Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Mining
2.2. Organisms, Media, and Growth Conditions
2.3. OSMAC Experiments
2.4. Preparation of Biotic Additives
2.5. Cell Removal
2.6. Extraction of Cell-Free Fermentation Broth
2.7. HPLC-MS Sample Preparation
2.8. HPLC-MS/MS Measurement
2.9. Evaluation of HPLC-MS Data
2.10. Utilized Databases
3. Results and Discussion
3.1. Genome-Mining for Selection of Bacteria
3.2. Verification of AntiSMASH-Predicted SM Compounds
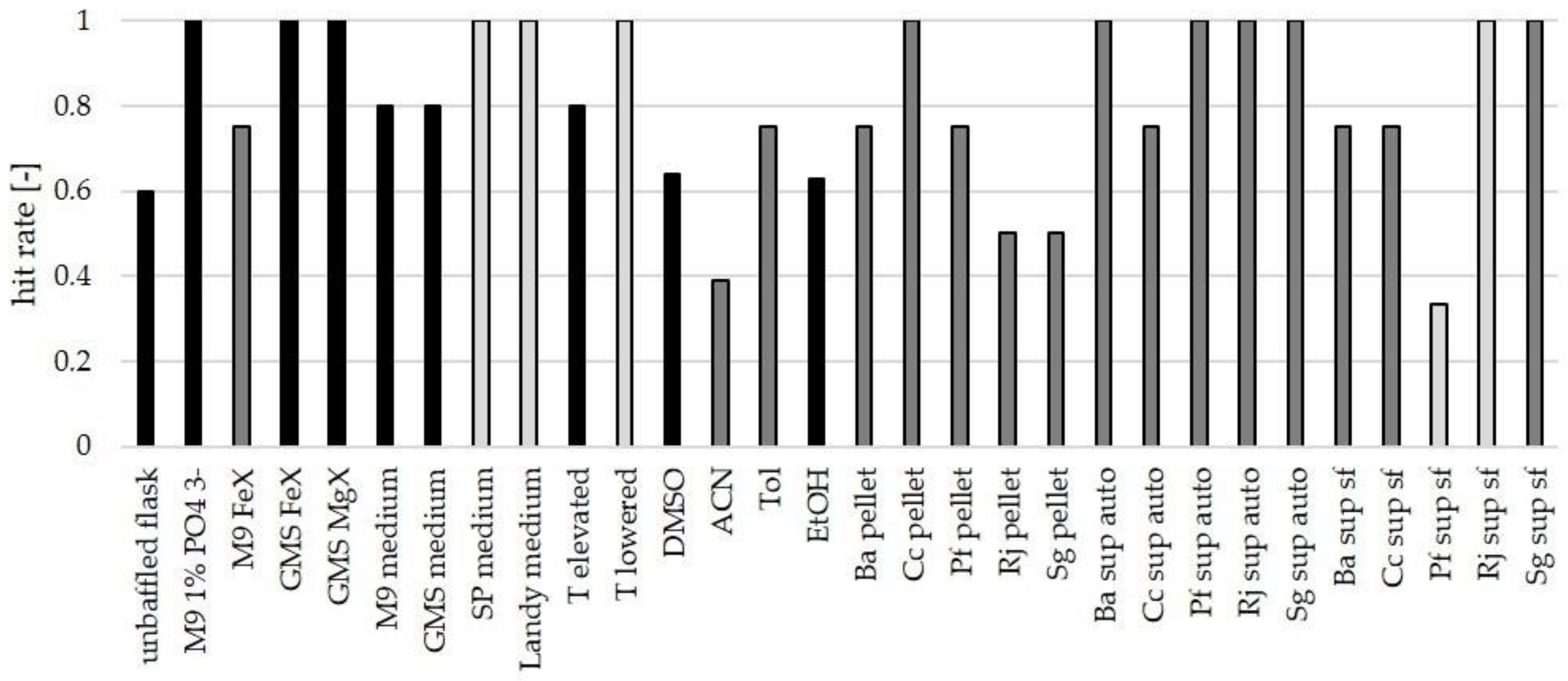
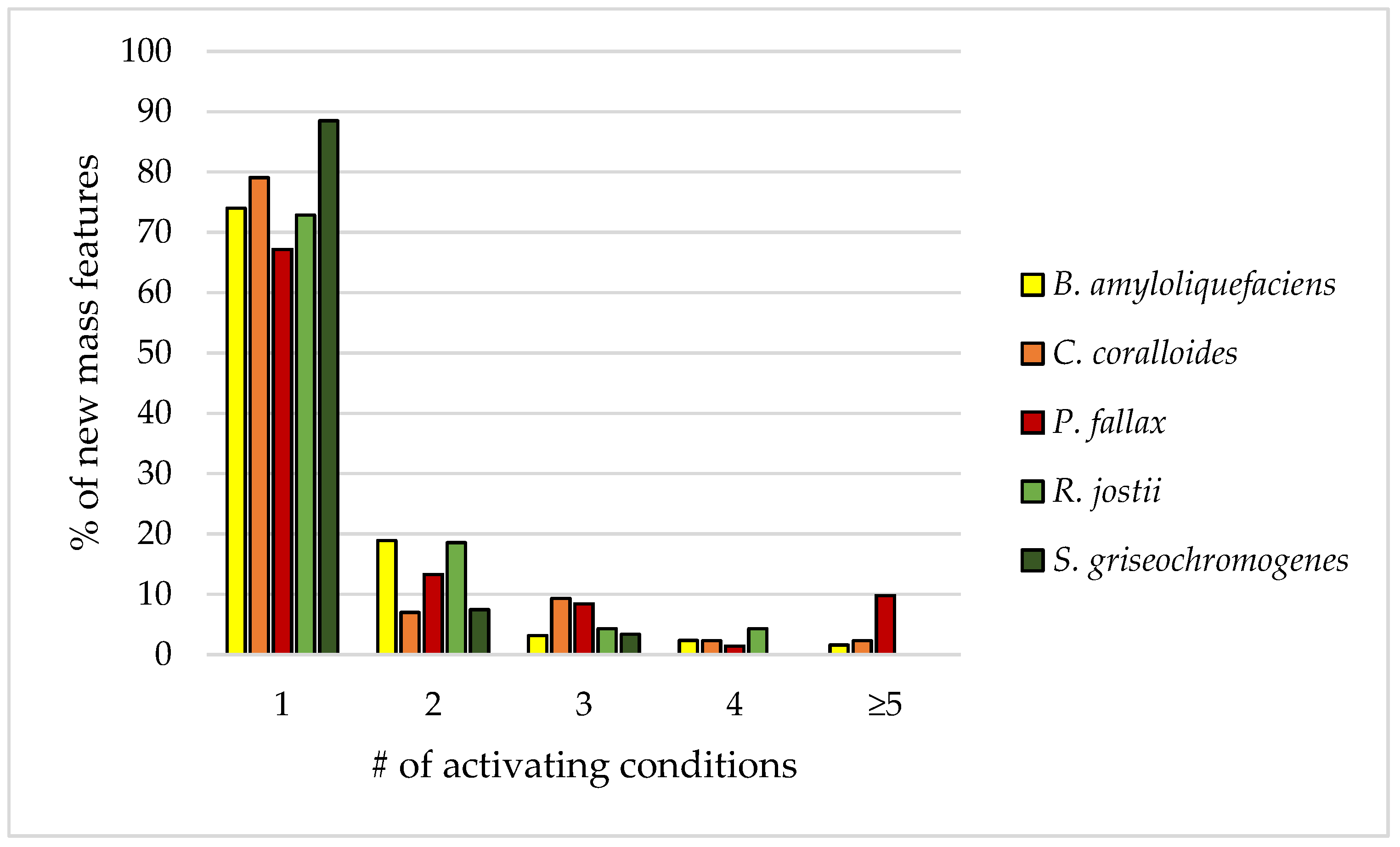
3.3. Influence of Culture Conditions on the Number of New Mass Features
3.4. Suitability of Culture Conditions for the Generation of New Mass Features
3.5. Putative Annotation of New Mass Features
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McMurry, J. Secondary Metabolites: An Introduction to Natural Products Chemistry. In Organic Chemistry with Biological Applications; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1016–1046. ISBN 9781285842912. [Google Scholar]
- Luckner, M. What is secondary metabolism? In Secondary Metabolism in Microorganisms, Plants, and Animals, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 1990; pp. 15–23. [Google Scholar]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Luckner, M. Expression and Control, Secondary Metabolism in Microorganisms, Plants, and Animals, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 1990; pp. 44–62. [Google Scholar]
- Wu, C.; Zacchetti, B.; Ram, A.F.J.; Van Wezel, G.P.; Claessen, D.; Choi, Y.H. Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef]
- Surup, F.; Viehrig, K.; Mohr, K.I.; Herrmann, J.; Jansen, R.; Müller, R. Disciformycins A and B: 12-membered macrolide glycoside antibiotics from the myxobacterium pyxidicoccus fallax active against multiresistant staphylococci. Angew. Chem. Int. Ed. 2014, 53, 13588–13591. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Houssen, W.E.; Harrison, W.T.A.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; et al. Diverse Metabolic Profiles of a Streptomyces Strain Isolated from a Hyper-arid Environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Jamil, B.; Hasan, F.; Hameed, A.; Ahmed, S. Isolation of bacillus subtilis MH-4 from soil and its potential of polypeptidic antibiotic production. Pak. J. Pharm. Sci. 2007, 20, 26–31. [Google Scholar] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0-A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Wilkinson, B.; Micklefield, J. Mining and engineering natural-product biosynthetic pathways. Nat. Chem. Biol. 2007, 3, 379–386. [Google Scholar] [CrossRef]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef]
- Zarins-Tutt, J.S.; Barberi, T.T.; Gao, H.; Mearns-Spragg, A.; Zhang, L.; Newman, D.J.; Goss, R.J.M. Prospecting for new bacterial metabolites: A glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2016, 33, 54–72. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef]
- Rackham, E.J.; Grüschow, S.; Ragab, A.E.; Dickens, S.; Goss, R.J.M. Pacidamycin biosynthesis: Identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. ChemBioChem 2010, 11, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Bode, B.H.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Bode, H.B.; Walker, M.; Zeeck, A. Secondary metabolites by chemical screening. 42 Cladospirones B to I from Sphaeropsidales sp. F-24’707 by variation of culture conditions. Eur. J. Org. Chem. 2000, 3185–3193. [Google Scholar] [CrossRef]
- Machushynets, N.V.; Wu, C.; Elsayed, S.S.; Hankemeier, T.; van Wezel, G.P. Discovery of novel glycerolated quinazolinones from Streptomyces sp. MBT27. J. Ind. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Li, W.; Ding, L.; Wang, N.; Xu, J.; Zhang, W.; Zhang, B.; He, S.; Wu, B.; Jin, H. Isolation and Characterization of Two New Metabolites from the Sponge-Derived Fungus Aspergillus sp. LS34 by OSMAC Approach. Mar. Drugs 2019, 17, 283. [Google Scholar] [CrossRef]
- Urem, M.; Świątek-Połatyńska, M.A.; Rigali, S.; van Wezel, G.P. Intertwining nutrient-sensory networks and the control of antibiotic production in Streptomyces. Mol. Microbiol. 2016, 102, 183–195. [Google Scholar] [CrossRef]
- Chen, G.; Wang, G.; Li, X.; Waters, B.; Davies, J. Enhanced Production of Microbial Metabolites in the Presence of Dimethyl Sulfoxide streptomycin (phenotypic suppression, for example) can be removed, and the pellet washed twice by resuspending in Morphological Observations Streptomyces glaucescens wa. J. Antibiot. (Tokyo) 2000, 53, 1145–1153. [Google Scholar] [CrossRef]
- Lindquist, S. The Heat-Shock Response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Doull, J.L.; Singh, A.K.; Hoare, M.; Ayer, S.W. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: Effects of heat shock, ethanol treatment and phage infection. J. Ind. Microbiol. 1994, 13, 120–125. [Google Scholar] [CrossRef]
- Weinberg, E.D. Roles of trace metals in transcriptional control of microbial secondary metabolism. Biol. Met. 1990, 2, 191–196. [Google Scholar] [CrossRef]
- Paranagama, P.A.; Wijeratne, E.M.; Gunatilaka, A.A. Uncovering Biosynthetic Potential of Plant-Associated Fungi: Effect of Culture Conditions on Metabolite Production by Paraphaeosphaeria quadriseptata and Chaetomium chiversii(1). J. Nat. Prod. 2007, 21, 1939–1945. [Google Scholar] [CrossRef] [PubMed]
- Freemont, P.; Polizzi, K.M.; Goers, L. Co-culture systems and technologies: Taking synthetic biology to the next level. J. R. Soc. Interface 2014, 11. [Google Scholar] [CrossRef]
- Moody, S.C. Microbial co-culture: Harnessing intermicrobial signaling for the production of novel antimicrobials. Future Microbiol. 2014, 9, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Mearns-Spragg, A.; Bregu, M.; Boyd, K.G.; Burgess, J.G. Cross-species, induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 1998, 27, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Rather, M.A.; Dar, M.S.; Aga, M.A.; Ahmad, N.; Manzoor, A.; Qayum, A.; Shah, A.; Mushtaq, S.; Ahmad, Z.; et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorganic Med. Chem. Lett. 2017, 27, 2579–2582. [Google Scholar] [CrossRef]
- Santos, J.D.; Vitorino, I.; De La Cruz, M.; Díaz, C.; Cautain, B.; Annang, F.; Pérez-Moreno, G.; Martinez, I.G.; Tormo, J.R.; Martín, J.M.; et al. Bioactivities and extract dereplication of actinomycetales isolated from marine sponges. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef]
- Amano, S.; Morota, T.; Kano, Y.; Narita, H.; Hashidzume, T.; Yamamoto, S.; Mizutani, K.; Sakuda, S.; Furihata, K.; Takano-Shiratori, H. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. (Tokyo) 2010, 63, 486–491. [Google Scholar] [CrossRef]
- Grond, S.; Papastavrou, I.; Zeeck, A. Novel α-L-rhamnopyranosides from a single strain of Streptomyces by supplement-induced biosynthetic steps. Eur. J. Org. Chem. 2002, 3237–3242. [Google Scholar] [CrossRef]
- Schäberle, T.F.; Orland, A.; König, G.M. Enhanced production of undecylprodigiosin in Streptomyces coelicolor by co-cultivation with the corallopyronin A-producing myxobacterium, Corallococcus coralloides. Biotechnol. Lett. 2014, 36, 641–648. [Google Scholar] [CrossRef]
- Angell, S.; Bench, B.J.; Williams, H.; Watanabe, C.M.H. Pyocyanin Isolated from a Marine Microbial Population: Synergistic Production between Two Distinct Bacterial Species and Mode of Action. Chem. Biol. 2006, 13, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Sola-Landa, A.; Santos-Beneit, F.; Fernández-Martínez, L.T.; Prieto, C.; Rodríguez-García, A. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 2011, 4, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Senges, C.H.R.; Al-Dilaimi, A.; Marchbank, D.H.; Wibberg, D.; Winkler, A.; Haltli, B.; Nowrousian, M.; Kalinowski, J.; Kerr, R.G.; Bandow, J.E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. USA 2018, 115, 2490–2495. [Google Scholar] [CrossRef] [PubMed]
- DSMZ GmbH Nutrient Medium Composition. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1.pdf (accessed on 11 September 2020).
- DSMZ GmbH SP Medium Composition. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium222.pdf (accessed on 11 September 2020).
- DSMZ GmbH MD1 Medium Composition. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium1118.pdf (accessed on 11 September 2020).
- DSMZ GmbH TSB Medium Composition. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium545.pdf (accessed on 11 September 2020).
- DSMZ GmbH GYM Medium Composition. Available online: https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium65.pdf (accessed on 11 September 2020).
- Seidel, V. Initial and Bulk Extraction of Natural Products Isolation. In Natural Products Isolation- Methods and Protocols; Sarker, S.D., Nahar, L., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 27–41. ISBN 9781617796241. [Google Scholar]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.O.; Sui, J.; Young, A.B.; Whittal, R.M. Interferences and contaminants encountered in modern mass spectrometry. Anal. Chim. Acta 2008, 71–81. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Cawoy, H.; Mariutto, M.; Henry, G.; Fisher, C.; Vasilyeva, N.; Thonart, P.; Dommes, J.; Ongena, M. Plant Defense Stimulation by Natural Isolates of Bacillus Depends on Efficient Surfactin Production. Mol. Plant-Microbe Interact. 2013, 27, 87–100. [Google Scholar] [CrossRef]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Mögel, G.; Kunz, S.; Borriss, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Bowman, M.J.; Schisler, D.A. Genomic analysis and secondary metabolite production in Bacillus amyloliquefaciens AS 43.3: A biocontrol antagonist of Fusarium head blight. Biol. Control 2013, 64, 166–175. [Google Scholar] [CrossRef]
- Korp, J.; König, S.; Schieferdecker, S.; Dahse, H.M.; König, G.M.; Werz, O.; Nett, M. Harnessing Enzymatic Promiscuity in Myxochelin Biosynthesis for the Production of 5-Lipoxygenase Inhibitors. ChemBioChem 2015, 16, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Bosello, M.; Robbel, L.; Linne, U.; Xie, X.; Marahiel, M.A. Biosynthesis of the siderophore rhodochelin requires the coordinated expression of three independent gene clusters in Rhodococcus jostii RHA1. J. Am. Chem. Soc. 2011, 133, 4587–4595. [Google Scholar] [CrossRef] [PubMed]
- Cone, M.C.; Yin, X.; Grochowski, L.L.; Parker, M.R.; Zabriskie, T.M. The blasticidin S biosynthesis gene cluster from Streptomyces griseochromogenes: Sequence analysis, organization, and initial characterization. ChemBioChem 2003, 4, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Westers, H.; Blom, E.J.; Kuipers, O.P.; Marahiel, M.A. Iron starvation triggers the stringent response and induces amino acid biosynthesis for bacillibactin production in Bacillus subtilis. J. Bacteriol. 2006, 188, 8655–8657. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Masuma, R.; Tanaka, Y.; Tanaka, H.; Omura, S. Production of Nanaomycin and other Antibiotics by Phosphate-depressed Fermentation Using Phosphate-trapping Agents. J. Antibiot. (Tokyo) 1986, 39, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Snchez, M.; Rocha, D.; Snchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of microbial secondary metabolites: Regulation by the carbon source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, G.; Wang, B.; Li, X.; Yue, S.; Chen, J.; Zhang, H.; Wang, H. Production and Identification of Inthomycin B Produced by a Deep-Sea Sediment-Derived Streptomyces sp. YB104 Based on Cultivation-Dependent Approach. Curr. Microbiol. 2018, 75, 942–951. [Google Scholar] [CrossRef]
- Pinzón-Espinosa, A.; Martinez-Matamoros, D.; Castellanos, L.; Duque, C.; Rodríguez, J.; Jiménez, C.; Ramos, F.A. Cereusitin A, a cyclic tetrapeptide from a Bacillus cereus strain isolated from the soft coral Antillogorgia (syn. Pseudopterogorgia) elisabethae. Tetrahedron Lett. 2017, 58, 634–637. [Google Scholar] [CrossRef]
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K.M. Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Jakeman, D.L.; Graham, C.L.; Young, W.; Vining, L.C. Culture conditions improving the production of jadomycin B. J. Ind. Microbiol. Biotechnol. 2006, 33, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.; Somerville, G.A.; Heilmann, C.; Sahl, H.G.; Maurer, H.H.; Herrmann, M. Very low ethanol concentrations affect the viability and growth recovery in post-stationary-phase Staphylococcus aureus populations. Appl. Environ. Microbiol. 2006, 72, 2627–2636. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K. Mixed fermentation for natural product drug discovery. Appl. Microbiol. Biotechnol. 2009, 83, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Tobias, N.J.; Brehm, J.; Kresovic, D.; Brameyer, S.; Bode, H.B.; Heermann, R. New Vocabulary for Bacterial Communication. ChemBioChem 2019, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hamoen, L.W.; Venema, G.; Kuipers, O.P. Controlling competence in Bacillus subtilis: Shared use of regulators. Microbiology 2003, 149, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Milne, P.J.; Hunt, A.L.; Rostoll, K.; Van Der Walt, J.J.; Graz, C.J.M. The Biological Activity of Selected Cyclic Dipeptides. J. Pharm. Phamacol. 1998, 50, 1331–1337. [Google Scholar] [CrossRef]
- Pomilio, A.; Battista, M.; Vitale, A. Naturally-Occurring Cyclopeptides: Structures and Bioactivity. Curr. Org. Chem. 2006, 10, 2075–2121. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Henne, A.; Liesegang, H.; Hitezeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and Functional Characterization of Gene Clusters Directing Nonribosomal Synthesis of Bioactive Cyclic Lipopeptides in. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef]
- Rückert, C.; Blom, J.; Chen, X.H.; Reva, O.; Borriss, R. Genome sequence of B. amyloliquefaciens type strain DSM7T reveals differences to plant-associated B. amyloliquefaciens FZB42. J. Biotechnol. 2011, 155, 78–85. [Google Scholar] [CrossRef]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7 T and FZB42 T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete gen. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Hee, J.S.; Kyoung, H.J.; Tae, S.K.; Oh, K.B.; Shin, J. Cyclic peptides of the nocardamine class from a marine-derived bacterium of the genus Streptomyces. J. Nat. Prod. 2005, 68, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.O.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef]
- Liu, M.; Grkovic, T.; Liu, X.; Han, J.; Zhang, L.; Quinn, R.J. A systems approach using OSMAC, Log P, and NMR fingerprinting: An approach to novelty. Synth. Syst. Biotechnol. 2017, 2, 276–286. [Google Scholar] [CrossRef] [PubMed]
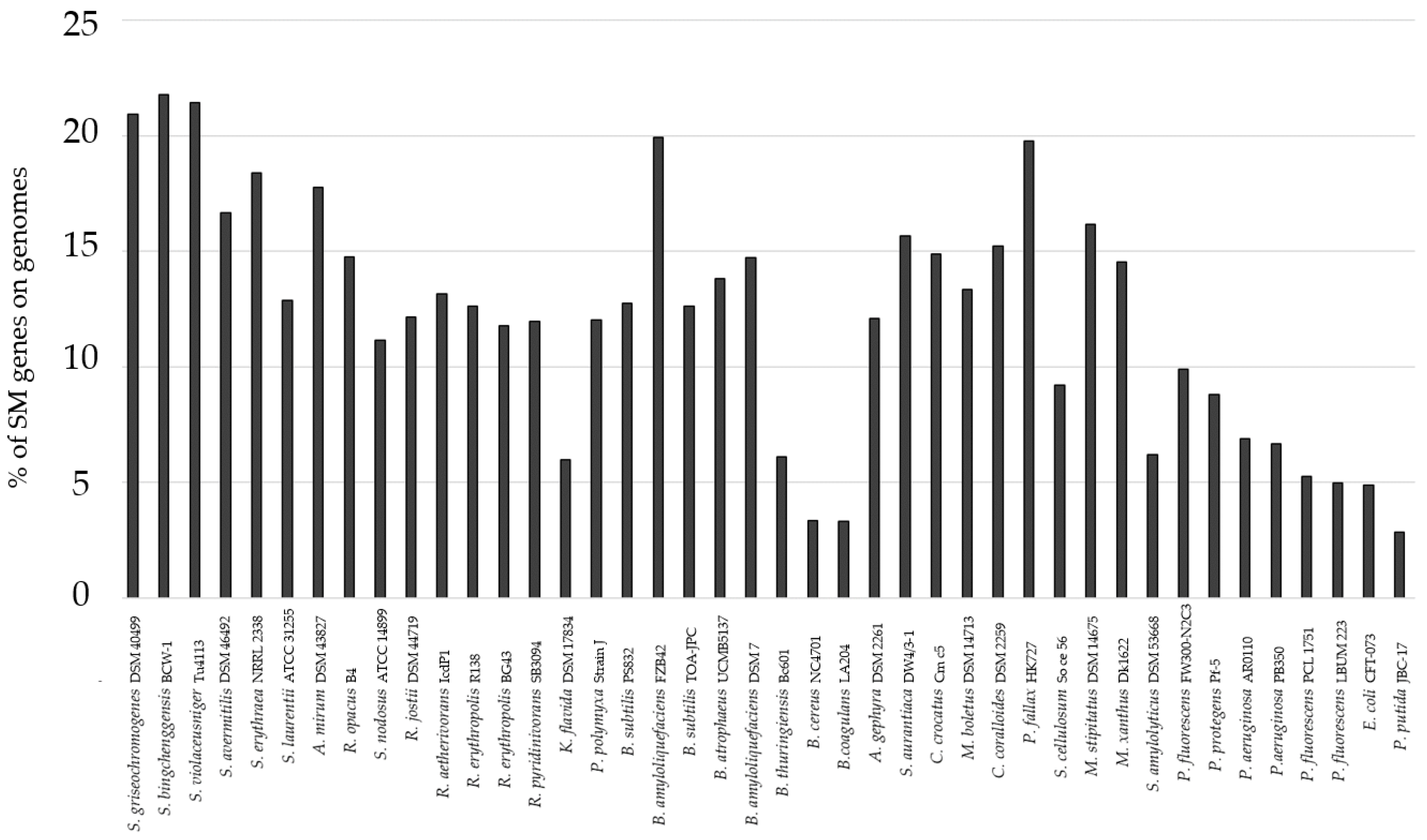
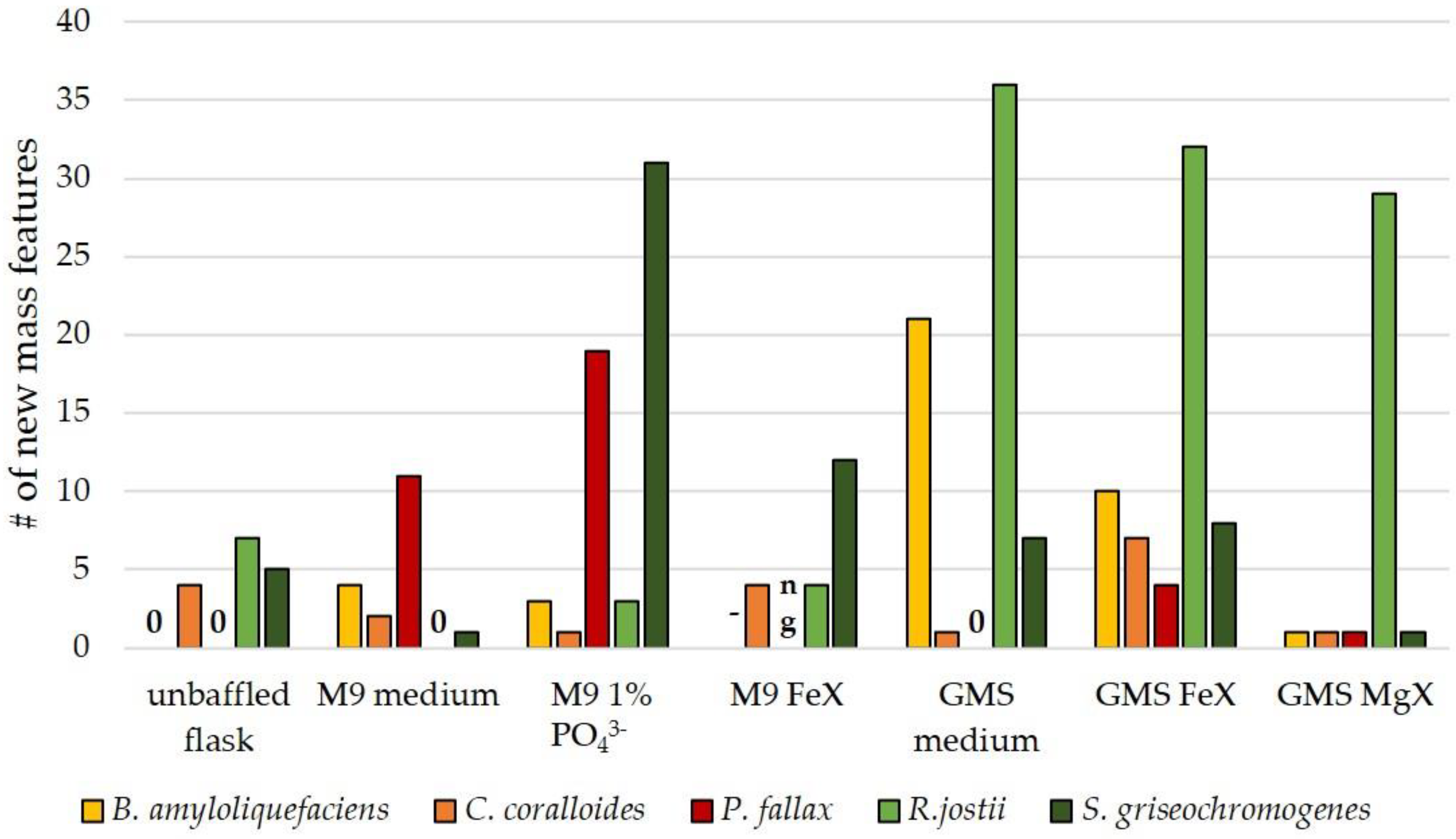

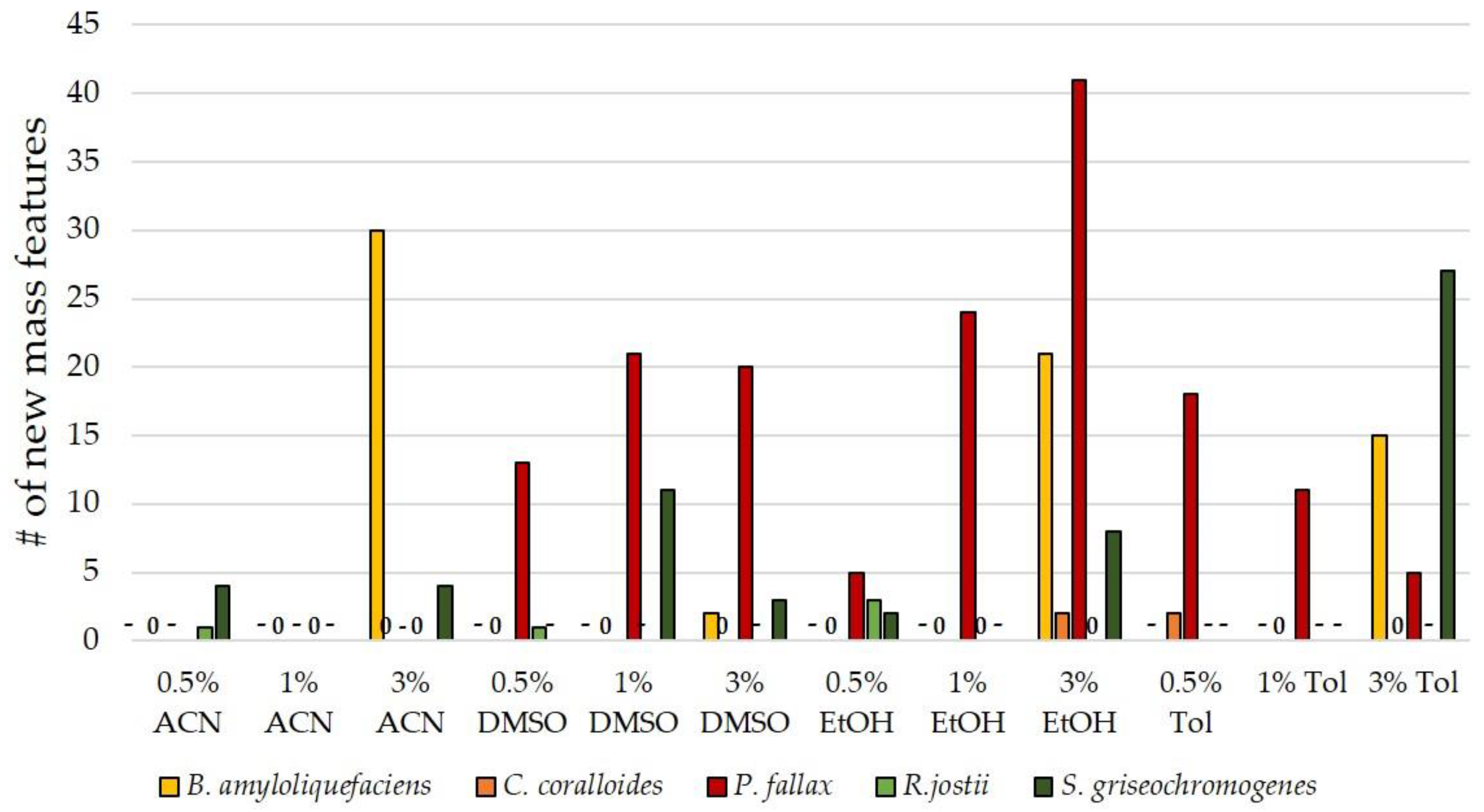
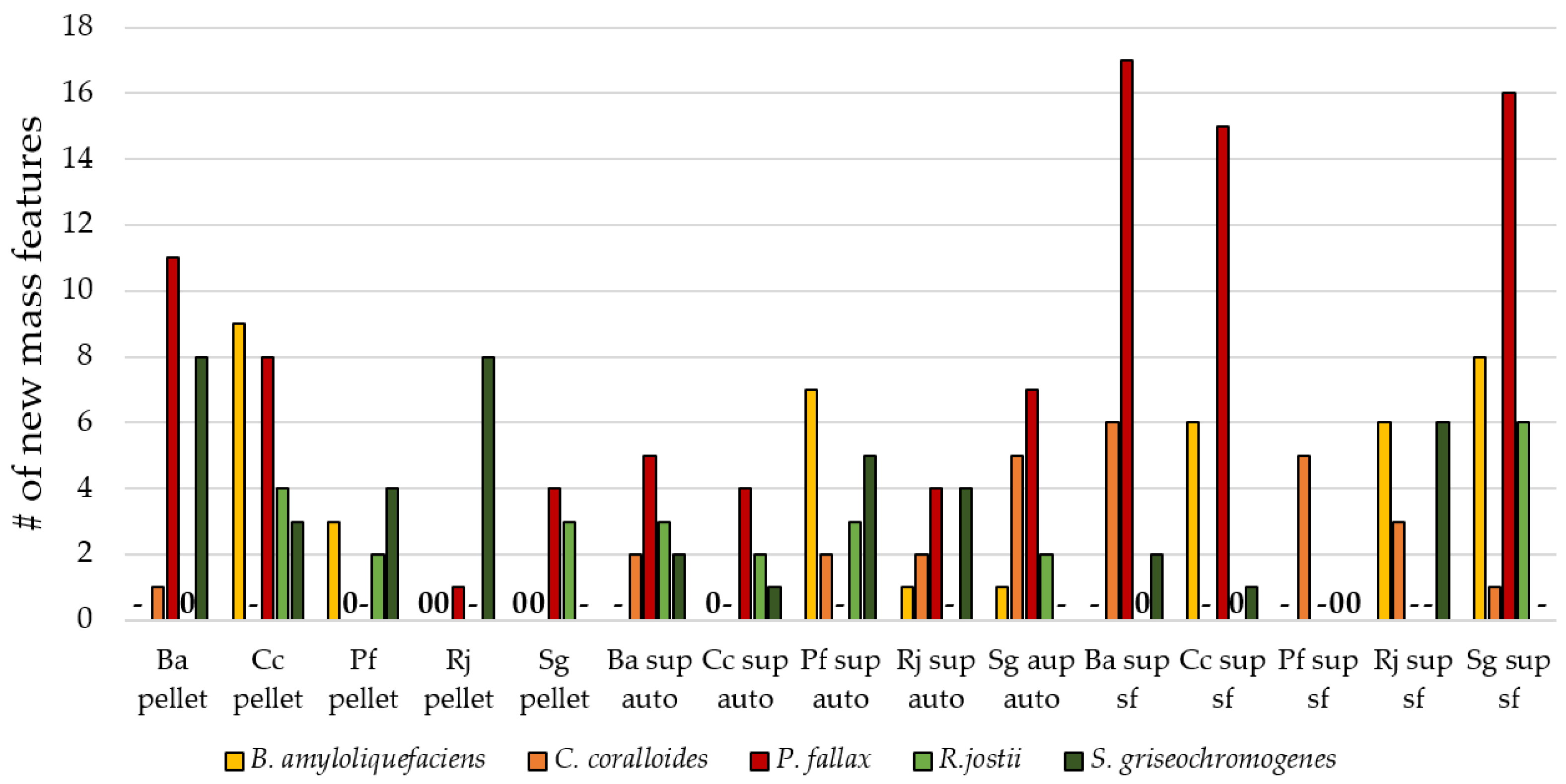
| Strain | Natural Product | Altered Conditions | Properties | Reference |
|---|---|---|---|---|
| Sphaeropsidales sp. F-24’707 | 42 cladospirones | variation of media | antibacterial and antifungal | [16] |
| Streptomyces sp. MBT27 | quinazolinone A and B | variation of carbon sources | anti-inflammatory, antitumor, antimicrobial, and anti-fungal properties | [17] |
| Aspergillus sp. LS34 | 9 new compounds | solid rice medium and potato dextrose medium | cytotoxic activity against cancer cell lines/active against pathogenic Staphylococcus aureus | [18] |
| Category | Tested Conditions |
|---|---|
| culture vessel | unbaffled flask instead of baffled flask |
| nutrient-rich media | 3 per bacterium out of SP medium, TSB medium, CY/H medium, MD1 medium, MD1+G medium, LB medium, GYM medium, NB medium, Landy medium, Luria medium |
| minimal media | M9 medium, GMS medium |
| limitations | Mg2+-, PO43−- and Fe2+ limitation (named MgX [no Mg2+-salts], FeX [no Fe3+-salts], 1% PO43− [1% of amount of phosphate salts in original recipe]) |
| organic solvents | acetonitrile (ACN), dimethyl sulfoxide (DMSO), ethanol (EtOH), toluene (Tol) at 0.5/1/3/6 Vol % |
| biotic additives | autoclaved supernatant, sterile-filtered supernatant, autoclaved cell pellet of B. amyloliqufaciens, C. coralloides, P. fallax, R. jostii, and S. griseochromogenes |
| Strain | Genome Size [Mbp] | % BGCs | Number of BGCs | Known Compounds |
|---|---|---|---|---|
| B. amyloliquefaciens | 3.98 | 14.7 | 11 | surfactin [47], bacillaene [48], fengycin [47], and bacillibactin [49] from B. amyloliquefaciens FZB42 |
| C. coralloides | 10.08 | 15.2 | 34 | - |
| P. fallax | 10.77 | 19.1 | 33 | myxochelin [50] from P. fallax HKI727 |
| R. jostii | 7.89 | 12.2 | 18 | rhodochelin [51] from R. jostii RHA1 |
| S. griseochromogenes | 10.76 | 20.9 | 49 | blasticidin [52] from S. griseochromogenes (not further specified) heterologously expressed in S. lividans |
| Strain | Number of BGCs According to AntiSMASH | Number of Searchable Compounds | Detected Compounds (% Sequence Similarity) | Means of Identification | ||
|---|---|---|---|---|---|---|
| Mass-Based | MS2- Fragmentation (MetFrag) | Reference Compound | ||||
| B. amyloliquefaciens | 11 | 6 | surfactin (82%) | ✓ Δppm = 2.2 (C15) Δppm = 4.9 (C14) | ||
| bacillibactin (100%) | ✓ (and [49]) Δppm = 1.4 | |||||
| bacillaene (100%) | ✓ Δppm = 0.7 | |||||
| C. coralloides | 34 | 12 | - | |||
| P. fallax | 33 | 22 | myxochelin A (75%) | ✓ Δppm = 10.1 | ||
| nostophycin (18%) | ✓ Δppm = 7.0 | |||||
| R. jostii | 18 | 13 | - | |||
| S. griseochromogenes | 49 | 38 | desferrioxamine B (100%) | ✓ Δppm = 4.6 | ||
| albaflavenone (100%) | ✓ Δppm = 38.8 | |||||
| Strain | New Mass Features on M9 FeX | New Mass Features on GMS FeX |
|---|---|---|
| C. coralloides | Cc243, Cc244, Cc249, Cc250 | Cc22, Cc181, Cc186, Cc187, Cc189, Cc193, Cc210 |
| S. griseochromogenes | Sg115, Sg117, Sg119, Sg120, Sg135, Sg198, Sg199, Sg201, Sg203, Sg206, Sg207, Sg214 | Sg96, Sg97, Sg101, Sg103, Sg104, Sg106, Sg107, Sg109 |
| Strain | B. amyloliquefaciens | C. coralloides | P. fallax | R. jostii | S. griseo-chromogenes |
|---|---|---|---|---|---|
| Genome size [Mbp] | 3.98 | 10.08 | 10.76 | 7.89 | 10.77 |
| Number of predicted BGCs | 11 | 34 | 33 | 18 | 49 |
| Number of new mass features in extracts | 127 | 35 | 143 | 138 | 147 |
| ID | m/z | tR [min] | Proposed Compound | Means of Identification | ||
|---|---|---|---|---|---|---|
| GNPS | MetFrag | Reference Compound | ||||
| Ba3 | 261.12 | 4.5 | cyclo (Tyr-Pro) | ✓ | ✓ Δppm = 0.7 | |
| Ba8 | 1057.57 | 7.2 | iturin A-4 | ✓ | ✓ | |
| Ba9 | 1071.58 | 7.6 | iturin A-6 | mass-based | ||
| Ba10 | 1085.6 | 7.9 | iturin A-8 | ✓ | ||
| Ba58 | 883.26 | 6.1 | bacillibactin | comparison to fragmentation pattern from literature, Δppm = 1.4 | ||
| Cc42 | 601.36 | 4.7 | nocardamin | ✓ | ✓ Δppm = 4.4 | |
| Pf336 | 405.16 | 5.4 | myxochelin A | ✓ Δppm = 10.1 | ||
| Sg117 | 587.35 | 4.5 | desmethyl enyl nocardamin Δppm = 0 (GNPS) | ✓ | ||
| Sg130 | 585.36 | 4.9 | desferrioxamine B + Al Δppm = 13 (GNPS) | ✓ | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarz, J.; Hubmann, G.; Rosenthal, K.; Lütz, S. Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria. Biomolecules 2021, 11, 193. https://doi.org/10.3390/biom11020193
Schwarz J, Hubmann G, Rosenthal K, Lütz S. Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria. Biomolecules. 2021; 11(2):193. https://doi.org/10.3390/biom11020193
Chicago/Turabian StyleSchwarz, Jenny, Georg Hubmann, Katrin Rosenthal, and Stephan Lütz. 2021. "Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria" Biomolecules 11, no. 2: 193. https://doi.org/10.3390/biom11020193
APA StyleSchwarz, J., Hubmann, G., Rosenthal, K., & Lütz, S. (2021). Triaging of Culture Conditions for Enhanced Secondary Metabolite Diversity from Different Bacteria. Biomolecules, 11(2), 193. https://doi.org/10.3390/biom11020193






