Structural Insights into Substrate Recognition and Processing by the 20S Proteasome
Abstract
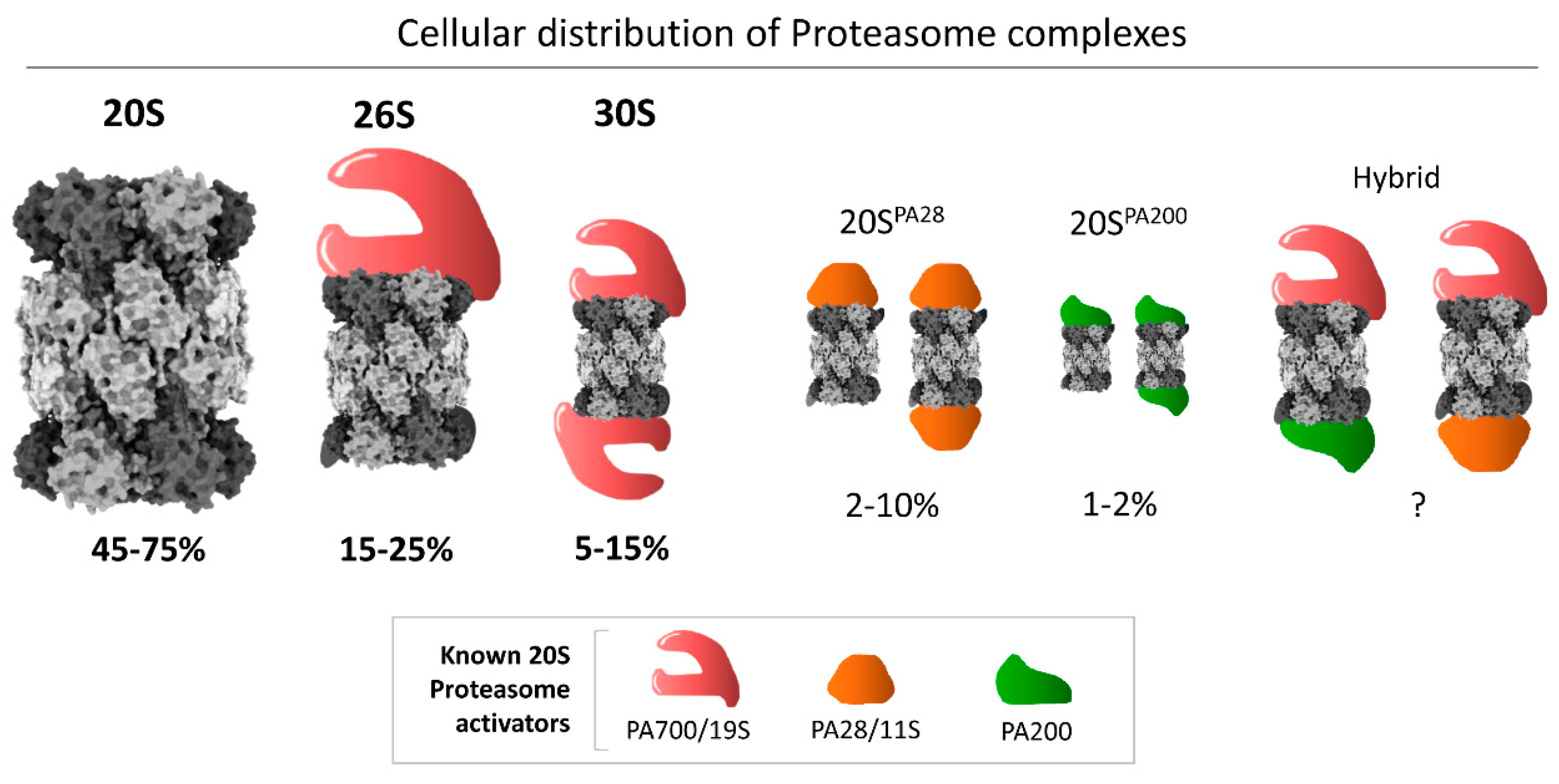
1. Introduction
2. Substrate Degradation Signals for 20S Proteasomes
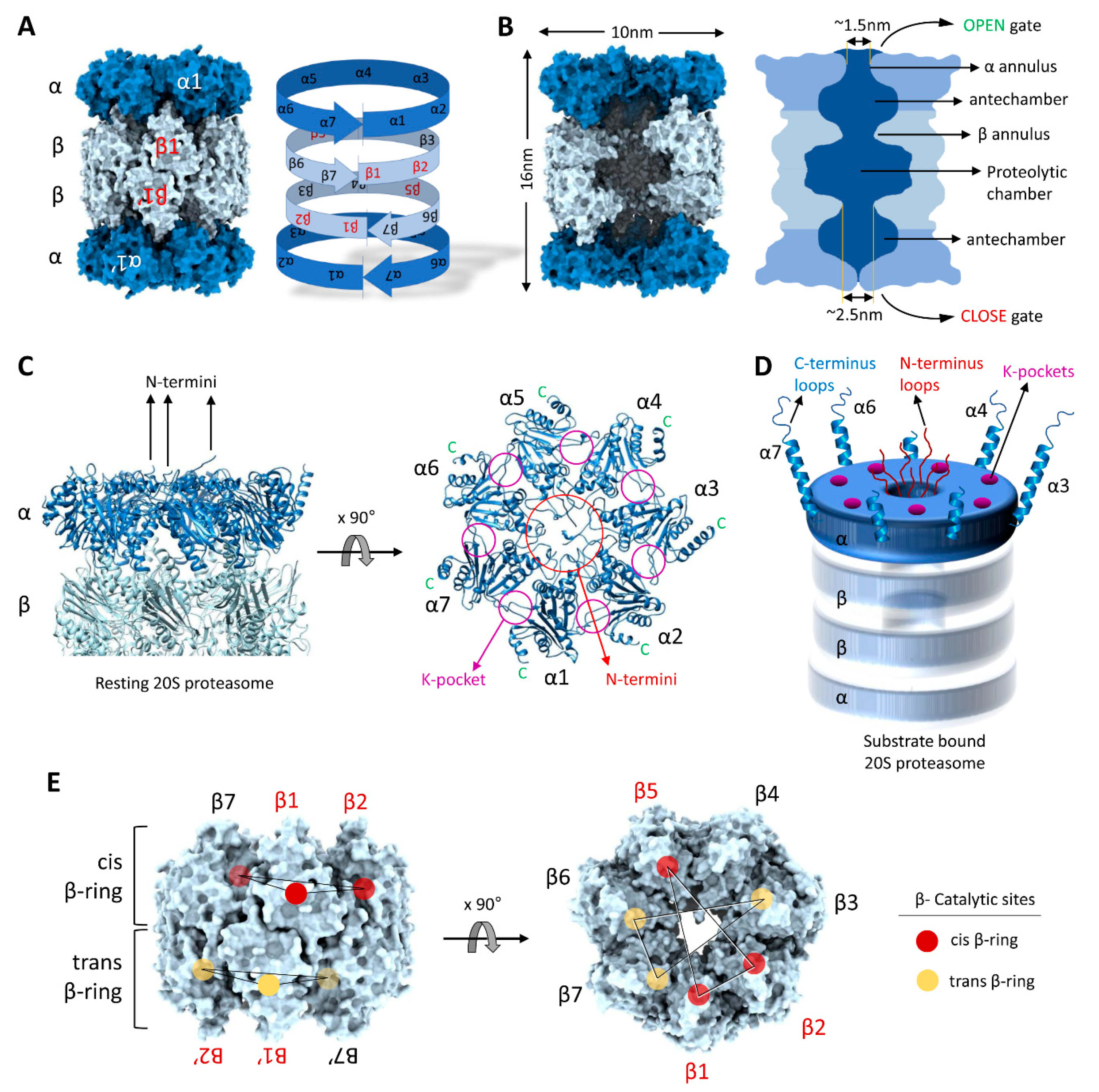
3. Structural Precision of 20S Proteasomes for Substrate Degradation
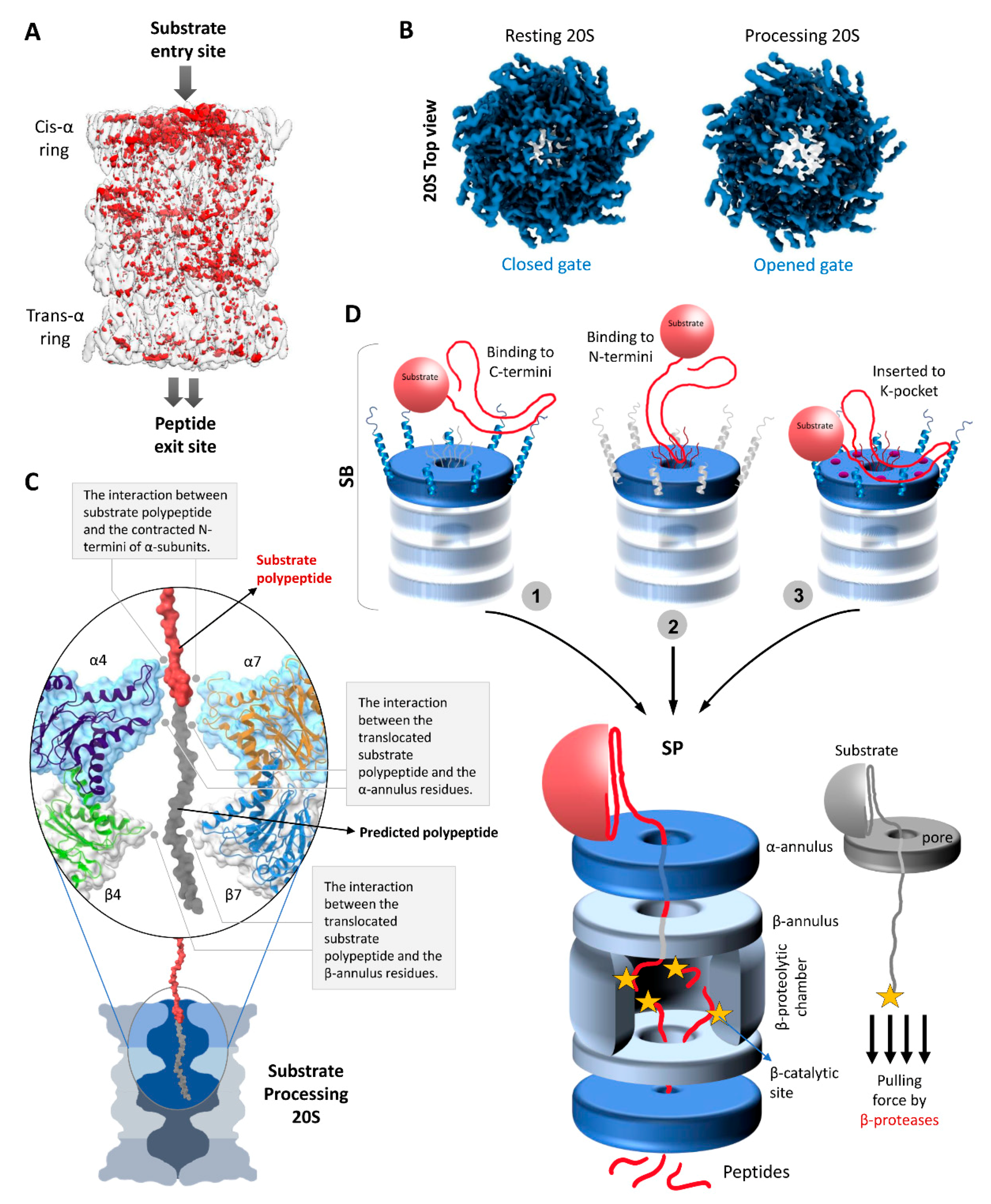
4. The Functional Cycle of 20S Proteasomes
4.1. Substrate Accepting State (SA)
4.2. Substrate Binding State (SB)
4.3. Substrate Processing State (SP)
5. Peptide Generation by the 20S Proteasome
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Bajorek, M.; Finley, D.; Glickman, M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 2003, 13, 1140–1144. [Google Scholar] [CrossRef]
- Livnat-Levanon, N.; Kevei, E.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.; Reis, N.; Hoppe, T.; et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Fabre, B.; Lambour, T.; Garrigues, L.; Ducoux-Petit, M.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Flanagan, J.M.; Tamura, T.; Baumeister, W. Self-compartmentalizing proteases. Trends Biochem. Sci. 1997, 22, 399–404. [Google Scholar] [CrossRef]
- Demasi, M.; da Cunha, F.M. The physiological role of the free 20S proteasome in protein degradation: A critical review. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2948–2954. [Google Scholar] [CrossRef]
- Pickering, A.M.; Davies, K.J. Degradation of damaged proteins: The main function of the 20S proteasome. Prog. Mol. Biol. Transl. Sci. 2012, 109, 227–248. [Google Scholar] [CrossRef]
- Kumar Deshmukh, F.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- Shringarpure, R.; Grune, T.; Mehlhase, J.; Davies, K.J. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003, 278, 311–318. [Google Scholar] [CrossRef]
- Eytan, E.; Ganoth, D.; Armon, T.; Hershko, A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc. Natl. Acad. Sci. USA 1989, 86, 7751–7755. [Google Scholar] [CrossRef]
- Liu, C.W.; Corboy, M.J.; DeMartino, G.N.; Thomas, P.J. Endoproteolytic activity of the proteasome. Science 2003, 299, 408–411. [Google Scholar] [CrossRef] [PubMed]
- McGuire, M.J.; DeMartino, G.N. The latent form of macropain (high molecular weight multicatalytic protease) restores ATP-dependent proteolysis to soluble extracts of BHK fibroblasts pretreated with anti-macropain antibodies. Biochem. Biophys. Res. Commun. 1989, 160, 911–916. [Google Scholar] [CrossRef]
- McGuire, M.J.; McCullough, M.L.; Croall, D.E.; DeMartino, G.N. The high molecular weight multicatalytic proteinase, macropain, exists in a latent form in human erythrocytes. Biochim. Biophys. Acta 1989, 995, 181–186. [Google Scholar] [CrossRef]
- Raynes, R.; Pomatto, L.C.; Davies, K.J. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Aspects Med. 2016, 50, 41–55. [Google Scholar] [CrossRef]
- Majumder, P.; Rudack, T.; Beck, F.; Danev, R.; Pfeifer, G.; Nagy, I.; Baumeister, W. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc. Natl. Acad Sci. USA 2019, 116, 534–539. [Google Scholar] [CrossRef]
- Lowe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef]
- Maupin-Furlow, J.A.; Humbard, M.A.; Kirkland, P.A.; Li, W.; Reuter, C.J.; Wright, A.J.; Zhou, G. Proteasomes from structure to function: Perspectives from Archaea. Curr. Top. Dev. Biol. 2006, 75, 125–169. [Google Scholar]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of proteasome regulators in eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef]
- Barthelme, D.; Sauer, R.T. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science 2012, 337, 843–846. [Google Scholar] [CrossRef]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Bajorek, M.; Glickman, M.H. Proteasome regulatory particles: Keepers of the gates. Cell. Mol. Life Sci. 2004, 61, 1579–1588. [Google Scholar] [PubMed]
- Choi, W.H.; de Poot, S.A.; Lee, J.H.; Kim, J.H.; Han, D.H.; Kim, Y.K.; Finley, D.; Lee, M.J. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat. Commun. 2016, 7, 10963. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, E.; Bajorek, M.; Glickman, M.; Bar-Nun, S. Proteasome Channel Opening as a Rate-Limiting Step in the Ubiquitin-Proteasome System. Isr. J. Chem. 2006, 46, 219–224. [Google Scholar] [CrossRef]
- Groll, M.; Huber, R. Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 2003, 35, 606–616. [Google Scholar] [CrossRef]
- Myers, N.; Olender, T.; Savidor, A.; Levin, Y.; Reuven, N.; Shaul, Y. The Disordered Landscape of the 20S Proteasome Substrates Reveals Tight Association with Phase Separated Granules. Proteomics 2018, 18, e1800076. [Google Scholar] [CrossRef]
- Chondrogianni, N.; Georgila, K.; Kourtis, N.; Tavernarakis, N.; Gonos, E.S. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J. 2015, 29, 611–622. [Google Scholar] [CrossRef]
- Arrigo, A.P.; Tanaka, K.; Goldberg, A.L.; Welch, W.J. Identity of the 19S ‘prosome’ particle with the large multifunctional protease complex of mammalian cells (the proteasome). Nature 1988, 331, 192–194. [Google Scholar] [CrossRef]
- Shringarpure, R.; Grune, T.; Davies, K.J. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell. Mol. Life Sci. 2001, 58, 1442–1450. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Sussman, J.L.; Silman, I.; Shaul, Y. Context-dependent resistance to proteolysis of intrinsically disordered proteins. Protein Sci. 2011, 20, 1285–1297. [Google Scholar] [CrossRef]
- Tonoki, A.; Kuranaga, E.; Tomioka, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Miura, M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009, 29, 1095–1106. [Google Scholar] [CrossRef]
- Davies, K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie 2001, 83, 301–310. [Google Scholar] [CrossRef]
- Chu-Ping, M.; Vu, J.H.; Proske, R.J.; Slaughter, C.A.; DeMartino, G.N. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J. Biol. Chem. 1992, 269, 3539–3547. [Google Scholar] [CrossRef]
- Hoffman, L.; Pratt, G.; Rechsteiner, M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992, 267, 22362–22368. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef] [PubMed]
- Greene, E.R.; Dong, K.C.; Martin, A. Understanding the 26S proteasome molecular machine from a structural and conformational dynamics perspective. Curr. Opin. Struct. Biol. 2020, 61, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Spaller, B.L.; Matouschek, A. Mechanisms of substrate recognition by the 26S proteasome. Curr. Opin. Struct. Biol. 2020, 67, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Baumeister, W. Proteasomes: Unfoldase-assisted protein degradation machines. Biol. Chem. 2019, 401, 183–199. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Mao, Y. Structure, Dynamics and Function of the 26S Proteasome. Sub-Cell. Biochem. 2021, 96, 1–151. [Google Scholar] [CrossRef]
- Bard, J.A.M.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Cascio, P. PA28alphabeta: The enigmatic magic ring of the proteasome? Biomolecules 2014, 4, 566–584. [Google Scholar] [CrossRef] [PubMed]
- Savulescu, A.F.; Glickman, M.H. Proteasome activator 200: The heat is on. Mol. Cell. Proteom. 2011, 10, R110.006890. [Google Scholar] [CrossRef] [PubMed]
- Baldin, V.; Militello, M.; Thomas, Y.; Doucet, C.; Fic, W.; Boireau, S.; Jariel-Encontre, I.; Piechaczyk, M.; Bertrand, E.; Tazi, J.; et al. A novel role for PA28gamma-proteasome in nuclear speckle organization and SR protein trafficking. Mol. Biol. Cell. 2008, 19, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Moriishi, K.; Fukuda, K.; Shirakura, M.; Ishii, K.; Shoji, I.; Wakita, T.; Miyamura, T.; Matsuura, Y.; Suzuki, T. Proteasomal turnover of hepatitis C virus core protein is regulated by two distinct mechanisms: A ubiquitin-dependent mechanism and a ubiquitin-independent but PA28gamma-dependent mechanism. J. Virol. 2009, 83, 2389–2392. [Google Scholar] [CrossRef]
- Dubiel, W.; Pratt, G.; Ferrell, K.; Rechsteiner, M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992, 267, 22369–22377. [Google Scholar] [CrossRef]
- Ma, C.P.; Slaughter, C.A.; DeMartino, G.N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 1992, 267, 10515–10523. [Google Scholar] [CrossRef]
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef]
- Fehlker, M.; Wendler, P.; Lehmann, A.; Enenkel, C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003, 4, 959–963. [Google Scholar] [CrossRef]
- Sahu, I.; Mali, S.M.; Sulkshane, P.; Rozenberg, A.; Xu, C.; Morag, R.; Sahoo, M.P.; Singh, S.K.; Ding, Z.; Wang, Y.; et al. Signature activities of 20S proteasome include degradation of the ubiquitin-tag with the protein under hypoxia. bioRxiv 2019. [Google Scholar] [CrossRef]
- Armon, T.; Ganoth, D.; Hershko, A. Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J. Biol. Chem. 1990, 265, 20723–20726. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Mendillo, M.L.; Zhao, J.; Carette, J.E.; Merrill, P.H.; Cikes, D.; Varadarajan, M.; van Diemen, F.R.; Penninger, J.M.; Goldberg, A.L.; et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Hendil, K.B.; Kriegenburg, F.; Tanaka, K.; Murata, S.; Lauridsen, A.M.; Johnsen, A.H.; Hartmann-Petersen, R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J. Mol. Biol. 2009, 394, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Sahara, K.; Kogleck, L.; Yashiroda, H.; Murata, S. The mechanism for molecular assembly of the proteasome. Adv. Biol. Regul. 2014, 54, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Tomko, R.J., Jr.; Hochstrasser, M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 2013, 82, 415–445. [Google Scholar] [CrossRef] [PubMed]
- Njomen, E.; Osmulski, P.A.; Jones, C.L.; Gaczynska, M.; Tepe, J.J. Small Molecule Modulation of Proteasome Assembly. Biochemistry 2018, 57, 4214–4224. [Google Scholar] [CrossRef]
- Jung, T.; Grune, T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 2008, 60, 743–752. [Google Scholar] [CrossRef]
- Hohn, T.J.; Grune, T. The proteasome and the degradation of oxidized proteins: Part III-Redox regulation of the proteasomal system. Redox Biol. 2014, 2, 388–394. [Google Scholar] [CrossRef]
- Mayor, T.; Sharon, M.; Glickman, M.H. Tuning the proteasome to brighten the end of the journey. Am. J. Physiol. Cell Physiol. 2016, 311, C793–C804. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Myers, N.; Moscovitz, O.; Sharon, M.; Prilusky, J.; Shaul, Y. Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates. Mol. Biosyst. 2012, 8, 368–373. [Google Scholar] [CrossRef]
- Kurepa, J.; Smalle, J.A. To misfold or to lose structure?: Detection and degradation of oxidized proteins by the 20S proteasome. Plant Signal. Behav. 2008, 3, 386–388. [Google Scholar] [CrossRef]
- Jung, T.; Hohn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II - protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Enenkel, C. The paradox of proteasome granules. Curr. Genet. 2018, 64, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Peters, L.Z.; Hazan, R.; Breker, M.; Schuldiner, M.; Ben-Aroya, S. Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J. Cell Biol. 2013, 201, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yen, J.; Kaiser, P.; Huang, L. Regulation of the 26S proteasome complex during oxidative stress. Sci. Signal. 2010, 3, ra88. [Google Scholar] [CrossRef] [PubMed]
- Besche, H.C.; Sha, Z.; Kukushkin, N.V.; Peth, A.; Hock, E.M.; Kim, W.; Gygi, S.; Gutierrez, J.A.; Liao, H.; Dick, L.; et al. Autoubiquitination of the 26S Proteasome on Rpn13 Regulates Breakdown of Ubiquitin Conjugates. EMBO J. 2014, 33, 1159–1176. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Wen, X.; Klionsky, D.J. The proteasome subunit RPN10 functions as a specific receptor for degradation of the 26S proteasome by macroautophagy in Arabidopsis. Autophagy 2016, 12, 905–906. [Google Scholar] [CrossRef]
- Cohen-Kaplan, V.; Livneh, I.; Avni, N.; Fabre, B.; Ziv, T.; Kwon, Y.T.; Ciechanover, A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, E7490–E7499. [Google Scholar] [CrossRef]
- Reinheckel, T.; Sitte, N.; Ullrich, O.; Kuckelkorn, U.; Davies, K.J.; Grune, T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998, 335 (Pt 3), 637–642. [Google Scholar] [CrossRef]
- Okonko, D.O.; Shah, A.M. Heart failure: Mitochondrial dysfunction and oxidative stress in CHF. Nat. Rev. Cardiol. 2015, 12, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Day, S.M.; Divald, A.; Wang, P.; Davis, F.; Bartolone, S.; Jones, R.; Powell, S.R. Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure. Circ. Heart Fail. 2013, 6, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Predmore, J.M.; Wang, P.; Davis, F.; Bartolone, S.; Westfall, M.V.; Dyke, D.B.; Pagani, F.; Powell, S.R.; Day, S.M. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 2010, 121, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Waku, T. New addiction to the NRF2-related factor NRF3 in cancer cells: Ubiquitin-independent proteolysis through the 20S proteasome. Cancer Sci. 2020, 111, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, V.; Yaniv, K.; Sharon, M. Beyond cells: The extracellular circulating 20S proteasomes. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1867, 166041. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Groll, M.; Nazif, T.; Huber, R.; Bogyo, M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chem. Biol. 2002, 9, 655–662. [Google Scholar] [CrossRef]
- Wenzel, T.; Baumeister, W. Conformational constraints in protein degradation by the 20S proteasome. Nat. Struct. Biol. 1995, 2, 199–204. [Google Scholar] [CrossRef]
- Osmulski, P.A.; Gaczynska, M. Nanoenzymology of the 20S proteasome: Proteasomal actions are controlled by the allosteric transition. Biochemistry 2002, 41, 7047–7053. [Google Scholar] [CrossRef]
- Grune, T.; Merker, K.; Sandig, G.; Davies, K.J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003, 305, 709–718. [Google Scholar] [CrossRef]
- Grune, T.; Botzen, D.; Engels, M.; Voss, P.; Kaiser, B.; Jung, T.; Grimm, S.; Ermak, G.; Davies, K.J. Tau protein degradation is catalyzed by the ATP/ubiquitin-independent 20S proteasome under normal cell conditions. Arch Biochem. Biophys. 2010, 500, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Pickering, A.M.; Staab, T.A.; Tower, J.; Sieburth, D.; Davies, K.J. A conserved role for the 20S proteasome and Nrf2 transcription factor in oxidative stress adaptation in mammals, Caenorhabditis elegans and Drosophila melanogaster. J. Exp. Biol. 2013, 216, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Touitou, R.; Richardson, J.; Bose, S.; Nakanishi, M.; Rivett, J.; Allday, M.J. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 2001, 20, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Ruff, K.M. Predicting Conformational Properties of Intrinsically Disordered Proteins from Sequence. Methods Mol. Biol. 2020, 2141, 347–389. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Reuven, N.; Shaul, Y. The nanny model for IDPs. Nat. Chem. Biol. 2009, 5, 778–781. [Google Scholar] [CrossRef]
- Demasi, M.; Hand, A.; Ohara, E.; Oliveira, C.L.; Bicev, R.N.; Bertoncini, C.A.; Netto, L.E. 20S proteasome activity is modified via S-glutathionylation based on intracellular redox status of the yeast Saccharomyces cerevisiae: Implications for the degradation of oxidized proteins. Arch Biochem. Biophys. 2014, 557, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Demasi, M.; Netto, L.E.; Silva, G.M.; Hand, A.; de Oliveira, C.L.; Bicev, R.N.; Gozzo, F.; Barros, M.H.; Leme, J.M.; Ohara, E. Redox regulation of the proteasome via S-glutathionylation. Redox Biol. 2014, 2, 44–51. [Google Scholar] [CrossRef]
- Ng, A.H.; Fang, N.N.; Comyn, S.A.; Gsponer, J.; Mayor, T. System-wide analysis reveals intrinsically disordered proteins are prone to ubiquitylation after misfolding stress. Mol. Cell Proteom. 2013, 12, 2456–2467. [Google Scholar] [CrossRef]
- Arendt, C.S.; Hochstrasser, M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl. Acad. Sci. USA 1997, 94, 7156–7161. [Google Scholar] [CrossRef]
- Groll, M.; Heinemeyer, W.; Jager, S.; Ullrich, T.; Bochtler, M.; Wolf, D.H.; Huber, R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc. Nat. Acad. Sci. USA 1999, 96, 10976–10983. [Google Scholar] [CrossRef]
- Huber, E.M.; Heinemeyer, W.; Li, X.; Arendt, C.S.; Hochstrasser, M.; Groll, M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat. Commun. 2016, 7, 10900. [Google Scholar] [CrossRef] [PubMed]
- Volker, C.; Lupas, A. Molecular Evolution of Proteasomes. In Current Topics in Microbiology and Immunology; Zwickl, P., Baumeister, W., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2002; Volume 268, pp. 1–22. [Google Scholar]
- da Fonseca, P.C.; Morris, E.P. Structure of the human 26S proteasome: Subunit radial displacements open the gate into the proteolytic core. J. Biol. Chem. 2008, 283, 23305–23314. [Google Scholar] [CrossRef] [PubMed]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Köhler, A.; Cascio, P.; Leggett, D.S.; Woo, K.M.; Goldberg, A.L.; Finley, D. The Axial Channel of the Proteasome Core Particle Is Gated by the Rpt2 ATPase and Controls Both Substrate Entry and Product Release. Mol. Cell 2001, 7, 1143–1152. [Google Scholar] [CrossRef]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T.; Hill, C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 2010, 37, 728–735. [Google Scholar] [CrossRef]
- Osmulski, P.A.; Hochstrasser, M.; Gaczynska, M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 2009, 17, 1137–1147. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55. [Google Scholar] [CrossRef]
- Sdek, P.; Ying, H.; Chang, D.L.; Qiu, W.; Zheng, H.; Touitou, R.; Allday, M.J.; Xiao, Z.X. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 2005, 20, 699–708. [Google Scholar] [CrossRef]
- Giono, L.E.; Resnick-Silverman, L.; Carvajal, L.A.; St Clair, S.; Manfredi, J.J. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase. Oncogene 2017, 36, 6762–6773. [Google Scholar] [CrossRef]
- Barthelme, D.; Chen, J.Z.; Grabenstatter, J.; Baker, T.A.; Sauer, R.T. Architecture and assembly of the archaeal Cdc48*20S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, E1687–E1694. [Google Scholar] [CrossRef]
- Gillette, T.G.; Kumar, B.; Thompson, D.; Slaughter, C.A.; DeMartino, G.N. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 2008, 283, 31813–31822. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Kim, Y.C.; DeMartino, G.N. The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation. J. Biol. Chem. 2010, 285, 39523–39535. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell. 2007, 27, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Toste Rego, A.; da Fonseca, P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell. 2019, 76, 138–147.e135. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Kaganovich, D.; Goldberg, A.L. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20 S proteasomes. Evidence for peptide-induced channel opening in the alpha-rings. J. Biol. Chem. 2002, 277, 22260–22270. [Google Scholar] [CrossRef]
- Gaczynska, M.; Osmulski, P.A. Harnessing proteasome dynamics and allostery in drug design. Antioxid. Redox Signal. 2014, 21, 2286–2301. [Google Scholar] [CrossRef]
- Silva, G.M.; Netto, L.; Simoes, V.; Santos, L.F.; Gozzo, F.C.; Demasi, M.A.; Oliveira, C.L.; Bicev, R.N.; Klitzke, C.F.; Sogayar, M.C.; et al. Redox Control of 20s Proteasome Gating. Antioxid. Redox Signal. 2012. [Google Scholar] [CrossRef]
- Chen, P.; Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 1996, 86, 961–972. [Google Scholar] [CrossRef]
- Dick, T.P.; Nussbaum, A.K.; Deeg, M.; Heinemeyer, W.; Groll, M.; Schirle, M.; Keilholz, W.; Stevanovic, S.; Wolf, D.H.; Huber, R.; et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J. Biol. Chem. 1998, 273, 25637–25646. [Google Scholar] [CrossRef]
- Heinemeyer, W.; Fischer, M.; Krimmer, T.; Stachon, U.; Wolf, D.H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 1997, 272, 25200–25209. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.C.; Enenkel, C. Proteasome assembly. Cell. Mol. Life Sci. 2014, 71, 4729–4745. [Google Scholar] [CrossRef] [PubMed]
- Hutschenreiter, S.; Tinazli, A.; Model, K.; Tampe, R. Two-substrate association with the 20S proteasome at single-molecule level. EMBO J. 2004, 23, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Rennella, E.; Huang, R.; Yu, Z.; Kay, L.E. Exploring long-range cooperativity in the 20S proteasome core particle from Thermoplasma acidophilum using methyl-TROSY-based NMR. Proc. Natl. Acad. Sci. USA 2020, 117, 5298–5309. [Google Scholar] [CrossRef] [PubMed]
- de la Pena, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Huynh, T.; Li, J.; Zhou, R. Nanomechanics of Protein Unfolding Outside a Generic Nanopore. ACS Nano 2016, 10, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Akopian, T.N.; Woo, K.M.; Goldberg, A.L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999, 274, 3363–3371. [Google Scholar] [CrossRef]
- Nussbaum, A.K.; Dick, T.P.; Keilholz, W.; Schirle, M.; Stevanovic, S.; Dietz, K.; Heinemeyer, W.; Groll, M.; Wolf, D.H.; Huber, R.; et al. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. USA 1998, 95, 12504–12509. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Akopian, T.N.; Goldberg, A.L. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 1998, 273, 1982–1989. [Google Scholar] [CrossRef]
- Emmerich, N.P.; Nussbaum, A.K.; Stevanovic, S.; Priemer, M.; Toes, R.E.; Rammensee, H.G.; Schild, H. The human 26 S and 20 S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J. Biol. Chem. 2000, 275, 21140–21148. [Google Scholar] [CrossRef]
- Toes, R.E.; Nussbaum, A.K.; Degermann, S.; Schirle, M.; Emmerich, N.P.; Kraft, R.; Zwinderman, A.; Dick, T.P.; Muller, J.; Schonfisch, B.; et al. Discrete cleavage motifs of constitutive and immunoproteasomes: Quantitive analysis of cleavage products. J. Exp. Med. 2001, 194, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28alphabeta reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, G.; Emch, S.; Groettrup, M.; Holzhutter, H.G. Evidence for the existence of a non-catalytic modifier site of peptide hydrolysis by the 20 S proteasome. J. Biol. Chem. 2000, 275, 22056–22063. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H. Getting in and out of the proteasome. Semin. Cell Dev. Biol. 2000, 11, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Realini, C.; Ustrell, V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem. J. 2000, 345 Pt 1, 1–15. [Google Scholar] [CrossRef]
- Dasgupta, S.; Castro, L.M.; Dulman, R.; Yang, C.; Schmidt, M.; Ferro, E.S.; Fricker, L.D. Proteasome inhibitors alter levels of intracellular peptides in HEK293T and SH-SY5Y cells. PLoS One 2014, 9, e103604. [Google Scholar] [CrossRef]
- Dasgupta, S.; Yang, C.; Castro, L.M.; Tashima, A.K.; Ferro, E.S.; Moir, R.D.; Willis, I.M.; Fricker, L.D. Analysis of the Yeast Peptidome and Comparison with the Human Peptidome. PLoS ONE 2016, 11, e0163312. [Google Scholar] [CrossRef]
- de Araujo, C.B.; Heimann, A.S.; Remer, R.A.; Russo, L.C.; Colquhoun, A.; Forti, F.L.; Ferro, E.S. Intracellular Peptides in Cell Biology and Pharmacology. Biomolecules 2019, 9, 150. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, I.; Glickman, M.H. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules 2021, 11, 148. https://doi.org/10.3390/biom11020148
Sahu I, Glickman MH. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules. 2021; 11(2):148. https://doi.org/10.3390/biom11020148
Chicago/Turabian StyleSahu, Indrajit, and Michael H. Glickman. 2021. "Structural Insights into Substrate Recognition and Processing by the 20S Proteasome" Biomolecules 11, no. 2: 148. https://doi.org/10.3390/biom11020148
APA StyleSahu, I., & Glickman, M. H. (2021). Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules, 11(2), 148. https://doi.org/10.3390/biom11020148





