AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family
Abstract
1. Introduction
2. Materials and Methods
2.1. Three-Dimensional Structures of KCTD Proteins and Their Analysis
2.2. Comparison of the Three-Dimensional Structures
2.3. Generation of Structural Tree
3. Results
3.1. KCTDs in the PDB: The State of the Art
| Protein | Domain/Complex | PDB Code(s)/References | Resolution(s) (Å) | RMSD (Å) (#) 1 |
|---|---|---|---|---|
| KCTD1 | BTB | 5BXD/5BXB [17] | 1.8/2.2 | 0.6 (103)/0.4 (103) |
| Full-length | 6S4L | 2.4 | 0.8 (205) | |
| KCTD5 | BTB | 3DRZ [16] | 1.9 | 0.8 (102) |
| Full-length | 3DRX/3DRY [16] | 3.1/3.3 | 2.1 (163)/2.4(133) | |
| KCTD8 | CTD | 6G57 | 2.8 | 1.8 (88) |
| KCTD9 | BTB | 5BXH [17] | 2.8 | 0.7 (97) |
| KCTD10 | BTB | 5FTA [15] | 2.6 | 0.4 (96) |
| KCTD12 | CTD | 6QZL | 2.0 | 0.7 (100) |
| CTD-Gβ1γ2 | 6M8S [22] | 3.7 | 0.8 (103) | |
| KCTD13 | BTB | 4UIJ [15] | 2.7 | 0.3 (102) |
| KCTD16 | BTB | 5A15/6OCR/6I0Q/6OCT [15,34,35] | 2.8/2.3/2.3 | 0.5 (91)/0.4 (90)/0.7 (95)/0.5 (91) |
| CTD | 6QB7 | 2.2 | 0.7 (110) | |
| BTB-GABAB2 pept | 6OCP/6M8R [22,35] | 2.3/3.2 | 0.5 (91)/0.5 (93) | |
| BTB | 5A6R [15] | 2.8 | 0.7 (101) | |
| BTB | 4CRH [15] | 1.7 | 1.0 (90) |
3.2. AlphaFold Versus PDB KCTD Structures
3.3. Detecting Analogies among AlphaFold Structures of KCTD
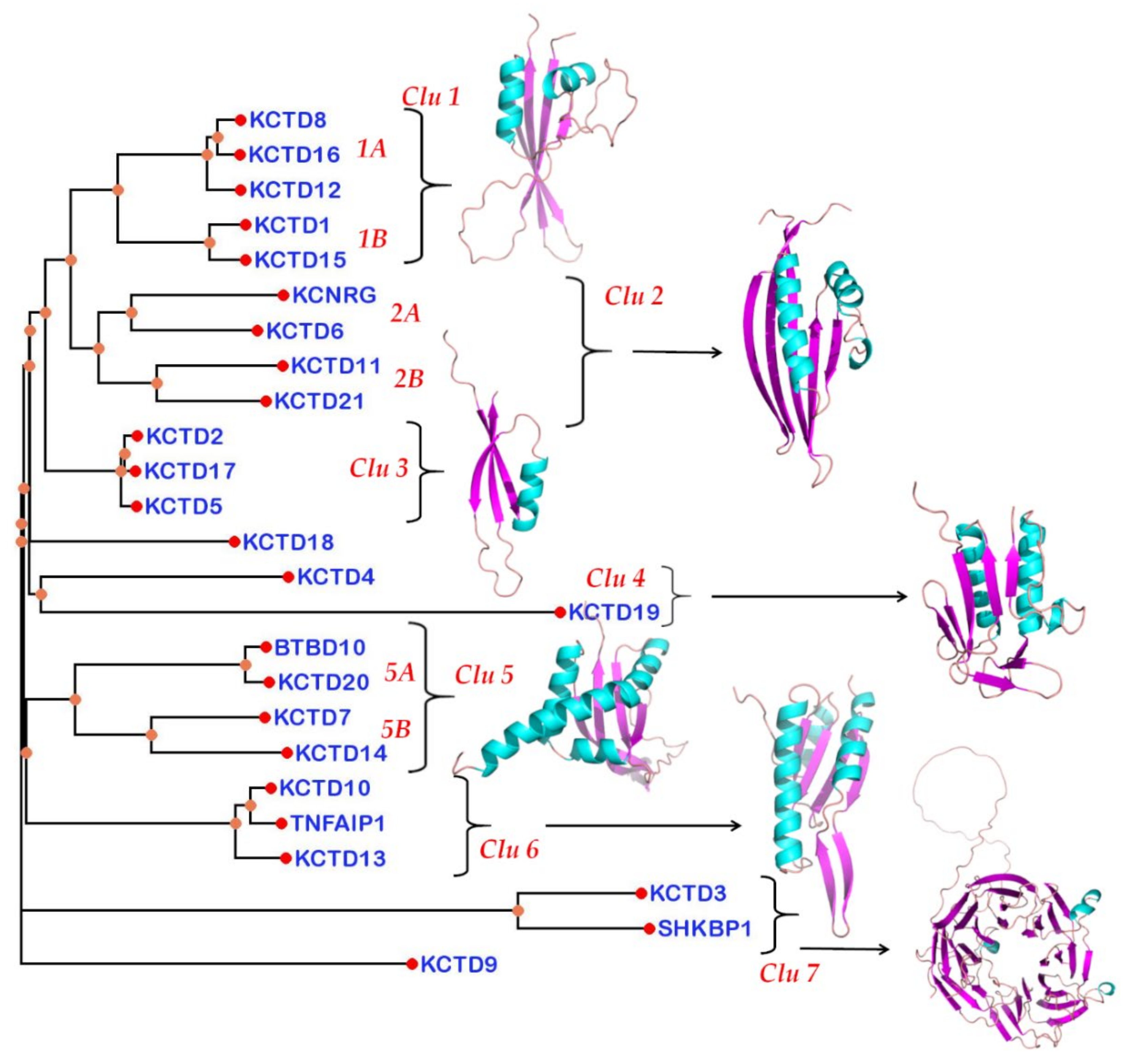
3.4. Structure-Based Pseudo-Phylogenetic Tree
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Canettieri, G.; Di Marcotullio, L.; Greco, A.; Coni, S.; Antonucci, L.; Infante, P.; Pietrosanti, L.; De Smaele, E.; Ferretti, E.; Miele, E.; et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 2010, 12, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, M.; Blomen, V.A.; Nieuwenhuis, J.; Stickel, E.; Raaben, M.; Bleijerveld, O.B.; Altelaar, A.F.M.; Jae, L.T.; Brummelkamp, T.R. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature 2017, 546, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Escamilla, C.O.; Filonova, I.; Walker, A.K.; Xuan, Z.X.; Holehonnur, R.; Espinosa, F.; Liu, S.; Thyme, S.B.; Lopez-Garcia, I.A.; Mendoza, D.B.; et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature 2017, 551, 227–231. [Google Scholar] [CrossRef]
- Golzio, C.; Willer, J.; Talkowski, M.E.; Oh, E.C.; Taniguchi, Y.; Jacquemont, S.; Reymond, A.; Sun, M.; Sawa, A.; Gusella, J.F.; et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 2012, 485, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, J.; Metz, M.; Zolles, G.; Turecek, R.; Fritzius, T.; Bildl, W.; Tarusawa, E.; Kulik, A.; Unger, A.; Ivankova, K.; et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature 2010, 465, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Aouacheria, A.; Lionnard, L.; Metz, K.A.; Soane, L.; Kamiya, A.; Hardwick, J.M. KCTD: A new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci. Ther. 2019, 25, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Angrisani, A.; Di Fiore, A.; De Smaele, E.; Moretti, M. The emerging role of the KCTD proteins in cancer. Cell Commun. Signal. 2021, 19, 56. [Google Scholar] [CrossRef]
- Marneros, A.G.; Beck, A.E.; Turner, E.H.; McMillin, M.J.; Edwards, M.J.; Field, M.; de Macena Sobreira, N.L.; Perez, A.B.; Fortes, J.A.; Lampe, A.K.; et al. Mutations in KCTD1 cause scalp-ear-nipple syndrome. Am. J. Hum. Genet. 2013, 92, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Balasco, N.; Pirone, L.; Caruso, D.; Di Gaetano, S.; Pedone, E.M.; Vitagliano, L. Molecular basis of the scalp-ear-nipple syndrome unraveled by the characterization of disease-causing KCTD1 mutants. Sci. Rep. 2019, 9, 10519. [Google Scholar] [CrossRef] [PubMed]
- Willer, C.J.; Speliotes, E.K.; Loos, R.J.; Li, S.; Lindgren, C.M.; Heid, I.M.; Berndt, S.I.; Elliott, A.L.; Jackson, A.U.; Lamina, C.; et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009, 41, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Pirone, L.; Smaldone, G.; Spinelli, R.; Barberisi, M.; Beguinot, F.; Vitagliano, L.; Miele, C.; Di Gaetano, S.; Raciti, G.A.; Pedone, E. KCTD1: A novel modulator of adipogenesis through the interaction with the transcription factor AP2alpha. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 158514. [Google Scholar] [CrossRef] [PubMed]
- Zollman, S.; Godt, D.; Prive, G.G.; Couderc, J.L.; Laski, F.A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 1994, 91, 10717–10721. [Google Scholar] [CrossRef]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Prive, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Pirone, L.; Pedone, E.; Marlovits, T.; Vitagliano, L.; Ciccarelli, L. The BTB domains of the potassium channel tetramerization domain proteins prevalently assume pentameric states. FEBS Lett. 2016, 590, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Pinkas, D.M.; Sanvitale, C.E.; Bufton, J.C.; Sorrell, F.J.; Solcan, N.; Chalk, R.; Doutch, J.; Bullock, A.N. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem. J. 2017, 474, 3747–3761. [Google Scholar] [CrossRef] [PubMed]
- Dementieva, I.S.; Tereshko, V.; McCrossan, Z.A.; Solomaha, E.; Araki, D.; Xu, C.; Grigorieff, N.; Goldstein, S.A. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J. Mol. Biol. 2009, 387, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.X.; Chu, A.; Nielsen, T.K.; Benlekbir, S.; Rubinstein, J.L.; Prive, G.G. Structural Insights into KCTD Protein Assembly and Cullin3 Recognition. J. Mol. Biol. 2016, 428, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Balasco, N.; Smaldone, G.; Vitagliano, L. The Structural Versatility of the BTB Domains of KCTD Proteins and Their Recognition of the GABAB Receptor. Biomolecules 2019, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Pirone, L.; Balasco, N.; Di Gaetano, S.; Pedone, E.M.; Vitagliano, L. Cullin 3 Recognition Is Not a Universal Property among KCTD Proteins. PLoS ONE 2015, 10, e0126808. [Google Scholar] [CrossRef] [PubMed]
- Balasco, N.; Pirone, L.; Smaldone, G.; Di Gaetano, S.; Esposito, L.; Pedone, E.M.; Vitagliano, L. Molecular recognition of Cullin3 by KCTDs: Insights from experimental and computational investigations. Biochim. Biophys. Acta 2014, 1844, 1289–1298. [Google Scholar] [CrossRef]
- Zarelli, V.E.; Dawid, I.B. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc. Natl. Acad. Sci. USA 2013, 110, 2870–2875. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Abreu, N.; Levitz, J.; Kruse, A.C. Structural basis for KCTD-mediated rapid desensitization of GABAB signalling. Nature 2019, 567, 127–131. [Google Scholar] [CrossRef]
- Skoblov, M.; Marakhonov, A.; Marakasova, E.; Guskova, A.; Chandhoke, V.; Birerdinc, A.; Baranova, A. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. Bioessays 2013, 35, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiang, Y.; Sun, G. The KCTD family of proteins: Structure, function, disease relevance. Cell Biosci. 2013, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- Smaldone, G.; Beneduce, G.; Incoronato, M.; Pane, K.; Franzese, M.; Coppola, L.; Cordella, A.; Parasole, R.; Ripaldi, M.; Nassa, G.; et al. KCTD15 is overexpressed in human childhood B-cell acute lymphoid leukemia. Sci. Rep. 2019, 9, 20108. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zidek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Correale, S.; Pirone, L.; Di Marcotullio, L.; De Smaele, E.; Greco, A.; Mazza, D.; Moretti, M.; Alterio, V.; Vitagliano, L.; Di Gaetano, S.; et al. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie 2011, 93, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laakso, L.M. Dali server update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Rosenstrom, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Holm, L.; Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993, 233, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Fredslund, J. PHY.FI: Fast and easy online creation and manipulation of phylogeny color figures. BMC Bioinform. 2006, 7, 315. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sereikaite, V.; Fritzius, T.; Kasaragod, V.B.; Bader, N.; Maric, H.M.; Schindelin, H.; Bettler, B.; Stromgaard, K. Targeting the gamma-Aminobutyric Acid Type B (GABAB) Receptor Complex: Development of Inhibitors Targeting the K(+) Channel Tetramerization Domain (KCTD) Containing Proteins/GABAB Receptor Protein-Protein Interaction. J. Med. Chem. 2019, 62, 8819–8830. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.; Glaaser, I.; Zhao, Y.; Kurinov, I.; Mosyak, L.; Wang, H.; Liu, J.; Park, J.; Frangaj, A.; Sturchler, E.; et al. Structural basis for auxiliary subunit KCTD16 regulation of the GABAB receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 8370–8379. [Google Scholar] [CrossRef] [PubMed]
- Barone, D.; Balasco, N.; Vitagliano, L. KCTD5 is endowed with large, functionally relevant, interdomain motions. J. Biomol. Struct. Dyn. 2016, 34, 1725–1735. [Google Scholar] [CrossRef]
- Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 2020, 588, 203–204. [Google Scholar] [CrossRef] [PubMed]
- Service, R.F. ‘The game has changed.’ AI triumphs at protein folding. Science 2020, 370, 1144–1145. [Google Scholar] [CrossRef] [PubMed]
- De Smaele, E.; Di Marcotullio, L.; Moretti, M.; Pelloni, M.; Occhione, M.A.; Infante, P.; Cucchi, D.; Greco, A.; Pietrosanti, L.; Todorovic, J.; et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia 2011, 13, 374–385. [Google Scholar] [CrossRef]
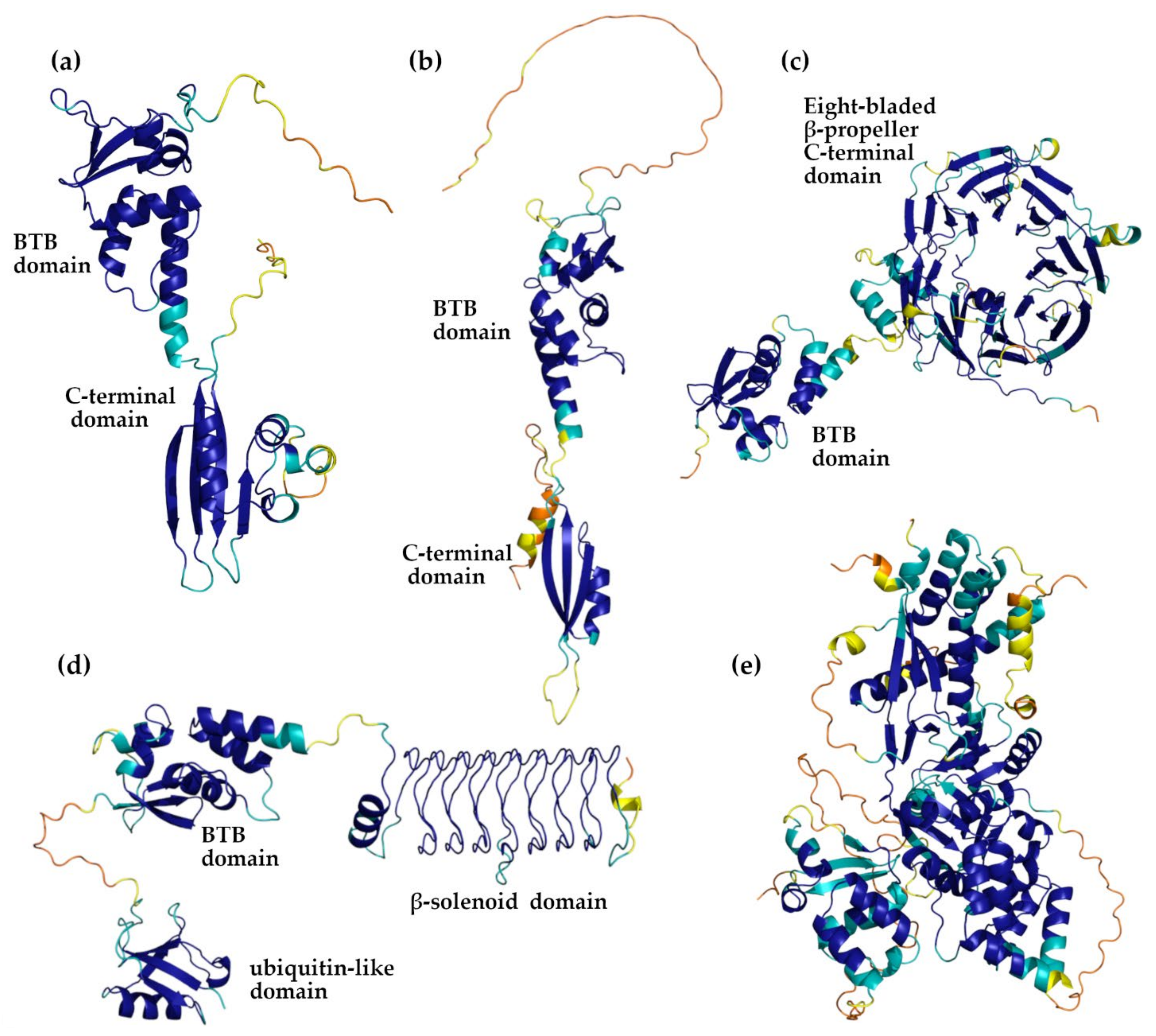
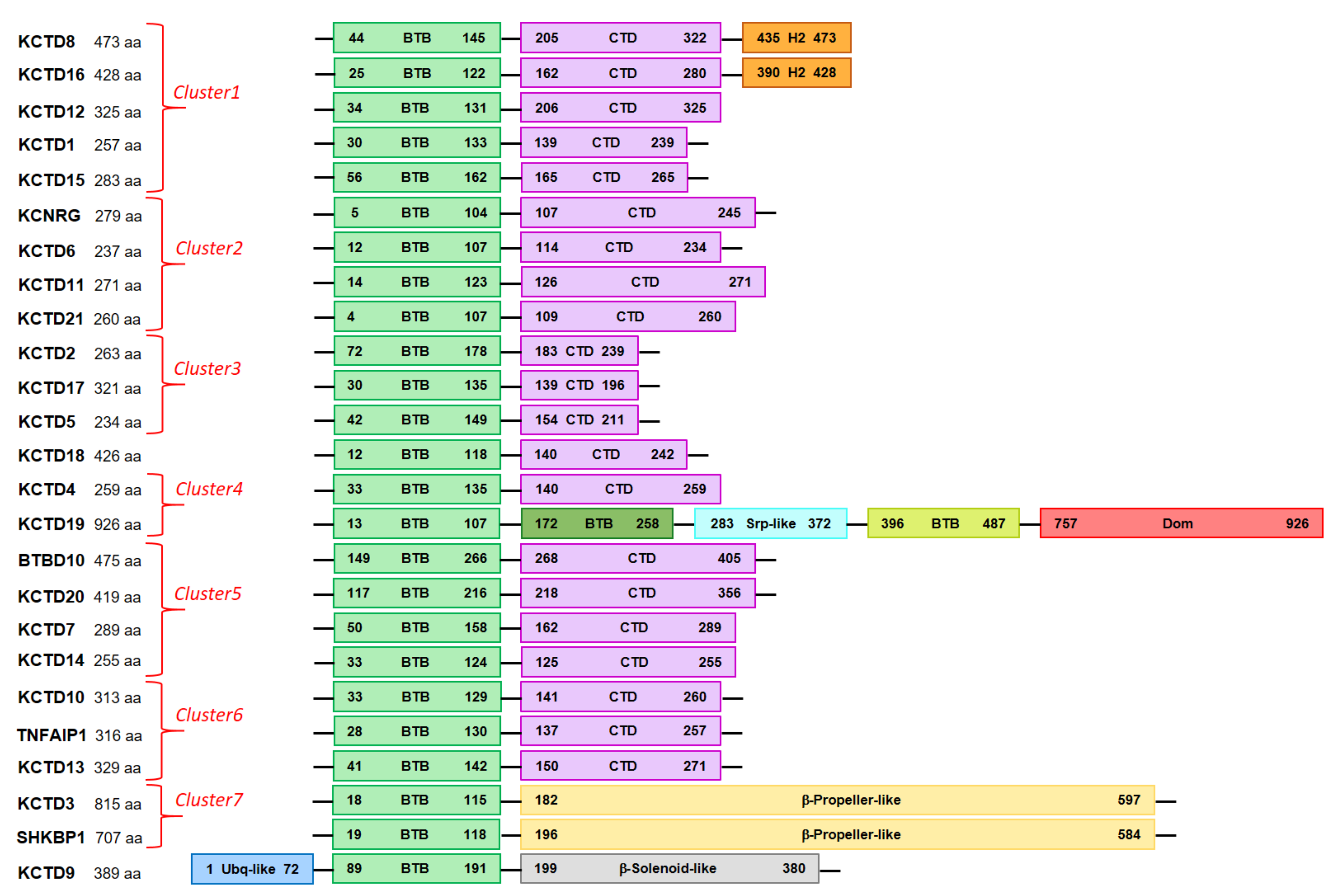
| Protein | AlphaFold Code | Low Structured and/or Low Confident Regions 1 | BTB Domain | CTD Domain |
|---|---|---|---|---|
| KCTD1 | AF-Q719H9 | 1–17/242–257 | A30-T133 | P139-L239 |
| KCTD2 | AF-Q14681 | 1–71/239–263 | R72-T178 | V183-N239 |
| KCTD3 | AF-Q9Y597 | 1–17/139–181/598–815 | E18-L115 | - 4 |
| KCTD4 | AF-Q8WVF5 | 1–32/198–217 | T33-L135 | T140-K259 |
| KCTD5 | AF-Q9NXV2 | 1–41/211–234 | V42-T149 | V154-N211 |
| KCTD6 | AF-Q8NC69 | - | D12-D107 | M114-K234 |
| KCTD7 | AF-Q96MP8 | 1–46/196–224 | P50-G158 | Y162-W289 |
| KCTD8 | AF-Q6ZWB6 | 1–41/148–203/325–434 | E44-L145 | R205-P322 |
| KCTD9 | AF-Q7L273 | 73–88 | D89-S191 | - 4 |
| KCTD10 | AF-Q9H3F6 | 1–18/274–295 | Y33-Q129 | P141-E260 |
| KCTD11 2 | AF-Q693B1 | - | G14-A123 | A126-H271 |
| KCTD12 | AF-Q96CX2 | 1–29/132–204/ | P34-A131 | R206-E325 |
| KCTD13 | AF-Q8WZ19 | 1–40/272–303/311–329 | K41-E142 | I150-T271 |
| KCTD14 | AF-Q9BQ13 | 1–28 | T33-D124 | M125-W255 |
| KCTD15 | AF-Q96SI1 | 1–43/266–283 | A56-R162 | A165-E265 |
| KCTD16 | AF-Q68DU8 | 1–21/124–161/281–389 | E25-T122 | K162-P280 |
| KCTD17 | AF-Q8N5Z5 | 1–29/197–260/265–321 | G30-V135 | P139-H196 |
| KCTD18 | AF-Q6PI47 | 242–426 | D12-S118 | P140-L242 |
| KCTD19 | AF-Q17RG1 | 108–171/259–282/ 501–756/788–803 | D13-E107/ V172-M258/ Q396-Q487 3 | - 4 |
| KCD20 | AF-Q7Z5Y7 | 1–115/359–419 | E117-C216 | D218-E356 |
| KCTD21 | AF-Q4G0X4 | - | P4-K107 | N109-R260 |
| KCNRG | AF-Q8N5I3 | 246–272 | E5- Q104 | P107-I245 |
| SH3KBP1 | AF-Q8TBC3 | 139–192/604–707 | E19-R118 | - 4 |
| TNFAIP1 | AF-Q13829 | 1–27/258–290/299–316 | K28-S130 | I137-E257 |
| BTBD10 | AF-Q9BSF8 | 1–148/409–475 | M149-C266 | D268-W405 |
| K1 | K15 | K8 | K12 | K16 | K6 | K11 | K21 | K4 | K20 | B10 | K18 | K19 | KCN | K7 | K14 | K10 | K13 | TNF | K2 | K5 | K17 | K3 | SHK | K9 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade A | K1 | 23.3 | 19.6 | 10.1 | 9.6 | 9.8 | 8.1 | 5.9 | 2.7 | 2.2 | - | - | 2.9 | - | 9.5 | 2.3 | 2.5 | - | - | - | 4.1 | 3.9 | 3.7 | - | - | - |
| K15 | 23.3 | 10.4 | 9.9 | 10.1 | 7.9 | 5.1 | 2.1 | 2.0 | - | - | 2.7 | - | 9.8 | 2.6 | 3.6 | - | - | - | 4.9 | 4.8 | 5.0 | - | - | - | ||
| Clade F | K8 | 22.8 | 19.5 | 20.4 | 7.8 | 5.8 | - | 2.0 | - | - | - | - | 8.1 | 2.3 | - | - | - | - | 2.7 | 2.8 | 2.8 | - | - | - | ||
| K12 | 22.8 | 19.2 | 7.6 | 5.7 | - | - | - | - | - | - | 8.9 | - | - | - | - | - | 2.5 | 2.5 | 2.6 | - | - | - | ||||
| K16 | 22.7 | 7.5 | 5.0 | - | 2.1 | - | - | 2.2 | - | 8.0 | - | 2.3 | - | - | - | 3.2 | 3.0 | 3.3 | - | - | - | |||||
| Clade B | K6 | 24.4 | 7.8 | 5.8 | 3.3 | - | - | 3.1 | 2.2 | 11.6 | 2.2 | 2.3 | - | - | - | 3.7 | 3.8 | 3.5 | - | - | - | |||||
| K11 | 26.9 | 14.2 | 5.3 | - | - | - | - | 8.7 | - | - | - | - | - | - | - | - | - | - | - | |||||||
| K21 | 25.2 | 2.1 | - | - | - | - | 9.2 | - | - | - | - | - | - | - | - | - | 2.7 | - | ||||||||
| K4 | 27.3 | - | - | - | 2.3 | 3.3 | - | - | - | - | - | - | - | - | - | - | - | |||||||||
| Clade G | K20 | 25.3 | 22.9 | - | - | - | 5.6 | 6.6 | - | - | - | - | - | - | - | - | - | |||||||||
| B10 | 25.0 | - | 2.2 | 2.1 | 5.7 | 6.6 | - | - | - | - | - | - | - | - | - | |||||||||||
| K18 | 21.8 | - | 2.5 | - | - | - | - | - | - | - | - | - | - | - | ||||||||||||
| K19 | 54.7 | - | - | 2.0 | - | - | - | - | - | - | - | - | - | |||||||||||||
| KCN | 27.0 | 3.8 | 3.4 | - | - | - | 4.8 | 4.5 | 4.8 | - | - | - | ||||||||||||||
| K7 | 24.9 | 13.4 | - | - | - | 3.0 | 3.0 | - | - | - | - | |||||||||||||||
| K14 | 27.1 | 2.4 | 2.2 | 2.4 | 3.0 | 3.0 | 3.0 | - | - | - | ||||||||||||||||
| Clade C | K10 | 25.6 | 21.4 | 23.5 | - | - | - | - | - | - | ||||||||||||||||
| K13 | 27.0 | 22.4 | - | - | - | - | - | - | ||||||||||||||||||
| TNF | 26.7 | - | - | - | - | - | - | |||||||||||||||||||
| Clade E | K2 | 12.1 | 10.7 | 10.8 | - | - | - | |||||||||||||||||||
| K5 | 12.0 | 10.2 | - | - | - | |||||||||||||||||||||
| K17 | 11.9 | - | - | - | ||||||||||||||||||||||
| Clade D | K3 | 62.7 | 50.3 | - | ||||||||||||||||||||||
| SHK | 63.6 | - | ||||||||||||||||||||||||
| K9 | 39.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, L.; Balasco, N.; Smaldone, G.; Berisio, R.; Ruggiero, A.; Vitagliano, L. AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family. Biomolecules 2021, 11, 1862. https://doi.org/10.3390/biom11121862
Esposito L, Balasco N, Smaldone G, Berisio R, Ruggiero A, Vitagliano L. AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family. Biomolecules. 2021; 11(12):1862. https://doi.org/10.3390/biom11121862
Chicago/Turabian StyleEsposito, Luciana, Nicole Balasco, Giovanni Smaldone, Rita Berisio, Alessia Ruggiero, and Luigi Vitagliano. 2021. "AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family" Biomolecules 11, no. 12: 1862. https://doi.org/10.3390/biom11121862
APA StyleEsposito, L., Balasco, N., Smaldone, G., Berisio, R., Ruggiero, A., & Vitagliano, L. (2021). AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family. Biomolecules, 11(12), 1862. https://doi.org/10.3390/biom11121862











