Reiterating the Emergence of Noncoding RNAs as Regulators of the Critical Hallmarks of Gall Bladder Cancer
Abstract
1. Introduction
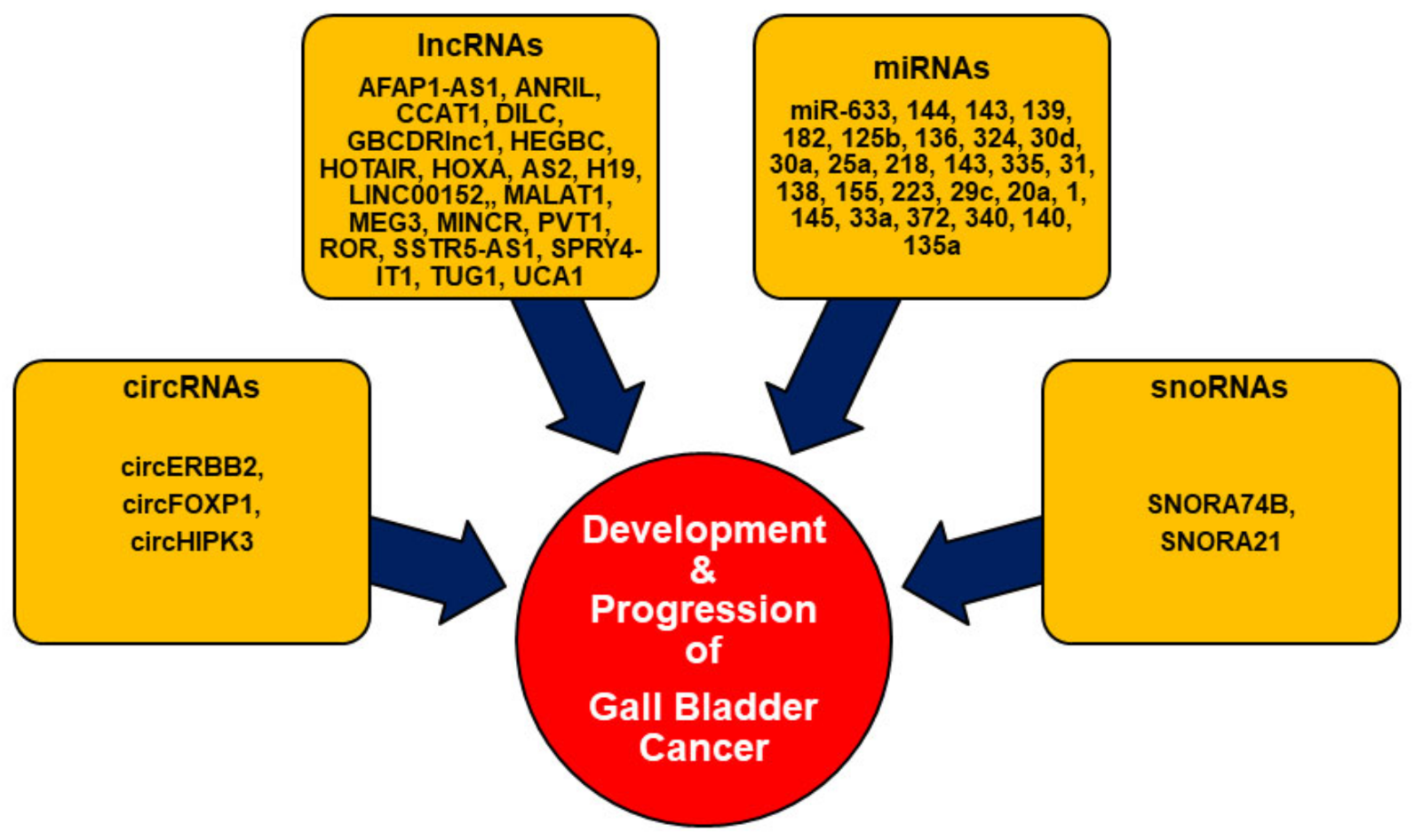
2. Noncoding RNAs (ncRNAs) in GBC
2.1. Circular RNAs (circRNAs)
2.2. Long Noncoding RNAs (lncRNAs)
2.3. MicroRNAs (miRNAs)
2.4. Small Nucleolar RNAs (snoRNAs)
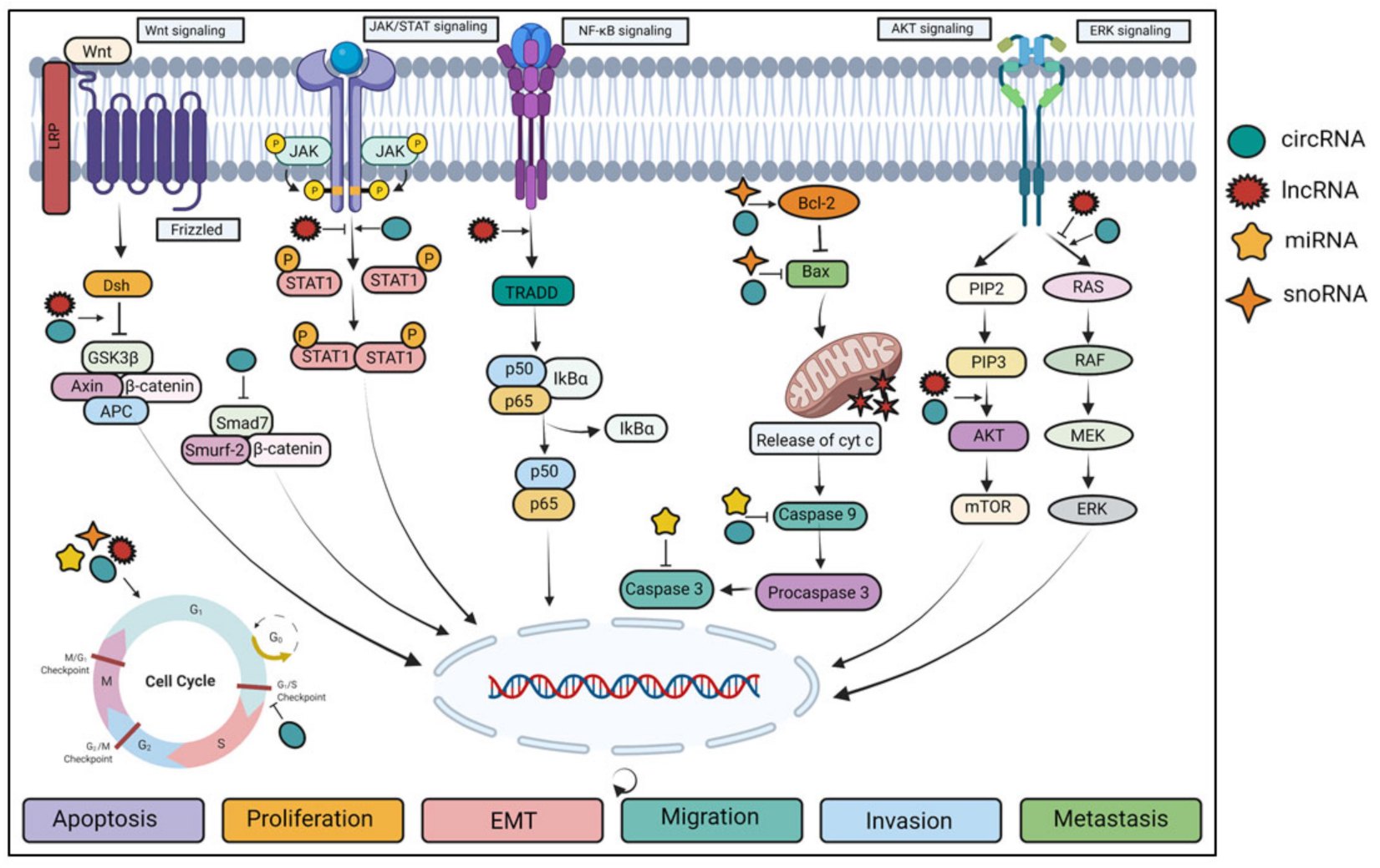
3. Deregulation of ncRNAs in Different Signaling Pathways
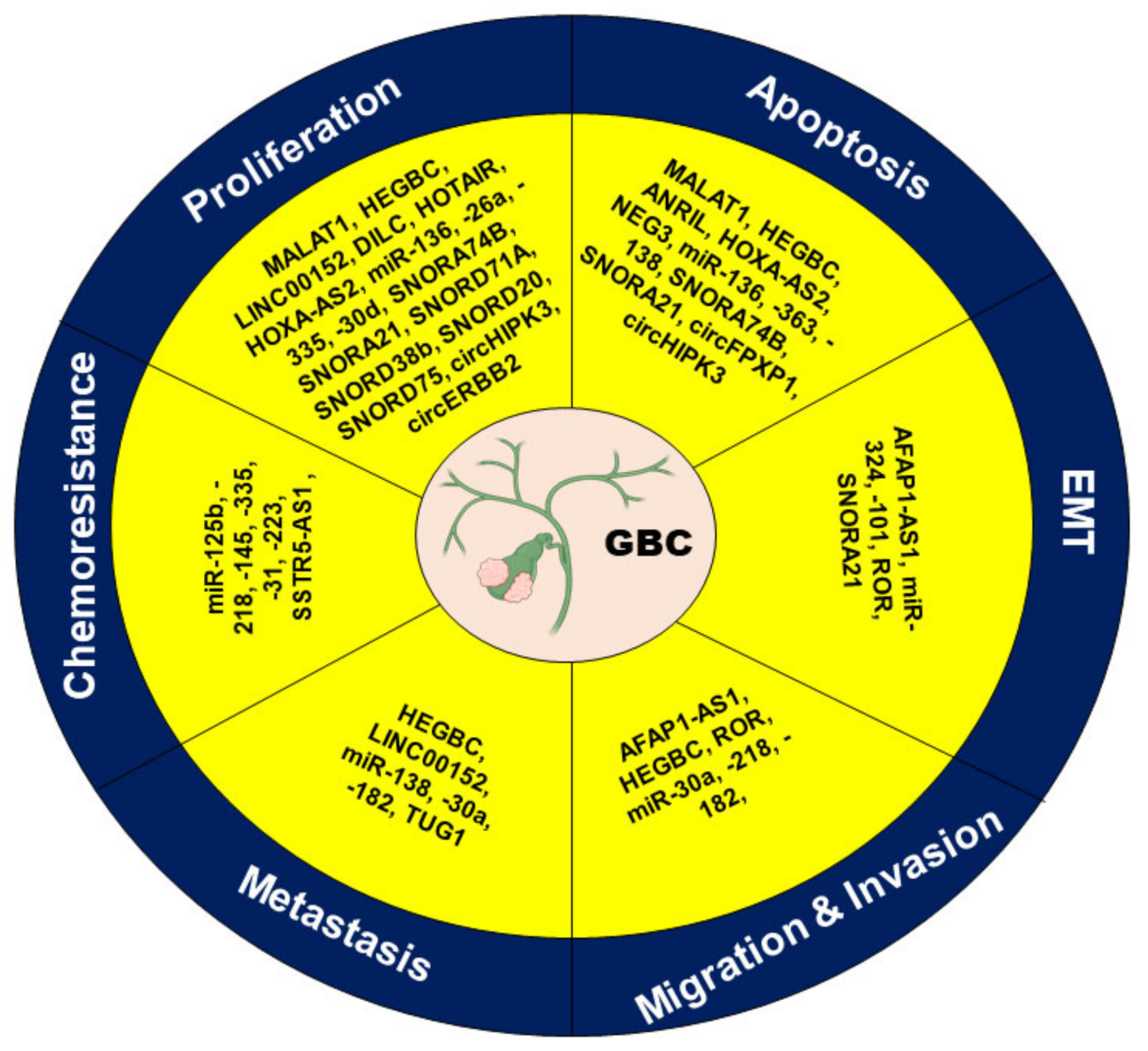
4. Effect of ncRNAs in Different Hallmarks of Cancer
4.1. Apoptosis
4.2. Cell Proliferation
4.3. EMT
4.4. Invasion and Migration
4.5. Metastasis
4.6. Chemoresistance
5. Challenges Associated with Clinical Applications of ncRNAs
6. Future Prospects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Girisa, S.; Saikia, Q.; Bordoloi, D.; Banik, K.; Monisha, J.; Daimary, U.D.; Verma, E.; Ahn, K.S.; Kunnumakkara, A.B. Xanthohumol from Hop: Hope for cancer prevention and treatment. IUBMB Life 2021, 73, 1016–1044. [Google Scholar] [CrossRef]
- Henamayee, S.; Banik, K.; Sailo, B.L.; Shabnam, B.; Harsha, C.; Srilakshmi, S.; Vgm, N.; Baek, S.H.; Ahn, K.S.; Kunnumakkara, A.B. Therapeutic Emergence of Rhein as a Potential Anticancer Drug: A Review of Its Molecular Targets and Anticancer Properties. Molecules 2020, 25, 2278. [Google Scholar] [CrossRef]
- Daimary, U.D.; Parama, D.; Rana, V.; Banik, K.; Kumar, A.; Harsha, C.; Kunnumakkara, A.B. Emerging roles of cardamonin, a multitargeted nutraceutical in the prevention and treatment of chronic diseases. Curr. Res. Pharmacol. Drug Discov. 2020, 2, 100008. [Google Scholar] [CrossRef]
- Girisa, S.; Shabnam, B.; Monisha, J.; Fan, L.; Halim, C.E.; Arfuso, F.; Ahn, K.S.; Sethi, G.; Kunnumakkara, A.B. Potential of Zerumbone as an Anti-Cancer Agent. Molecules 2019, 24, 734. [Google Scholar] [CrossRef]
- Ramachandran, A.; Srivastava, D.N.; Madhusudhan, K.S. Gallbladder cancer revisited: The evolving role of a radiologist. Br. J. Radiol. 2021, 94, 20200726. [Google Scholar] [CrossRef]
- Lin, H.Z.; Zhang, T.; Chen, M.Y.; Shen, J.L. Novel biomarkers for the diagnosis and prognosis of gallbladder cancer. J. Dig. Dis. 2021, 22, 62–71. [Google Scholar] [CrossRef]
- Parama, D.; Boruah, M.; Yachna, K.; Rana, V.; Banik, K.; Harsha, C.; Thakur, K.K.; Dutta, U.; Arya, A.; Mao, X.; et al. Diosgenin, a steroidal saponin, and its analogs: Effective therapies against different chronic diseases. Life Sci. 2020, 260, 118182. [Google Scholar] [CrossRef]
- Ertel, A.E.; Bentrem, D.; Abbott, D.E. Gall Bladder Cancer. Cancer Treat. Res. 2016, 168, 101–120. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, K.; Gupta, A.; Yadav, A.; Kumar, A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J. Gastroenterol. 2017, 23, 3978–3998. [Google Scholar] [CrossRef]
- Pilgrim, C.H.C.; Groeschl, R.T.; Christians, K.K.; Gamblin, T.C. Modern perspectives on factors predisposing to the development of gallbladder cancer. HPB 2013, 15, 839–844. [Google Scholar] [CrossRef]
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Saleh, S.H.S.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80. [Google Scholar] [CrossRef]
- Mishra, S.; Verma, S.S.; Rai, V.; Awasthee, N.; Chava, S.; Hui, K.M.; Kumar, A.P.; Challagundla, K.B.; Sethi, G.; Gupta, S.C. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell. Mol. Life Sci. 2019, 76, 1947–1966. [Google Scholar] [CrossRef]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, Y.-Y.; Xin, H.-W.; Wang, L.; Arfuso, F.; Dharmarajan, A.; Kumar, A.P.; Wang, H.; Tang, F.R.; Warrier, S.; et al. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int. J. Biochem. Cell Biol. 2019, 108, 17–20. [Google Scholar] [CrossRef]
- Cheng, J.-T.; Wang, L.; Wang, H.; Tang, F.-R.; Cai, W.-Q.; Sethi, G.; Xin, H.-W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef]
- Chen, X.; Tang, F.-R.; Arfuso, F.; Cai, W.-Q.; Ma, Z.; Yang, J.; Sethi, G. The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules 2019, 10, 66. [Google Scholar] [CrossRef]
- Qin, Y.; Zhou, Y.; Ge, A.; Chang, L.; Shi, H.; Fu, Y.; Luo, Q. Overexpression of SNORA21 suppresses tumorgenesis of gallbladder cancer in vitro and in vivo. Biomed. Pharmacother. 2019, 118, 109266. [Google Scholar] [CrossRef]
- Kai, D.; Yannian, L.; Yitian, C.; Dinghao, G.; Xin, Z.; Wu, J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem. Biophys. Res. Commun. 2018, 503, 863–869. [Google Scholar] [CrossRef]
- Wu, X.-S.; Wang, X.-A.; Wu, W.-G.; Hu, Y.-P.; Li, M.-L.; Ding, Q.; Weng, H.; Shu, Y.-J.; Liu, T.-Y.; Jiang, L.; et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol. Ther. 2014, 15, 806–814. [Google Scholar] [CrossRef]
- Kono, H.; Nakamura, M.; Ohtsuka, T.; Nagayoshi, Y.; Mori, Y.; Takahata, S.; Aishima, S.; Tanaka, M. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncol. Rep. 2013, 30, 17–24. [Google Scholar] [CrossRef]
- Yu, C.-Y.; Kuo, H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef]
- Huang, X.; He, M.; Huang, S.; Lin, R.; Zhan, M.; Yang, D.; Shen, H.; Xu, S.; Cheng, W.; Yu, J.; et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol. Cancer 2019, 18, 166. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Y.K.; Lu, Z.L.; Li, J. Circular RNA circ-MTO1 serves as a novel potential diagnostic and prognostic biomarker for gallbladder cancer. Eur. Rev. Med. Pharmacol. Sci 2020, 24, 8359–8366. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; Cai, Q.; Ma, M.; Jin, L.Y.; Weng, M.; Zhou, D.; Tang, Z.; Wang, J.D.; Quan, Z. Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Mol. Cancer 2019, 18, 145. [Google Scholar] [CrossRef]
- Thakur, K.K.; Kumar, A.; Banik, K.; Verma, E.; Khatoon, E.; Harsha, C.; Sethi, G.; Gupta, S.C.; Kunnumakkara, A.B. Long noncoding RNAs in triple-negative breast cancer: A new frontier in the regulation of tumorigenesis. J. Cell. Physiol. 2021, 519, 1. [Google Scholar] [CrossRef]
- Revythis, A.; Shah, S.; Kutka, M.; Moschetta, M.; Ozturk, M.; Pappas-Gogos, G.; Ioannidou, E.; Sheriff, M.; Rassy, E.; Boussios, S. Unraveling the Wide Spectrum of Melanoma Biomarkers. Diagnostics (Basel) 2021, 11, 1341. [Google Scholar] [CrossRef]
- Garg, M.; Sethi, G. Emerging role of long non-coding RNA (lncRNA) in human malignancies: A unique opportunity for precision medicine. Cancer Lett. 2021, 519, 1. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Z.; Wang, S.; Weng, M.; Zhou, D.; Li, C.; Wang, J.; Chen, E.; Quan, Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial–mesenchymal transition by regulating HIF-1α via miR-138. Open Biol. 2017, 7. [Google Scholar] [CrossRef]
- Cai, Q.; Jin, L.; Wang, S.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 2017, 8, 47957–47968. [Google Scholar] [CrossRef]
- Rishabh, K.; Khadilkar, S.; Kumar, A.; Kalra, I.; Kumar, A.; Kunnumakkara, A. MicroRNAs as Modulators of Oral Tumorigenesis—A Focused Review. Int. J. Mol. Sci. 2021, 22, 2561. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.; Anderson, T. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Kent, O.; Mendell, J.T. A small piece in the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes. Oncogene 2006, 25, 6188–6196. [Google Scholar] [CrossRef]
- Pidíkova, P.; Reis, R.; Herichova, I. miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation. Int. J. Mol. Sci. 2020, 21, 4633. [Google Scholar] [CrossRef]
- Kansara, S.; Pandey, V.; Lobie, P.E.; Sethi, G.; Garg, M.; Pandey, A.K. Mechanistic Involvement of Long Non-Coding RNAs in Oncotherapeutics Resistance in Triple-Negative Breast Cancer. Cells 2020, 9, 1511. [Google Scholar] [CrossRef]
- Wang, C.; Kar, S.; Lai, X.; Cai, W.; Arfuso, F.; Sethi, G.; Lobie, P.E.; Goh, B.C.; Lim, L.H.; Hartman, M.; et al. Triple negative breast cancer in Asia: An insider’s view. Cancer Treat. Rev. 2018, 62, 29–38. [Google Scholar] [CrossRef]
- Fabris, L.; Ceder, Y.; Chinnaiyan, A.M.; Jenster, G.W.; Sørensen, K.D.; Tomlins, S.; Visakorpi, T.; Calin, G.A. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur. Urol. 2016, 70, 312–322. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Boussios, S.; Ozturk, M.A.; Moschetta, M.; Karathanasi, A.; Zakynthinakis-Kyriakou, N.; Katsanos, K.H.; Christodoulou, D.K.; Pavlidis, N. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J. Pers. Med. 2019, 9, 12. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, J.-J.; Zhao, L.-J.; Yang, X.-R.; Yu, Y.-L. Exosomal miR-182 regulates the effect of RECK on gallbladder cancer. World J. Gastroenterol. 2020, 26, 933–946. [Google Scholar] [CrossRef]
- Niu, J.; Li, Z.; Li, F. Overexpressed microRNA-136 works as a cancer suppressor in gallbladder cancer through suppression of JNK signaling pathway via inhibition of MAP2K4. Am. J. Physiol. Gastrointest Liver Physiol. 2019, 317, G670–G681. [Google Scholar] [CrossRef]
- Ye, Y.-Y.; Mei, J.-W.; Xiang, S.-S.; Li, H.-F.; Ma, Q.; Song, X.-L.; Wang, Z.; Zhang, Y.-J.; Liu, Y.-C.; Jin, Y.-P.; et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 2018, 9, 410. [Google Scholar] [CrossRef]
- Yang, L.; Huang, S.; Ma, H.; Wu, X.; Feng, F. MicroRNA-125b predicts clinical outcome and suppressed tumor proliferation and migration in human gallbladder cancer. Tumor Biol. 2017, 39, 1010428317692249. [Google Scholar] [CrossRef]
- Qin, Y.; Meng, L.; Fu, Y.; Quan, Z.; Ma, M.; Weng, M.; Zhang, Z.; Gao, C.; Shi, X.; Han, K. SNORA74B gene silencing inhibits gallbladder cancer cells by inducing PHLPP and suppressing Akt/mTOR signaling. Oncotarget 2017, 8, 19980–19996. [Google Scholar] [CrossRef]
- Ma, F.; Wang, S.-H.; Cai, Q.; Zhang, M.-D.; Yang, Y.; Ding, J. Overexpression of LncRNA AFAP1-AS1 predicts poor prognosis and promotes cells proliferation and invasion in gallbladder cancer. Biomed. Pharmacother. 2016, 84, 1249–1255. [Google Scholar] [CrossRef]
- Liu, B.; Shen, E.-D.; Liao, M.-M.; Hu, Y.-B.; Wu, K.; Yang, P.; Zhou, L.; Chen, W.-D. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumor Biol. 2016, 37, 9875–9886. [Google Scholar] [CrossRef]
- Ma, M.-Z.; Chu, B.-F.; Zhang, Y.; Weng, M.-Z.; Qin, Y.-Y.; Gong, W.; Quan, Z.-W. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015, 6, e1583. [Google Scholar] [CrossRef]
- Liang, C.; Yang, P.; Han, T.; Wang, R.-Y.; Xing, X.-L.; Si, A.-F.; Ma, Q.-Y.; Chen, Z.; Li, H.-Y.; Zhang, B. Long non-coding RNA DILC promotes the progression of gallbladder carcinoma. Gene 2019, 694, 102–110. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82. [Google Scholar] [CrossRef]
- Yang, L.; Gao, Q.; Wu, X.; Feng, F.; Xu, K. Long noncoding RNA HEGBC promotes tumorigenesis and metastasis of gallbladder cancer via forming a positive feedback loop with IL-11/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 186. [Google Scholar] [CrossRef]
- Ma, M.-Z.; Li, C.-X.; Zhang, Y.; Weng, M.-Z.; Zhang, M.-D.; Qin, Y.-Y.; Gong, W.; Quan, Z.-W. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol. Cancer 2014, 13, 156. [Google Scholar] [CrossRef]
- Zhang, P.; Cao, P.; Zhu, X.; Pan, M.; Zhong, K.; He, R.; Li, Y.; Jiao, X.; Gao, Y. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget 2017, 8, 33137–33143. [Google Scholar] [CrossRef]
- Wang, S.-H.; Ma, F.; Tang, Z.-H.; Wu, X.-C.; Xiao-Cai, W.; Zhang, M.-D.; Weng, M.-Z.; Zhou, D.; Wang, J.-D.; Quan, Z.-W. Long non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 160. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, Z.-Q.; Wang, S.-H.; Li, C.; Zhu, Z.-G.; Quan, Z.-W.; Zhang, W.-J. Upregulation of long non-coding RNA LINC00152 by SP1 contributes to gallbladder cancer cell growth and tumor metastasis via PI3K/AKT pathway. Am. J. Transl. Res. 2016, 8, 4068–4081. [Google Scholar]
- Wang, S.-H.; Zhang, W.-J.; Wu, X.-C.; Zhang, M.-D.; Weng, M.-Z.; Zhou, D.; Wang, J.-D.; Quan, Z.-W. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 2016, 7, 37857–37867. [Google Scholar] [CrossRef]
- Lin, N.; Yao, Z.; Xu, M.; Chen, J.; Lu, Y.; Yuan, L.; Zhou, S.; Zou, X.; Xu, R. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 244. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, W.; Wu, X.; Weng, M.; Zhang, M.; Cai, Q.; Zhou, D.; Wang, J.; Quan, Z. The lnc RNA MALAT 1 functions as a competing endogenous RNA to regulate MCL -1 expression by sponging miR-363-3p in gallbladder cancer. J. Cell. Mol. Med. 2016, 20, 2299–2308. [Google Scholar] [CrossRef]
- Jin, L.; Cai, Q.; Wang, S.; Wang, S.; Mondal, T.; Wang, J.; Quan, Z. Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 2018, 9, 1017. [Google Scholar] [CrossRef]
- Wang, S.-H.; Yang, Y.; Wu, X.-C.; Zhang, M.-D.; Weng, M.-Z.; Zhou, D.; Wang, J.-D.; Quan, Z.-W. Long non-coding RNA MINCR promotes gallbladder cancer progression through stimulating EZH2 expression. Cancer Lett. 2016, 380, 122–133. [Google Scholar] [CrossRef]
- Liu, K.; Xu, Q. LncRNA PVT1 regulates gallbladder cancer progression through miR-30d-5p. J. Biol. Regul. Homeost. Agents 2020, 34, 875–883. [Google Scholar] [CrossRef]
- Wang, S.-H.; Zhang, M.-D.; Wu, X.-C.; Weng, M.-Z.; Zhou, D.; Quan, Z.-W. Overexpression of LncRNA-ROR predicts a poor outcome in gallbladder cancer patients and promotes the tumor cells proliferation, migration, and invasion. Tumor Biol. 2016, 37, 12867–12875. [Google Scholar] [CrossRef]
- Xue, Z.; Yang, B.; Xu, Q.; Zhu, X.; Qin, G. Long non-coding RNA SSTR5-AS1 facilitates gemcitabine resistance via stabilizing NONO in gallbladder carcinoma. Biochem. Biophys. Res. Commun. 2020, 522, 952–959. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, X.; Ge, N.; Guo, W.; Feng, F.; Wan, F. Long non-coding RNA SPRY4-IT1 promotes gallbladder carcinoma progression. Oncotarget 2017, 8, 3104–3110. [Google Scholar] [CrossRef]
- Ma, F.; Wang, S.-H.; Cai, Q.; Jin, L.-Y.; Zhou, D.; Ding, J.; Quan, Z.-W. Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma. Biomed. Pharmacother. 2017, 88, 863–869. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, Y.; Liang, H.; Zhang, F.; Liu, F.; Chen, S.; Hu, Y.; Jiang, L.; Hao, Y.; Li, M.; et al. EMP3, which is regulated by miR-663a, suppresses gallbladder cancer progression via interference with the MAPK/ERK pathway. Cancer Lett. 2018, 430, 97–108. [Google Scholar] [CrossRef]
- Jin, Y.-P.; Hu, Y.-P.; Wu, X.-S.; Wu, Y.-S.; Ye, Y.-Y.; Li, H.-F.; Liu, Y.-C.; Jiang, L.; Liu, F.-T.; Zhang, Y.-J.; et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018, 9, 182. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Chen, X.; He, Y.; Hu, Q.; Li, H.; Han, Q.; Ren, F.; Li, J.; Li, C.; et al. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018, 51, e12510. [Google Scholar] [CrossRef]
- He, M.; Zhan, M.; Chen, W.; Xu, S.; Long, M.; Shen, H.; Shi, Y.; Liu, Q.; Mohan, M.; Wang, J. MiR-143-5p Deficiency Triggers EMT and Metastasis by Targeting HIF-1α in Gallbladder Cancer. Cell. Physiol. Biochem. 2017, 42, 2078–2092. [Google Scholar] [CrossRef]
- Yang, D.; Zhan, M.; Chen, T.; Chen, W.; Zhang, Y.; Xu, S.; Yan, J.; Huang, Q.; Wang, J. miR-125b-5p enhances chemotherapy sensitivity to cisplatin by down-regulating Bcl2 in gallbladder cancer. Sci. Rep. 2017, 7, srep43109. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Chen, M.; Liu, D. miR-324-5p inhibits gallbladder carcinoma cell metastatic behaviours by downregulation of transforming growth factor beta 2 expression. Artif. Cells, Nanomedicine, Biotechnol. 2020, 48, 315–324. [Google Scholar] [CrossRef]
- He, Y.; Chen, X.; Yu, Y.; Li, J.; Hu, Q.; Xue, C.; Chen, J.; Shen, S.; Luo, Y.; Ren, F.; et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol. Carcinog. 2018, 57, 772–783. [Google Scholar] [CrossRef]
- Zhou, H.; Guo, W.; Zhao, Y.; Wang, Y.; Zha, R.; Ding, J.; Liang, L.; Hu, J.; Shen, H.; Chen, Z.; et al. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int. J. Oncol. 2014, 44, 2050–2058. [Google Scholar] [CrossRef]
- Wang, H.; Zhan, M.; Xu, S.; Chen, W.; Long, M.-M.; Shi, Y.-H.; Liu, Q.; Mohan, M.; Wang, J. miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell Death Dis. 2017, 8, e2770. [Google Scholar] [CrossRef]
- Zhan, M.; Zhao, X.; Wang, H.; Chen, W.; Xu, S.; Wang, W.; Shen, H.; Huang, S.; Wang, J. miR-145 sensitizes gallbladder cancer to cisplatin by regulating multidrug resistance associated protein 1. Tumor Biol. 2016, 37, 10553–10562. [Google Scholar] [CrossRef]
- Wang, W.; Chen, L.-C.; Qian, J.-Y.; Zhang, Q. MiR-335 promotes cell proliferation by inhibiting MEF2D and sensitizes cells to 5-Fu treatment in gallbladder carcinoma. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 9829–9839. [Google Scholar]
- Li, M.; Chen, W.; Zhang, H.; Zhang, Y.; Ke, F.; Wu, X.; Zhang, Y.; Weng, M.; Liu, Y.; Gong, W. MiR-31 regulates the cisplatin resistance by targeting Src in gallbladder cancer. Oncotarget 2016, 7, 83060–83070. [Google Scholar] [CrossRef][Green Version]
- Ma, F.; Zhang, M.; Gong, W.; Weng, M.; Quan, Z. MiR-138 Suppresses Cell Proliferation by Targeting Bag-1 in Gallbladder Carcinoma. PLoS ONE 2015, 10, e0126499. [Google Scholar] [CrossRef]
- Lu, W.; Hu, Y.; Ma, Q.; Zhou, L.; Jiang, L.; Li, Z.; Zhao, S.; Xu, Y.; Shi, W.; Li, S.; et al. miR-223 increases gallbladder cancer cell sensitivity to docetaxel by downregulating STMN1. Oncotarget 2016, 7, 62364–62376. [Google Scholar] [CrossRef]
- Shu, Y.-J.; Bao, R.-F.; Jiang, L.; Wang, Z.; Wang, X.-A.; Zhang, F.; Liang, H.-B.; Li, H.-F.; Ye, Y.-Y.; Xiang, S.-S.; et al. MicroRNA-29c-5p suppresses gallbladder carcinoma progression by directly targeting CPEB4 and inhibiting the MAPK pathway. Cell Death Differ. 2017, 24, 445–457. [Google Scholar] [CrossRef]
- Chang, Y.; Liu, C.; Yang, J.; Liu, G.; Feng, F.; Tang, J.; Hu, L.; Li, L.; Jiang, F.; Chen, C.; et al. miR-20a triggers metastasis of gallbladder carcinoma. J. Hepatol. 2013, 59, 518–527. [Google Scholar] [CrossRef]
- Letelier, P.; García, P.; Leal, P.; Álvarez, H.; Ili, C.; López, J.; Castillo, J.; Brebi, P.; Roa, J.C. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 1849–1867. [Google Scholar]
- Zhang, M.; Gong, W.; Zuo, B.; Chu, B.; Tang, Z.; Zhang, Y.; Yang, Y.; Zhou, D.; Weng, M.; Qin, Y.; et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget 2016, 7, 78640–78652. [Google Scholar] [CrossRef]
- Hua, C.B.; Song, S.B.; Ma, H.L.; Li, X.Z. MiR-1-5p is down-regulated in gallbladder carcinoma and suppresses cell proliferation, migration and invasion by targeting Notch2. Pathol. Res. Pract. 2019, 215, 200–208. [Google Scholar] [CrossRef]
- Zhou, N.; Cheng, W.; Peng, C.; Liu, Y.; Jiang, B. Decreased expression of hsa-miR-372 predicts poor prognosis in patients with gallbladder cancer by affecting chloride intracellular channel 1. Mol. Med. Rep. 2017, 16, 7848–7854. [Google Scholar] [CrossRef]
- Wang, N.; Xiang, X.; Chen, K.; Liu, P.; Zhu, A. Targeting of NT5E by miR-30b and miR-340 attenuates proliferation, invasion and migration of gallbladder carcinoma. Biochimie 2018, 146, 56–67. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, W.; Tang, H.; Qian, H.; Yang, J.; Zhu, Z.; Ren, P.; Lu, B. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene 2016, 589, 20–26. [Google Scholar] [CrossRef]
- Zhou, H.; Guo, W.; Zhao, Y.; Wang, Y.; Zha, R.; Ding, J.; Liang, L.; Yang, G.; Chen, Z.; Ma, B.; et al. Micro RNA -135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014, 105, 956–965. [Google Scholar] [CrossRef]
- Shen, S.; Liu, H.; Wang, Y.; Wang, J.; Ni, X.; Ai, Z.; Pan, H.; Liu, H.; Shao, Y. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget 2016, 7, 72833–72844. [Google Scholar] [CrossRef]
- Liu, X.F.; Wang, K.; Du, H.C. LncRNA SNHG6 regulating Hedgehog signaling pathway and affecting the biological function of gallbladder carcinoma cells through targeting miR-26b-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7598–7611. [Google Scholar] [CrossRef]
- Bao, D.; Yuan, R.X.; Zhang, Y. Effects of lncRNA MEG3 on proliferation and apoptosis of gallbladder cancer cells through regulating NF-kappaB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6632–6638. [Google Scholar] [CrossRef]
- Goeppert, B.; Truckenmueller, F.; Ori, A.; Fritz, V.; Albrecht, T.; Fraas, A.; Scherer, D.; Silos, R.G.; Sticht, C.; Gretz, N.; et al. Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p. Sci. Rep. 2019, 9, 4796. [Google Scholar] [CrossRef]
- Ishigami, K.; Nosho, K.; Kanno, S.; Mitsuhashi, K.; Igarashi, H.; Shitani, M.; Motoya, M.; Kimura, Y.; Hasegawa, T.; Kaneto, H.; et al. MicroRNA-31 reflects IL-6 expression in cancer tissue and is related with poor prognosis in bile duct cancer. Carcinogenesis 2018, 39, 1127–1134. [Google Scholar] [CrossRef]
- Xu, G.; Wei, X.; Tu, Q.; Zhou, C. Up-regulated microRNA-33b inhibits epithelial–mesenchymal transition in gallbladder cancer through down-regulating CROCC. Biosci. Rep. 2020, 40, BSR20190108. [Google Scholar] [CrossRef]
- Bao, R.-F.; Shu, Y.-J.; Hu, Y.-P.; Wang, X.-A.; Zhang, F.; Liang, H.-B.; Ye, Y.-Y.; Li, H.-F.; Xiang, S.-S.; Weng, H.; et al. miR-101 targeting ZFX suppresses tumor proliferation and metastasis by regulating the MAPK/Erk and Smad pathways in gallbladder carcinoma. Oncotarget 2016, 7, 22339–22354. [Google Scholar] [CrossRef][Green Version]
- Ahmed, S.A.; Parama, D.; Daimari, E.; Girisa, S.; Banik, K.; Harsha, C.; Dutta, U.; Kunnumakkara, A.B. Rationalizing the therapeutic potential of apigenin against cancer. Life Sci. 2021, 267, 118814. [Google Scholar] [CrossRef]
- Banik, K.; Ranaware, A.M.; Deshpande, V.; Nalawade, S.P.; Padmavathi, G.; Bordoloi, D.; Sailo, B.L.; Shanmugam, M.K.; Fan, L.; Arfuso, F.; et al. Honokiol for cancer therapeutics: A traditional medicine that can modulate multiple oncogenic targets. Pharmacol. Res. 2019, 144, 192–209. [Google Scholar] [CrossRef]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A Target for Anticancer Therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef]
- Lee, J.H.; Chiang, S.Y.; Nam, D.; Chung, W.-S.; Lee, J.; Na, Y.-S.; Sethi, G.; Ahn, K.S. Capillarisin inhibits constitutive and inducible STAT3 activation through induction of SHP-1 and SHP-2 tyrosine phosphatases. Cancer Lett. 2014, 345, 140–148. [Google Scholar] [CrossRef]
- Li, F.; Shanmugam, M.K.; Chen, L.; Chatterjee, S.; Basha, J.; Kumar, A.P.; Kundu, T.K.; Sethi, G. Garcinol, a Polyisoprenylated Benzophenone Modulates Multiple Proinflammatory Signaling Cascades Leading to the Suppression of Growth and Survival of Head and Neck Carcinoma. Cancer Prev. Res. 2013, 6, 843–854. [Google Scholar] [CrossRef]
- Tewari, D.; Nabavi, S.F.; Nabavi, S.M.; Sureda, A.; Farooqi, A.A.; Atanasov, A.G.; Vacca, R.A.; Sethi, G.; Bishayee, A. Targeting activator protein 1 signaling pathway by bioactive natural agents: Possible therapeutic strategy for cancer prevention and intervention. Pharmacol. Res. 2018, 128, 366–375. [Google Scholar] [CrossRef]
- Ranaware, A.M.; Banik, K.; Deshpande, V.; Padmavathi, G.; Roy, N.K.; Sethi, G.; Fan, L.; Kumar, A.P.; Kunnumakkara, A.B. Magnolol: A Neolignan from the Magnolia Family for the Prevention and Treatment of Cancer. Int. J. Mol. Sci. 2018, 19, 2362. [Google Scholar] [CrossRef]
- Siveen, K.S.; Nguyen, A.H.; Lee, J.H.; Li, F.; Singh, S.S.; Kumar, A.P.; Low, G.; Jha, S.K.; Tergaonkar, V.; Ahn, K.S.; et al. Negative regulation of signal transducer and activator of transcription-3 signalling cascade by lupeol inhibits growth and induces apoptosis in hepatocellular carcinoma cells. Br. J. Cancer 2014, 111, 1327–1337. [Google Scholar] [CrossRef]
- Harsha, C.; Banik, K.; Ang, H.L.; Girisa, S.; Vikkurthi, R.; Parama, D.; Rana, V.; Shabnam, B.; Khatoon, E.; Kumar, A.P.; et al. Targeting AKT/mTOR in Oral Cancer: Mechanisms and Advances in Clinical Trials. Int. J. Mol. Sci. 2020, 21, 3285. [Google Scholar] [CrossRef]
- Puar, Y.R.; Shanmugam, M.K.; Fan, L.; Arfuso, F.; Sethi, G.; Tergaonkar, V. Evidence for the Involvement of the Master Transcription Factor NF-kappaB in Cancer Initiation and Progression. Biomedicines 2018, 6, 82. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Cai, W.; Chen, Z.X.; Rane, G.; Singh, S.S.; Choo, Z.; Wang, C.; Yuan, Y.; Tan, T.Z.; Arfuso, F.; Yap, C.T.; et al. Wanted DEAD/H or Alive: Helicases Winding Up in Cancers. J. Natl. Cancer Inst. 2017, 109, djw278. [Google Scholar] [CrossRef]
- Chopra, P.; Sethi, G.; Dastidar, S.G.; Ray, A. Polo-like kinase inhibitors: An emerging opportunity for cancer therapeutics. Expert Opin. Investig. Drugs 2010, 19, 27–43. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Hushmandi, K.; Kalantari, M.; Mohammadinejad, R.; Javaheri, T.; Sethi, G. Association of the Epithelial–Mesenchymal Transition (EMT) with Cisplatin Resistance. Int. J. Mol. Sci. 2020, 21, 4002. [Google Scholar] [CrossRef]
- Lee, J.H.; Chinnathambi, A.; Alharbi, S.A.; Shair, O.H.; Sethi, G.; Ahn, K.S. Farnesol abrogates epithelial to mesenchymal transition process through regulating Akt/mTOR pathway. Pharmacol. Res. 2019, 150, 104504. [Google Scholar] [CrossRef]
- Buhrmann, C.; Brockmueller, A.; Harsha, C.; Kunnumakkara, A.B.; Kubatka, P.; Aggarwal, B.B.; Shakibaei, M. Evidence That Tumor Microenvironment Initiates Epithelial-To-Mesenchymal Transition and Calebin A can Suppress it in Colorectal Cancer Cells. Front. Pharmacol. 2021, 12, 699842. [Google Scholar] [CrossRef]
- Yang, M.H.; Lee, J.H.; Ko, J.-H.; Jung, S.H.; Sethi, G.; Ahn, K.S. Brassinin Represses Invasive Potential of Lung Carcinoma Cells through Deactivation of PI3K/Akt/mTOR Signaling Cascade. Molecules 2019, 24, 1584. [Google Scholar] [CrossRef]
- Buhrmann, C.; Yazdi, M.; Popper, B.; Kunnumakkara, A.B.; Aggarwal, B.B.; Shakibaei, M. Induction of the Epithelial-to-Mesenchymal Transition of Human Colorectal Cancer by Human TNF-beta (Lymphotoxin) and its Reversal by Resveratrol. Nutrients 2019, 11, 704. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Wang, L.Z.; Kim, C.; Deivasigamni, A.; Arfuso, F.; Um, J.-Y.; Kumar, A.P.; Chang, Y.-C.; Kumar, D.; et al. Ascochlorin Enhances the Sensitivity of Doxorubicin Leading to the Reversal of Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma. Mol. Cancer Ther. 2016, 15, 2966–2976. [Google Scholar] [CrossRef]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef]
- Saxby, H.; Mikropoulos, C.; Boussios, S. An Update on the Prognostic and Predictive Serum Biomarkers in Metastatic Prostate Cancer. Diagnostics 2020, 10, 549. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Ahn, K.S.; Hsu, A.; Woo, C.C.; Yuan, Y.; Tan, K.H.B.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Koh, A.P.F.; et al. Thymoquinone Inhibits Bone Metastasis of Breast Cancer Cells Through Abrogation of the CXCR4 Signaling Axis. Front. Pharmacol. 2018, 9, 1294. [Google Scholar] [CrossRef]
- Chua, A.W.L.; Hay, H.S.; Rajendran, P.; Shanmugam, M.K.; Li, F.; Bist, P.; Koay, E.S.; Lim, L.H.; Kumar, A.P.; Sethi, G. Butein downregulates chemokine receptor CXCR4 expression and function through suppression of NF-kappaB activation in breast and pancreatic tumor cells. Biochem. Pharmacol. 2010, 80, 1553–1562. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Huysentruyt, L.C. On the Origin of Cancer Metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef]
- Azizi, A.A.; Lamarca, A.; McNamara, M.G.; Valle, J.W. Chemotherapy for advanced gallbladder cancer (GBC): A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2021, 163, 103328. [Google Scholar] [CrossRef]
- Sirohi, B.; Singh, A.; Jagannath, P.; Shrikhande, S.V. Chemotherapy and Targeted Therapy for Gall Bladder Cancer. Indian J. Surg. Oncol. 2014, 5, 134–141. [Google Scholar] [CrossRef]
- Boutros, C.; Gary, M.; Baldwin, K.; Somasundar, P. Gallbladder cancer: Past, present and an uncertain future. Surg. Oncol. 2012, 21, e183–e191. [Google Scholar] [CrossRef]
- Khatoon, E.; Banik, K.; Harsha, C.; Sailo, B.L.; Thakur, K.K.; Khwairakpam, A.D.; Vikkurthi, R.; Devi, T.B.; Gupta, S.C.; Kunnumakkara, A.B. Phytochemicals in cancer cell chemosensitization: Current knowledge and future perspectives. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Monisha, J.; Padmavathi, G.; Roy, N.; Deka, A.; Bordoloi, D.; Anip, A.; Kunnumakkara, A.B. NF-kappaB Blockers Gifted by Mother Nature: Prospectives in Cancer Cell Chemosensitization. Curr. Pharm. Des. 2016, 22, 4173–4200. [Google Scholar] [CrossRef]
- Bordoloi, D.; Roy, N.; Monisha, J.; Padmavathi, G.; Kunnumakkara, A.B. Multi-Targeted Agents in Cancer Cell Chemosensitization: What We Learnt from Curcumin Thus Far. Recent Patents Anti-Cancer Drug Discov. 2016, 11, 67–97. [Google Scholar] [CrossRef]
- Manu, K.A.; Shanmugam, M.K.; Ramachandran, L.; Li, F.; Siveen, K.S.; Chinnathambi, A.; Zayed, M.; Alharbi, S.A.; Arfuso, F.; Kumar, A.P.; et al. Isorhamnetin augments the anti-tumor effect of capeciatbine through the negative regulation of NF-kappaB signaling cascade in gastric cancer. Cancer Lett. 2015, 363, 28–36. [Google Scholar] [CrossRef]
- Manu, K.A.; Shanmugam, M.K.; Li, F.; Chen, L.; Siveen, K.S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Simvastatin sensitizes human gastric cancer xenograft in nude mice to capecitabine by suppressing nuclear factor-kappa B-regulated gene products. J. Mol. Med. 2014, 92, 267–276. [Google Scholar] [CrossRef]
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Dwyer, R.M. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Vicentini, C.; Galuppini, F.; Corbo, V.; Fassan, M. Current role of non-coding RNAs in the clinical setting. Non-Coding RNA Res. 2019, 4, 82–85. [Google Scholar] [CrossRef]
- Chaudhary, V.; Jangra, S.; Yadav, N.R. Nanotechnology based approaches for detection and delivery of microRNA in healthcare and crop protection. J. Nanobiotechnol. 2018, 16, 40. [Google Scholar] [CrossRef]
- Sánchez, Y.; Huarte, M. Long Non-Coding RNAs: Challenges for Diagnosis and Therapies. Nucleic Acid Ther. 2013, 23, 15–20. [Google Scholar] [CrossRef]
- Kumar, L.D. EMT in breast cancer metastasis an interplay of microRNAs signaling pathways and circulating tumor cells. Front. Biosci. 2020, 25, 979–1010. [Google Scholar] [CrossRef]
- Jiang, N.; Meng, X.; Mi, H.; Chi, Y.; Li, S.; Jin, Z.; Tian, H.; He, J.; Shen, W.; Tian, H.; et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin. Chim. Acta 2018, 486, 26–33. [Google Scholar] [CrossRef]
- Khandelwal, A.; Malhotra, A.; Jain, M.; Vasquez, K.M.; Jain, A. The emerging role of long non-coding RNA in gallbladder cancer pathogenesis. Biochimie 2017, 132, 152–160. [Google Scholar] [CrossRef]
| ncRNAs | In Vitro/ In Vivo | Model | Target | Mechanism/Outcome | References |
|---|---|---|---|---|---|
| cirRNAs | |||||
| ↑circERBB2 | In vitro | GBC patient tissues, SGC-996, GBC-SD | - | ↑Cell proliferation | [26] |
| ↑circERBB2 | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [26] |
| ↑circFOXP1 | In vitro | GBC patient tissues, NOZ, GBC-SD, EHGB-1, SGC-996, OCUG-1 | ↓miR-370 | ↑Cell proliferation, ↓apoptosis, ↓caspase-3, ↑PCNA, ↑MMP-9, ↑Akt, ↑warburg effect, ↑ECAR, ↑glycolysis, ↑PKLR | [28] |
| ↑circFOXP1 | In vivo | NOZ xenograft | ↓miR-370 | ↑Tumor growth, ↑Ki67 | [28] |
| ↑circHIPK3 | In vitro | GBC patient tissues, QBC939, GBC-SD, Mz-ChA-1 | ↓miR-124 | ↑Cell survival, ↑cell proliferation, ↓apoptosis, ↑ROCK1, ↑CDK6 | [22] |
| lncRNAs | |||||
| ↑AFAP1-AS1 | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ | - | Poor prognosis, ↑cell proliferation, ↑invasion, ↑migration | [52] |
| ↑ANRIL | In vitro | GBC patient tissues, GBC-SD, QBC939 | - | ↓Patient survival, ↑proliferation, ↓apoptosis, ↓p53, ↑cyclin D1 | [53] |
| ↑CCAT1 | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ, EH-GB2 | ↓miR-218-5p | ↑Bmi1 mRNA level | [54] |
| ↑DILC | In vitro | GBC patient tissues, SGC-996, GBC-SD | - | ↑Cell growth, ↑metastasis, ↑Wnt/β-catenin activation | [55] |
| ↑DILC | In vivo | SGC-996 xenograft | - | ↑Lung metastasis | [55] |
| ↑GBCDRlnc1 | In vitro | GBC-SD, NOZ | ↑PGK1 | Poor prognosis, ↑autophagy, ↑chemoresistance | [56] |
| ↑HEGBC | In vitro | GBC patient tissues, SGC-996, EH-GB2, GBC-SD, NOZ | - | Poor survival, ↑cell proliferation, ↑migration, ↓apoptosis, ↑ IL-11, ↑STAT3 activation | [57] |
| ↑HEGBC | In vivo | NOZ xenograft | - | ↑Tumorigenesis, ↑metastasis | [57] |
| ↑HOTAIR | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ, EH-GB2 | ↓miR-130a | ↑Cell proliferation, ↑invasion, ↑mRNA expression of c-Myc | [58] |
| ↑HOXA-AS2 | In vitro | GBC patient tissues, EHGB-1, NOZ, SGC-996, OCUG, GBC-SD | - | ↑Cell proliferation, ↓apoptosis, ↑vimentin, ↑N-cadherin, ↓E-cadherin | [59] |
| ↑H19 | In vitro | GBC patient tissues, GBC-SD, EHGB-1, NOZ | ↓miR-342-3p | ↑Cell proliferation, ↑invasion, ↑FOXM1 | [60] |
| ↑H19 | In vivo | NOZ xenograft | ↓miR-342-3p | ↑Tumor volume, ↑FOXM1 | [60] |
| ↑LINC00152 | In vitro | GBC patient tissues, NOZ, SGC-996, EHGB-2, GBC-SD | - | ↑Cell proliferation, ↑metastasis, ↓apoptosis | [61] |
| ↑LINC00152 | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [61] |
| ↑LINC00152 | In vitro | GBC patient tissues, GBC-SD, NOZ | ↓miR-138 | ↓Patient survival, ↑lymph node metastasis, ↑migration, ↑EMT progression, ↑invasion, ↑HIF-1α, ↓E-cadherin, ↑vimentin | [36] |
| ↑LINC00152 | In vivo | GBC-SD xenograft | - | ↑Peritoneal spreading, ↑metastasis | [36] |
| ↑MALAT1 | In vitro | GBC patient tissues, SGC-996, NOZ | - | ↑Cell proliferation, ↑invasion, ↑migration | [23] |
| ↑MALAT1 | In vitro | GBC patient tissues, GBC-SD, SGC-99, NOZ | ↓miR-206 | ↑Tumor size, ↑lymphatic metastasis, ↑proliferation, ↑invasion, ↓apoptosis, ↑ANXA2, ↑KRAS, ↑vimentin, ↓E-cadherin, ↑Twist | [62] |
| ↑MALAT1 | In vivo | NOZ xenograft | - | ↑Tumor growth, ↑tumor volume, ↑ANXA2, ↑KRAS | [62] |
| ↑MALAT1 | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ, OCUG1 | - | ↑Cell proliferation, ↑invasion, ↑migration, ↓ABI3BP | [63] |
| ↑MALAT1 | In vitro | GBC patient tissues, SGC-996, NOZ | ↓miR-363-3p | ↑Cell proliferation, ↓apoptosis, ↑MCL-1 | [64] |
| ↑MALAT1 | In vivo | NOZ xenograft | ↓miR-363-3p | ↑Tumor volume, ↑MCL-1 | [64] |
| ↓MEG3 | In vitro | GBC patient tissues, GBC-SD, QBC939 | - | ↑Cell proliferation, ↓apoptosis, ↓p53, ↑cyclin D1 | [53] |
| ↓MEG3 | In vitro | GBC patient tissues, NOZ, GBC-SD, SGC-996, EH-GB1, OCUG-1 | ↓EZH2 | Poor prognosis, ↑cell proliferation, ↓apoptosis, ↑invasion, ↑EMT, ↓E-cadherin, ↑vimentin, ↑N-cadherin | [65] |
| ↑MINCR | In vitro | GBC patient tissues, NOZ | ↓miR-26a-5p | ↓Overall patient survival, ↑EZH2, ↑cell proliferation, ↓apoptosis, ↑invasion, ↓E-cadherin, ↑vimentin | [66] |
| ↑PVT1 | In vitro | GBC patient tissues, GBC cells | ↓miR-30d-5p | ↑Cell proliferation, ↑invasion | [67] |
| ↑PVT1 | In vitro | GBC patient tissues, GBC-SD, NOZ | ↓miR-143 | ↑cell proliferation, ↑migration, ↑invasion, ↑HK2 | [35] |
| ↑PVT1 | In vivo | GBC-SD xenograft | - | ↑Tumor growth rate | [35] |
| ↑ROR | In vitro | GBC patient tissues, SGC-996, GBC-SD, NOZ | - | Poor prognosis, ↑cell proliferation, ↑migration, ↑invasion, ↓E-cadherin, ↑Twist1, ↑N-cadherin | [68] |
| ↑SSTR5-AS1 | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ | ↑NONO | ↑Chemoresistance, ↓apoptosis | [69] |
| ↑SSTR5-AS1 | In vivo | GBC-SD xenograft | ↑NONO | ↑Chemoresistance | [69] |
| ↑SPRY4-IT1 | In vitro | EH-GB1, GBC-SD, SGC-996, NOZ | - | ↑Cell proliferation, ↑migration, ↑invasion, ↑E-cadherin, ↓vimentin | [70] |
| ↑TUG1 | In vitro | GBC patient tissues, EH-GB1, GBC-SD, NOZ, SGC-996 | ↓miR-300 | ↑Cell proliferation, ↑metastasis | [71] |
| ↑UCA1 | In vitro | GBC patient tissues, NOZ, GBC-SD | - | ↑Cell proliferation, ↑tumor size, ↑lymph node metastasis, ↓patient survival time, ↑TNM stage, ↓E-cadherin, ↓p21, ↑EZH2 binding | [37] |
| ↑UCA1 | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [37] |
| miRNAs | |||||
| ↑miR-663a | In vitro | GBC patient tissues, GBC-SD, NOZ | ↓EMP3 | Poor prognosis, ↑cell proliferation, ↑metastasis, ↑migration, ↑invasion | [72] |
| ↓miR-143-3p | In vitro | NOZ, GBC-SD | ↑ITGA6 | ↑Cell proliferation, ↑angiogenesis, ↓PLGF, Inactivation of ITAG6/PI3K/AKT pathways | [73] |
| ↓miR-143-3p | In vivo | NOZ xenograft | - | ↑Tumor growth, ↑angiogenesis | [73] |
| ↓miR-139-5p | In vitro | GBC patient tissues, NOZ, GBC-SD | ↑PKM2 | Poor prognosis, ↑tumor progression | [74] |
| ↓miR-143-5p | In vitro | GBC patient tissues, SGC-996, GBC-SD, NOZ | ↑HIF-1α | ↓E-cadherin, ↑vimentin, ↑Twist1, ↑VEGF | [75] |
| ↑miR-182 | In vitro | GBC-SD, EHGB1, NOZ | ↓RECK | ↑Cell proliferation, ↑migration, ↑invasion, ↓apoptosis, ↓caspase-3, ↓caspase-9, ↓E-cadherin, ↓Bax, ↑Bcl-2, ↑N-cadherin, ↑β-catenin | [47] |
| ↓miR-125b | In vitro | GBC patient tissues, TYGBK-1, OCUG-1, TYGBK-8, NOZ, G-415, TGBC1TKB, TGBC2TKB, TGBC14TKB, TGBC24TKB | - | Poor prognosis, ↑cell proliferation, ↑migration | [50] |
| ↓miR-125b-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ | ↑Bcl2 | Poor prognosis, ↑chemoresistance | [76] |
| ↓miR-136 | In vitro | GBC patient tissues, SGC-996, GBC-SD, Mz-ChA-1 | ↑MAP2K4 | ↑Cell proliferation, ↓apoptosis, Activation of JNK pathway | [48] |
| ↓miR-136 | In vivo | Mz-ChA-1 xenograft | ↑MAP2K4 | ↑Tumor growth, ↓apoptosis | [48] |
| ↓miR-324-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996 | ↑TGFβ2 | ↑Invasion, ↑migration, ↑EMT | [77] |
| ↓miR-30d-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ | ↑LDHA | ↑Cell proliferation, ↑invasion | [78] |
| ↓miR-30a-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996, OCUG-1, EH-GB1, NOZ | ↑E2F7 | ↑Cell proliferation, ↑invasion, ↑migration, ↓E-cadherin, ↑vimentin | [49] |
| ↓miR-30a-5p | In vivo | NOZ Xenograft | ↑E2F7 | ↑Cell proliferation, ↑invasion, ↑migration | [49] |
| ↓miR-26a | In vitro | GBC patient tissues, GBC-SD, EH-GB1, SGC-996 | ↑HMGA2 | ↑Cell proliferation | [79] |
| ↓miR-26a | In vivo | GBC-SD xenograft | ↑HMGA2 | ↑Cell proliferation | [79] |
| ↓miR-218-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ | ↑PRKCE | ↑Chemoresistance, ↑MDR1/P-gp | [80] |
| ↓miR-218-5p | In vivo | NOZ xenograft | ↑PRKCE | ↑Chemoresistance, ↑MDR1/P-gp | [80] |
| ↓miR-145 | In vitro | GBC patient tissues, GBC-SD, SGC-996 | ↑MRP1 | ↑Chemoresistance | [81] |
| ↓miR-145 | In vivo | GBC-SD xenograft | ↑MRP1 | ↑Chemoresistance | [81] |
| ↓miR-335 | In vitro | GBC patient tissues, GBC-SD, SGC-996 | ↑MEF2D | ↑Chemoresistance, ↑cell viability | [82] |
| ↓miR-31 | In vitro | GBC patient tissues, NOZ, NOZ/DDP, GBC-SD/DDP, GBC-SD | ↑Src | ↑Cell proliferation, ↑ cell viability, ↑invasion, ↓apoptosis, ↑chemoresistance | [83] |
| ↓miR-31 | In vivo | NOZ/DDP xenograft | ↑Src | ↑Chemoresistance | [83] |
| ↓miR-138 | In vitro | GBC patient tissues, OCUG-1, NOZ | ↑Bag-1 | ↑Cell proliferation, ↓apoptosis | [84] |
| ↑miR-155 | In vitro | GBC patient tissues, G-415, OCUG-1, NOZ | - | ↑Cell proliferation, ↑invasion | [24] |
| ↓miR-223 | In vitro | GBC patient tissues, GBC-SD, NOZ | ↑STMN1 | ↑Cell proliferation, ↑invasion, ↑chemoresistance | [85] |
| ↓miR-223 | In vivo | NOZ xenograft | ↑STMN1 | ↑Tumor growth | [85] |
| ↓miR-29c-5p | In vitro | GBC-SD, NOZ | ↑CPEB4 | ↑Invasion, ↑migration, ↑EMT | [86] |
| ↓miR-29c-5p | In vivo | NOZ xenograft | - | ↑Lung metastatic rate | [86] |
| ↑miR-20a | In vitro | GBC patient tissues, GBC-SD | ↓Smad7 | ↑Metastasis, ↑cell growth, ↑EMT | [87] |
| ↑miR-20a | In vivo | GBC-SD xenograft | ↓Smad7 | ↑Metastasis | [87] |
| ↓miR-1 | In vitro | GBC patient tissues, NOZ | - | ↑Cell growth | [88] |
| ↓miR-145 | In vitro | GBC patient tissues, NOZ | - | ↑Cell growth | [88] |
| ↓miR-33a | In vitro | GBC patient tissues, GBC-SD | ↑Twist1 | Poor prognosis, ↑cell proliferation, ↑metastasis | [89] |
| ↓miR-33a | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [89] |
| ↓miR-1-5p | In vitro | GBC patient tissues, GBC-SD, SGC-996, HUH28, TFK-1 | ↑Notch2 | ↑Cell growth, ↑invasion, ↑migration | [90] |
| ↓miR-1-5p | In vivo | SGC-996 xenograft | - | ↑Tumor growth | [90] |
| ↓miR-372 | In vitro | GBC patient tissues, G-415, OCUG-1, SGC-996 | ↑CLIC1 | Poor prognosis | [91] |
| ↓miR-30b | In vitro | GBC-SD | ↑NT5E | ↑Cell proliferation, ↑migration, ↑invasion | [92] |
| ↓miR-30b | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [92] |
| ↓miR-340 | In vitro | GBC-SD | ↑NT5E | ↑Cell proliferation, ↑migration, ↑invasion | [92] |
| ↓miR-340 | In vivo | GBC-SD xenograft | - | ↑Tumor growth | [92] |
| ↓miR-140-5p | In vitro | GBC patient tissues, GBC-SD | ↑SEPT2 | ↑Cell proliferation, ↑migration, ↑invasion | [93] |
| ↓miR-135a | In vitro | GBC patient tissues, GBC-SD, SGC-996, EH-GB1 | ↑VLDLR | ↑Cell proliferation, ↑p38 MAPK | [94] |
| ↓miR-135a | In vivo | GBC-SD xenograft | - | ↑Cell proliferation | [94] |
| snoRNA | |||||
| ↑SNORA74B | In vitro | GBC patient tissues, GBC-SD, SGC-996, NOZ, H69 | - | ↓Patient survival, ↑cell proliferation, ↓apoptosis, ↑Ki67, ↓PHLPP, ↓p21, ↓p27, ↑cyclin D1, ↓Bax, ↓cytosolic cyt C, ↓cleaved caspase-3, ↑Bcl-2, ↑mt cyt C, ↑Akt/mTOR | [51] |
| ↑SNORA74B | In vivo | GBC-SD xenograft | - | ↑Tumor growth, ↓apoptosis | [51] |
| ↓SNORA21 | In vitro | GBC patient tissues, GBC-SD, G415 | - | ↑Cell proliferation, ↓apoptosis, ↑migration, ↑invasion, ↓E-cadherin, ↑N-cadherin, ↑vimentin, ↓cleaved caspase-3, ↓Bax, ↑Bcl-2, ↓p21, ↑cyclin D1, ↑c-Myc | [21] |
| ↓SNORA21 | In vivo | GBC-SD xenograft | - | ↑Tumor growth, ↑c-Myc | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rana, V.; Parama, D.; Khatoon, E.; Girisa, S.; Sethi, G.; Kunnumakkara, A.B. Reiterating the Emergence of Noncoding RNAs as Regulators of the Critical Hallmarks of Gall Bladder Cancer. Biomolecules 2021, 11, 1847. https://doi.org/10.3390/biom11121847
Rana V, Parama D, Khatoon E, Girisa S, Sethi G, Kunnumakkara AB. Reiterating the Emergence of Noncoding RNAs as Regulators of the Critical Hallmarks of Gall Bladder Cancer. Biomolecules. 2021; 11(12):1847. https://doi.org/10.3390/biom11121847
Chicago/Turabian StyleRana, Varsha, Dey Parama, Elina Khatoon, Sosmitha Girisa, Gautam Sethi, and Ajaikumar B. Kunnumakkara. 2021. "Reiterating the Emergence of Noncoding RNAs as Regulators of the Critical Hallmarks of Gall Bladder Cancer" Biomolecules 11, no. 12: 1847. https://doi.org/10.3390/biom11121847
APA StyleRana, V., Parama, D., Khatoon, E., Girisa, S., Sethi, G., & Kunnumakkara, A. B. (2021). Reiterating the Emergence of Noncoding RNAs as Regulators of the Critical Hallmarks of Gall Bladder Cancer. Biomolecules, 11(12), 1847. https://doi.org/10.3390/biom11121847






