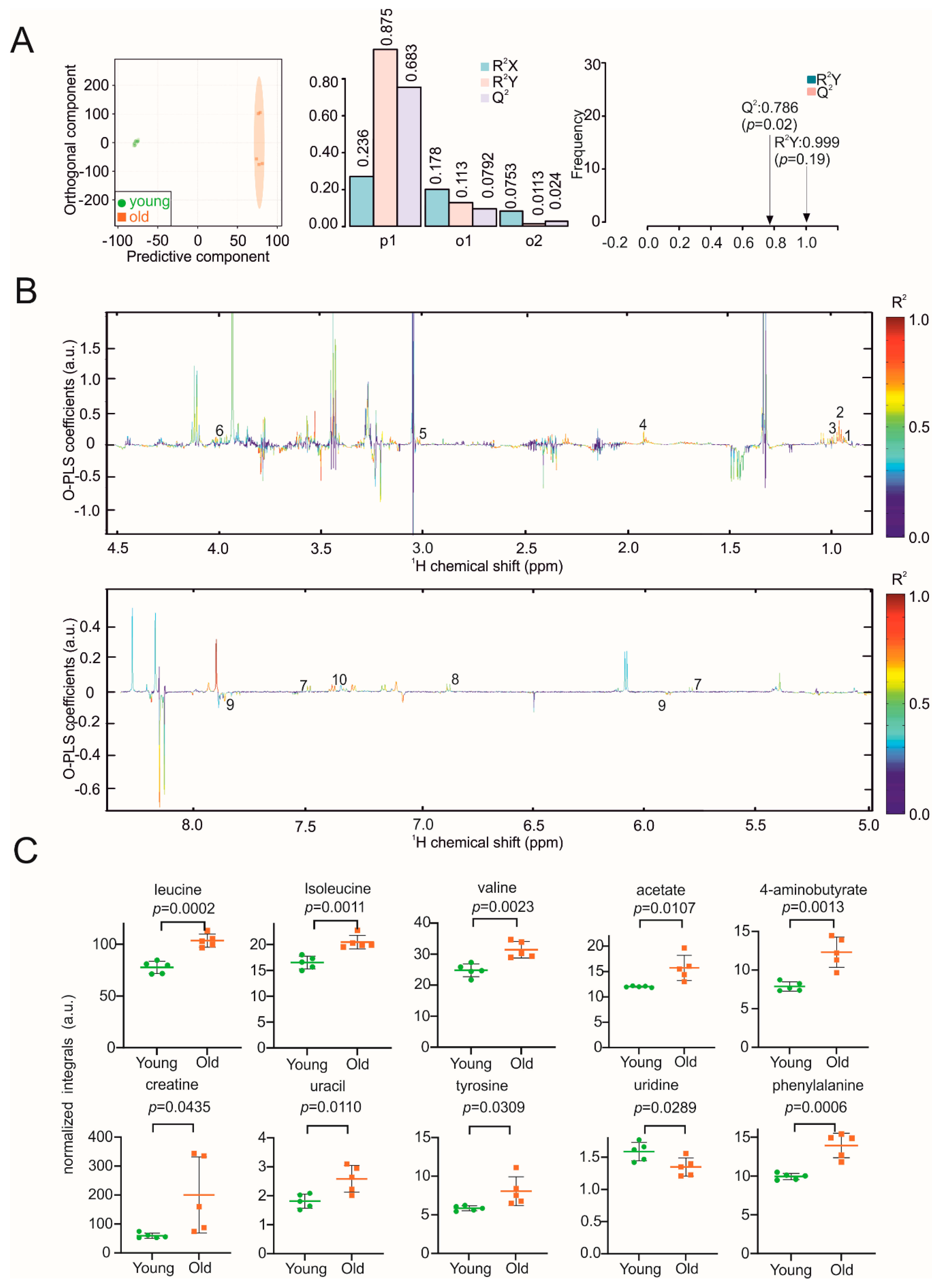
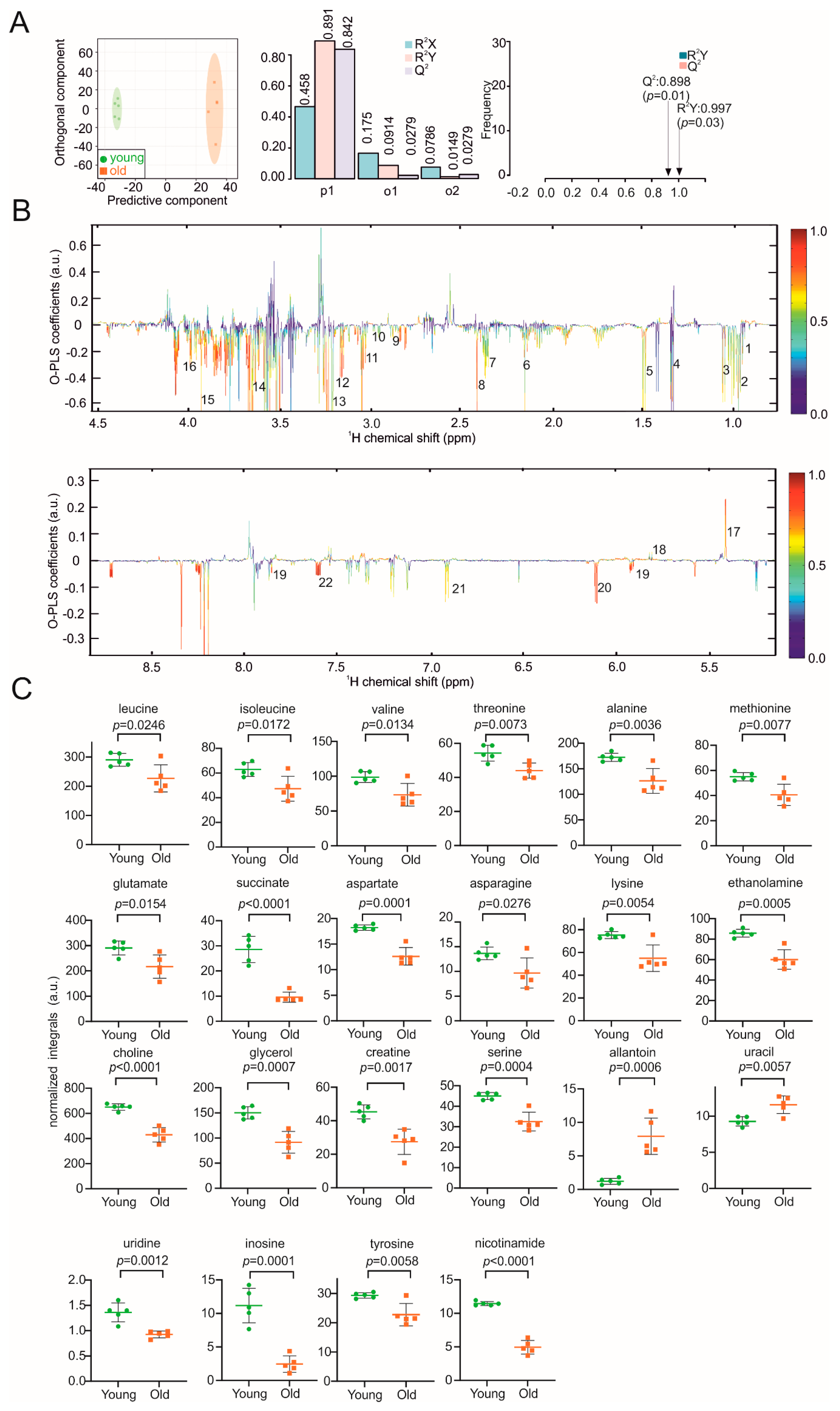
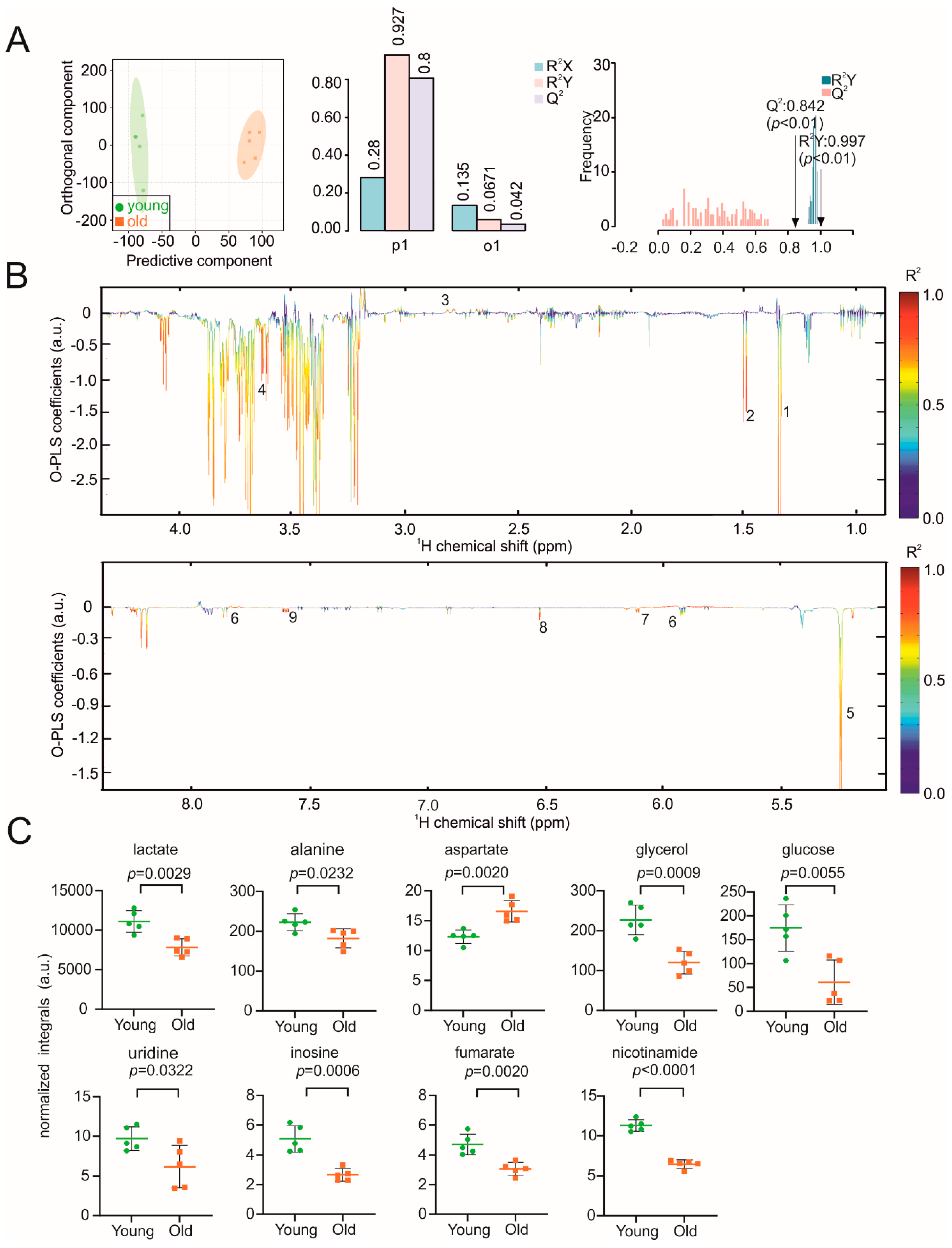
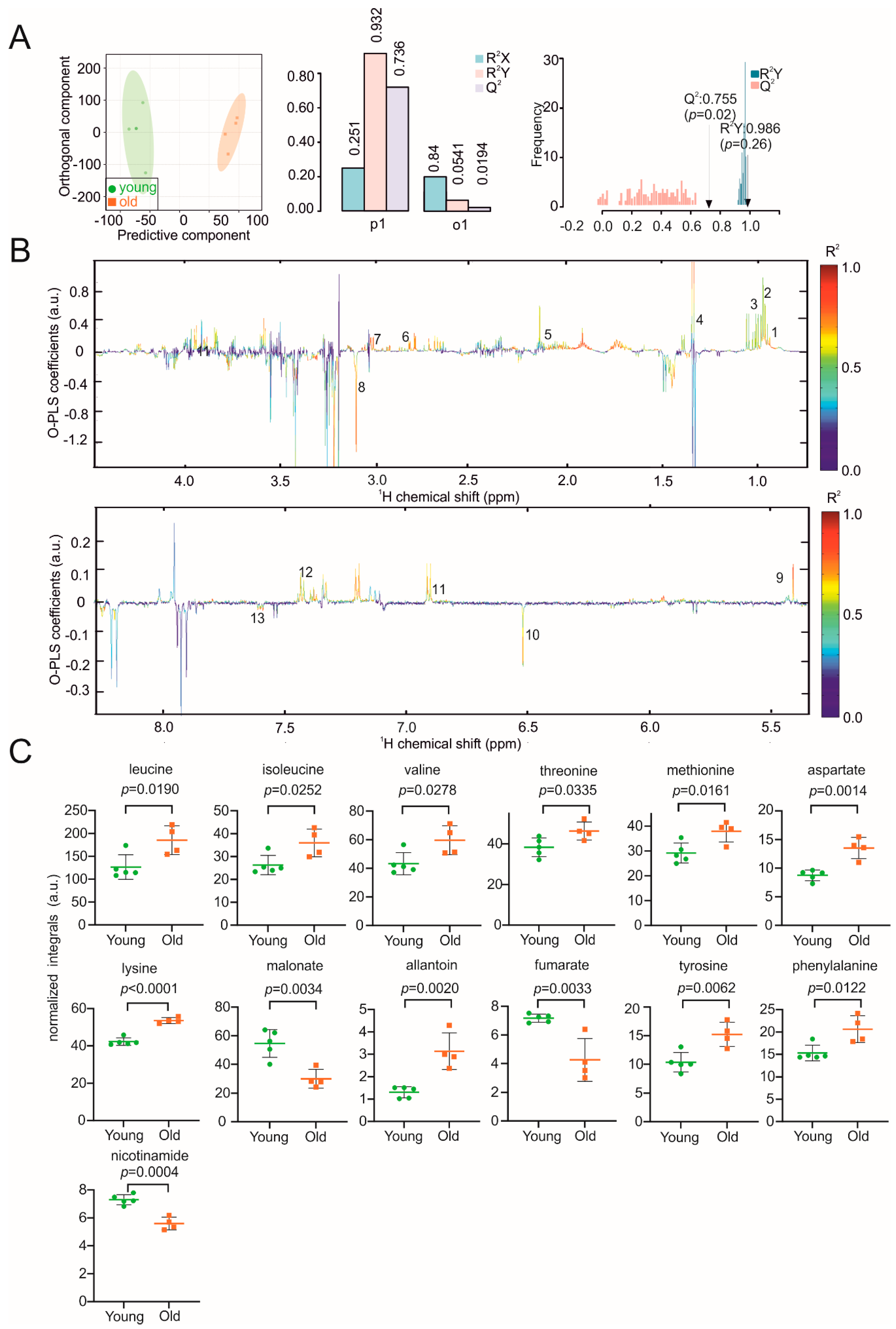
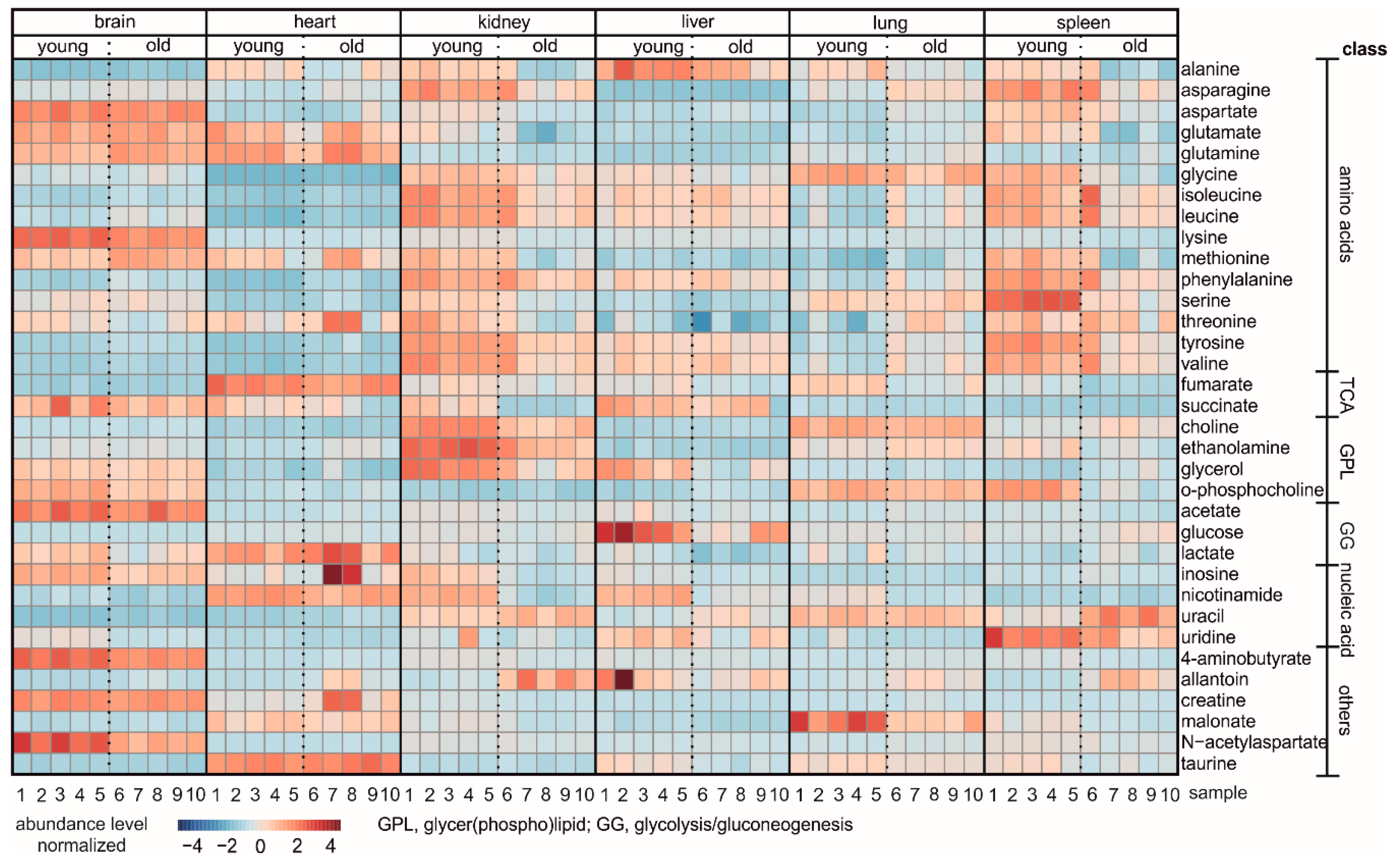
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Van Deursen, J.M. The role of senescent cells in ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Wang, Z.; Liu, J.-P. Roles of telomere biology in cell senescence, replicative and chronological ageing. Cells 2019, 8, 54. [Google Scholar] [CrossRef]
- Chapman, J.; Fielder, E.; Passos, J.F. Mitochondrial dysfunction and cell senescence: Deciphering a complex relationship. FEBS Lett. 2019, 593, 1566–1579. [Google Scholar] [CrossRef]
- Childs, B.G.; Baker, D.J.; Kirkland, J.L.; Campisi, J.; Van Deursen, J.M. Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 2014, 15, 1139–1153. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, L.; Giallauria, F.; Guralnik, J.M. Epidemiology of aging. Radiol. Clin. N. Am. 2008, 46, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Bennett, M. Aging and atherosclerosis: Mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ. Res. 2012, 111, 245–259. [Google Scholar] [CrossRef]
- Chang, A.M.; Halter, J.B. Aging and insulin secretion. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E7–E12. [Google Scholar] [CrossRef]
- De Lau, L.M.; Breteler, M.M. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Brookmeyer, R.; Gray, S.; Kawas, C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am. J. Public Health 1998, 88, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- Yazdanyar, A.; Newman, A.B. The burden of cardiovascular disease in the elderly: Morbidity, mortality, and costs. Clin. Geriatr. Med. 2009, 25, 563–577. [Google Scholar] [CrossRef]
- Farr, J.N.; Xu, M.; Weivoda, M.M.; Monroe, D.G.; Fraser, D.G.; Onken, J.L.; Negley, B.A.; Sfeir, J.G.; Ogrodnik, M.B.; Hachfeld, C.M. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017, 23, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Roos, C.M.; Zhang, B.; Palmer, A.K.; Ogrodnik, M.B.; Pirtskhalava, T.; Thalji, N.M.; Hagler, M.; Jurk, D.; Smith, L.A.; Casaclang-Verzosa, G. Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell 2016, 15, 973–977. [Google Scholar] [CrossRef]
- Catic, A. Cellular metabolism and aging. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2018; Volume 155, pp. 85–107. [Google Scholar]
- Zhang, L. Progress in Molecular Biology and Translational Science; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Guasch-Ferré, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martínez-González, M.A.; Salas-Salvadó, J.; Hu, F.B. Metabolomics in prediabetes and diabetes: A systematic review and meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef]
- Suhre, K.; Meisinger, C.; Döring, A.; Altmaier, E.; Belcredi, P.; Gieger, C.; Chang, D.; Milburn, M.V.; Gall, W.E.; Weinberger, K.M. Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE 2010, 5, e13953. [Google Scholar] [CrossRef] [PubMed]
- Trushina, E.; Dutta, T.; Persson, X.-M.T.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLoS ONE 2013, 8, e63644. [Google Scholar] [CrossRef]
- Wilkinson, D.J.; Rodriguez-Blanco, G.; Dunn, W.B.; Phillips, B.E.; Williams, J.P.; Greenhaff, P.L.; Smith, K.; Gallagher, I.J.; Atherton, P.J. Untargeted metabolomics for uncovering biological markers of human skeletal muscle ageing. Aging 2020, 12, 12517. [Google Scholar] [CrossRef] [PubMed]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An integrative metabolomics and transcriptomics study to identify metabolic alterations in aged skin of humans in vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The metabolic footprint of aging in mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Stauch, K.L.; Petrascheck, M.; Benton, H.P.; Epstein, A.A.; Fang, M.; Siuzdak, G. Metabolic drift in the aging brain. Aging (Albany NY) 2016, 8, 1000. [Google Scholar] [CrossRef]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Bandeen-Roche, K.; Xue, Q.-L.; Ferrucci, L.; Walston, J.; Guralnik, J.M.; Chaves, P.; Zeger, S.L.; Fried, L.P. Phenotype of frailty: Characterization in the women’s health and aging studies. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2006, 61, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Peters, L.L.; Paigen, B. Mice as a mammalian model for research on the genetics of aging. ILAR J. 2011, 52, 4–15. [Google Scholar] [CrossRef]
- Loria, J.P.; Rance, M.; Palmer, A.G. A relaxation-compensated Carr−Purcell−Meiboom−Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 1999, 121, 2331–2332. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Luchinat, C.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Stryeck, S.; Gastrager, M.; Degoricija, V.; Trbusic, M.; Potocnjak, I.; Radulovic, B.; Pregartner, G.; Berghold, A.; Madl, T.; Frank, S. Serum Concentrations of Citrate, Tyrosine, 2- and 3- Hydroxybutyrate are Associated with Increased 3-Month Mortality in Acute Heart Failure Patients. Sci. Rep. 2019, 9, 6743. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Raja, G.; Jung, Y.; Jung, S.H.; Kim, T.-J. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering–A review. Process Biochem. 2020, 99, 112–122. [Google Scholar] [CrossRef]
- Bingol, K. Recent advances in targeted and untargeted metabolomics by NMR and MS/NMR methods. High-Throughput 2018, 7, 9. [Google Scholar] [CrossRef]
- Park, D.C.; Reuter-Lorenz, P. The adaptive brain: Aging and neurocognitive scaffolding. Annu. Rev. Psychol. 2009, 60, 173–196. [Google Scholar] [CrossRef]
- Yin, F.; Boveris, A.; Cadenas, E. Mitochondrial energy metabolism and redox signaling in brain aging and neurodegeneration. Antioxid. Redox Signal. 2014, 20, 353–371. [Google Scholar] [CrossRef] [PubMed]
- Lesnefsky, E.J.; Chen, Q.; Hoppel, C.L. Mitochondrial metabolism in aging heart. Circ. Res. 2016, 118, 1593–1611. [Google Scholar] [CrossRef]
- Epstein, M. Effects of aging on the kidney. Fed. Proc. 1979, 38, 168–171. [Google Scholar] [PubMed]
- Sato, Y.; Yanagita, M. Immunology of the ageing kidney. Nat. Rev. Nephrol. 2019, 15, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Tas, E.; Yakar, S.; Muzumdar, R. Hepatic lipid metabolism and non-alcoholic fatty liver disease in aging. Mol. Cell. Endocrinol. 2017, 455, 115–130. [Google Scholar] [CrossRef]
- Brandenberger, C.; Mühlfeld, C. Mechanisms of lung aging. Cell Tissue Res. 2017, 367, 469–480. [Google Scholar] [CrossRef]
- Turner, V.M.; Mabbott, N.A. Influence of ageing on the microarchitecture of the spleen and lymph nodes. Biogerontology 2017, 18, 723–738. [Google Scholar] [CrossRef]
- Larsson, S.C.; Markus, H.S. Branched-chain amino acids and Alzheimer’s disease: A Mendelian randomization analysis. Sci. Rep. 2017, 7, 1–4. [Google Scholar] [CrossRef]
- Segovia, G.; Porras, A.; Del Arco, A.; Mora, F. Glutamatergic neurotransmission in aging: A critical perspective. Mech. Ageing Dev. 2001, 122, 1–29. [Google Scholar] [CrossRef]
- Kaiser, L.G.; Schuff, N.; Cashdollar, N.; Weiner, M.W. Age-related glutamate and glutamine concentration changes in normal human brain: 1H MR spectroscopy study at 4 T. Neurobiol. Aging. 2005, 26, 665–672. [Google Scholar] [CrossRef]
- Hashimoto, M.; Fukamizu, A.; Nakagawa, T.; Kizuka, Y. Roles of protein arginine methyltransferase 1 (PRMT1) in brain development and disease. Biochim. Biophys. Acta (BBA) Gen. Subj. 2020, 1865, 129776. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, M.; Sun, H.; Wang, Y. Branched-chain amino acid metabolism in heart disease: An epiphenomenon or a real culprit? Cardiovasc. Res. 2011, 90, 220–223. [Google Scholar] [CrossRef]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef]
- Uddin, G.M.; Zhang, L.; Shah, S.; Fukushima, A.; Wagg, C.S.; Gopal, K.; Al Batran, R.; Pherwani, S.; Ho, K.L.; Boisvenue, J. Impaired branched chain amino acid oxidation contributes to cardiac insulin resistance in heart failure. Cardiovasc. Diabetol. 2019, 18, 86. [Google Scholar] [CrossRef] [PubMed]
- Tobias, D.K.; Lawler, P.R.; Harada, P.H.; Demler, O.V.; Ridker, P.M.; Manson, J.E.; Cheng, S.; Mora, S. Circulating branched-chain amino acids and incident cardiovascular disease in a prospective cohort of US women. Circ. Genom. Precis. Med. 2018, 11, e002157. [Google Scholar] [CrossRef]
- DeFeudis, F. γ-Aminobutyric acid and cardiovascular function. Experientia 1983, 39, 845–849. [Google Scholar] [CrossRef]
- Pauwels, L.; Maes, C.; Swinnen, S.P. Aging, inhibition and GABA. Aging (Albany NY) 2018, 10, 3645. [Google Scholar] [CrossRef] [PubMed]
- Patterson, J.M.; Keevil, N.; McHenry, E. Choline and the prevention of hemorrhagic kidneys in the rat. 2. Phospholipid turnover determined with radioactive phosphorus. J. Biol. Chem. 1944, 153, 489–493. [Google Scholar] [CrossRef]
- Ossani, G.; Dalghi, M.; Repetto, M. Oxidative damage lipid peroxidation in the kidney of choline-deficient rats. Front. Biosci. 2007, 12, 1174–1183. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, G.; Gonzalez, F.J.; Park, S.-M.; Cai, D. Hypoxia-inducible factor directs POMC gene to mediate hypothalamic glucose sensing and energy balance regulation. PLoS Biol. 2011, 9, e1001112. [Google Scholar] [CrossRef]
- Pan, C.; Locasale, J.W. Targeting metabolism to influence aging. Science 2021, 371, 234–235. [Google Scholar] [CrossRef]
- Johmura, Y.; Yamanaka, T.; Omori, S.; Wang, T.-W.; Sugiura, Y.; Matsumoto, M.; Suzuki, N.; Kumamoto, S.; Yamaguchi, K.; Hatakeyama, S.; et al. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science 2021, 371, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Kannan, L.; Knudsen, J.; Jolly, C.A. Aging and acyl-CoA binding protein alter mitochondrial glycerol-3-phosphate acyltransferase activity. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2003, 1631, 12–16. [Google Scholar] [CrossRef]
- Sastre, J.; Pallardo, F.V.; Plá, R.; Pellín, A.; Juan, G.; O’Connor, J.E.; Estrela, J.M.; Miquel, J.; Vina, J. Aging of the liver: Age-associated mitochondrial damage in intact hepatocytes. Hepatology 1996, 24, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-F.; Wang, D.; Hwang, C.P.; Liu, H.W.; Wei, J.; Lee, R.P.; Chen, H.I. The protective effect of niacinamide on ischemia-reperfusion-induced liver injury. J. Biomed. Sci. 2001, 8, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Mármol, F.; Sánchez, J.; López, D.; Martínez, N.; Xaus, C.; Peralta, C.; Roselló-Catafau, J.; Mitjavila, M.T.; Puig-Parellada, P. Role of oxidative stress and adenosine nucleotides in the liver of aging rats. Physiol. Res. 2010, 59, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.A.; Wilund, K.R.; Martin, S.A.; Kistler, B.M. Exercise, inflammation and aging. Aging Dis. 2012, 3, 130. [Google Scholar]
- Lu, G.; Ren, S.; Korge, P.; Choi, J.; Dong, Y.; Weiss, J.; Koehler, C.; Chen, J.-N.; Wang, Y. A novel mitochondrial matrix serine/threonine protein phosphatase regulates the mitochondria permeability transition pore and is essential for cellular survival and development. Genes Dev. 2007, 21, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef]
- Nishio, S.-I.; Negoro, S.; Hosokawa, T.; Hara, H.; Tanaka, T.; Deguchi, Y.; Ling, J.; Awata, N.; Azuma, J.; Aoike, A. The effect of taurine on age-related immune decline in mice: The effect of taurine on T cell and B cell proliferative response under costimulation with ionomycin and phorbol myristate acetate. Mech. Ageing Dev. 1990, 52, 125–139. [Google Scholar] [CrossRef]
- Bircan, F.S.; Balabanli, B.; Turkozkan, N.; Ozan, G. Effects of taurine on nitric oxide and 3-nitrotyrosine levels in spleen during endotoxemia. Neurochem. Res. 2011, 36, 1978–1983. [Google Scholar] [CrossRef] [PubMed]
- Hunsberger, H.C.; Greenwood, B.P.; Tolstikov, V.; Narain, N.R.; Kiebish, M.A.; Denny, C.A. Divergence in the metabolome between natural aging and Alzheimer’s disease. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Delfarah, A.; Parrish, S.; Yang, J.; Seo, F.; Li, S.; Wang, P.; Graham, N.A. Inhibition of nucleotide synthesis mediates replicative senescence of human mammary epithelial cells. BioRxiv 2018, 423665. [Google Scholar] [CrossRef]
- Xia, X.; Chen, W.; McDermott, J.; Han, J.-D.J. Molecular and phenotypic biomarkers of aging. F1000Research 2017, 6, 860. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M. The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 2015, 14, 644–658. [Google Scholar] [CrossRef]
- Baar, M.P.; Brandt, R.M.; Putavet, D.A.; Klein, J.D.; Derks, K.W.; Bourgeois, B.R.; Stryeck, S.; Rijksen, Y.; van Willigenburg, H.; Feijtel, D.A. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 2017, 169, 132–147. [Google Scholar] [CrossRef]
- Chang, J.; Wang, Y.; Shao, L.; Laberge, R.-M.; Demaria, M.; Campisi, J.; Janakiraman, K.; Sharpless, N.E.; Ding, S.; Feng, W. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016, 22, 78–83. [Google Scholar] [CrossRef]
- Fuhrmann-Stroissnigg, H.; Ling, Y.Y.; Zhao, J.; McGowan, S.J.; Zhu, Y.; Brooks, R.W.; Grassi, D.; Gregg, S.Q.; Stripay, J.L. Identification of HSP90 inhibitors as a novel class of senolytics. Nat. Commun. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- LeBrasseur, N.K.; Tchkonia, T.; Kirkland, J.L. Cellular senescence and the biology of aging, disease, and frailty. In Frailty: Pathophysiology, Phenotype and Patient Care; Karger Publishers: Basel, Switzerland, 2015; Volume 83, pp. 11–18. [Google Scholar]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Kerbl-Knapp, J.; Akhmetshina, A.; Korbelius, M.; Kuentzel, K.B.; Vujić, N.; Hörl, G.; Paar, M.; Kratky, D.; Steyrer, E.; et al. Tissue-Specific Landscape of Metabolic Dysregulation during Ageing. Biomolecules 2021, 11, 235. https://doi.org/10.3390/biom11020235
Zhang F, Kerbl-Knapp J, Akhmetshina A, Korbelius M, Kuentzel KB, Vujić N, Hörl G, Paar M, Kratky D, Steyrer E, et al. Tissue-Specific Landscape of Metabolic Dysregulation during Ageing. Biomolecules. 2021; 11(2):235. https://doi.org/10.3390/biom11020235
Chicago/Turabian StyleZhang, Fangrong, Jakob Kerbl-Knapp, Alena Akhmetshina, Melanie Korbelius, Katharina Barbara Kuentzel, Nemanja Vujić, Gerd Hörl, Margret Paar, Dagmar Kratky, Ernst Steyrer, and et al. 2021. "Tissue-Specific Landscape of Metabolic Dysregulation during Ageing" Biomolecules 11, no. 2: 235. https://doi.org/10.3390/biom11020235
APA StyleZhang, F., Kerbl-Knapp, J., Akhmetshina, A., Korbelius, M., Kuentzel, K. B., Vujić, N., Hörl, G., Paar, M., Kratky, D., Steyrer, E., & Madl, T. (2021). Tissue-Specific Landscape of Metabolic Dysregulation during Ageing. Biomolecules, 11(2), 235. https://doi.org/10.3390/biom11020235






