Effects of Sulfur Assimilation in Pseudomonas fluorescens SS101 on Growth, Defense, and Metabolome of Different Brassicaceae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth
2.2. Bacterial Strains and Culture Conditions
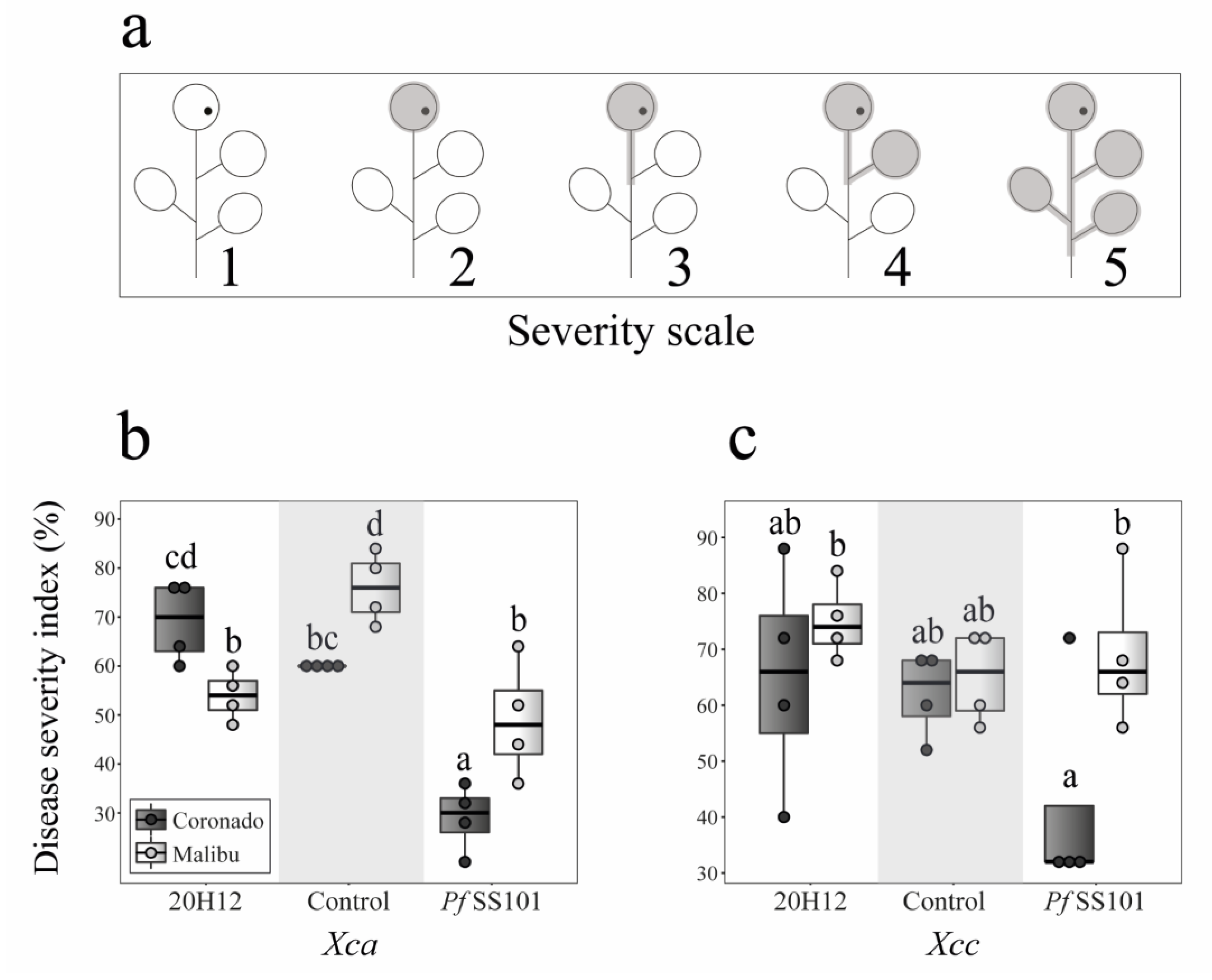
2.3. Bioassay for Induced Resistance against the Leaf Pathogen Xanthomonas
2.4. Plant Metabolite Analysis
2.4.1. Sample Preparation
2.4.2. Metabolite Analysis
2.4.3. Non-Targeted LC–MS Data Processing and Analysis
2.5. Statistical Analysis
3. Results
3.1. Role of the cysH Gene of P. fluorescens SS101 in Plant Growth Promotion
3.2. Role of the cysH Gene of P. fluorescens SS101 in Induced Resistance
3.3. Effect of P. fluorescens SS101 and the cysH Mutant on the Plant Metabolome
3.3.1. Arabidopsis
3.3.2. Broccoli
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Etalo, D.W.; Jeon, J.-S.; Raaijmakers, J.M. Modulation of plant chemistry by beneficial root microbiota. Nat. Prod. Rep. 2018, 35, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Cheng, X.; Etalo, D.W.; van de Mortel, J.E.; Dekkers, E.; Nguyen, L.; Medema, M.H.; Raaijmakers, J.M. Genome-wide analysis of bacterial determinants of plant growth promotion and induced systemic resistance byPseudomonas fluorescens. Environ. Microbiol. 2017, 19, 4638–4656. [Google Scholar] [CrossRef] [Green Version]
- Meldau, D.G.; Meldau, S.; Hoang, L.H.; Underberg, S.; Wunsche, H.; Baldwin, I.T. Dimethyl Disulfide Produced by the Naturally Associated Bacterium Bacillus sp B55 Promotes Nicotiana attenuata Growth by Enhancing Sulfur Nutrition. Plant Cell 2013, 25, 2731–2747. [Google Scholar] [CrossRef] [Green Version]
- Aziz, M.; Nadipalli, R.K.; Xie, X.; Sun, Y.; Surowiec, K.; Zhang, J.-L.; Paré, P.W. Augmenting Sulfur Metabolism and Herbivore Defense in Arabidopsis by Bacterial Volatile Signaling. Front. Plant Sci. 2016, 7, 458. [Google Scholar] [CrossRef] [Green Version]
- Saito, K. Sulfur Assimilatory Metabolism. The Long and Smelling Road. Plant Physiol. 2004, 136, 2443–2450. [Google Scholar] [CrossRef] [Green Version]
- Van De Mortel, J.E.; De Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; Van Loon, J.J.; Dicke, M.; Raaijmakers, J.M. Metabolic and Transcriptomic Changes Induced in Arabidopsis by the Rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef] [Green Version]
- Kopriva, S.; Malagoli, M.; Takahashi, H. Sulfur nutrition: Impacts on plant development, metabolism, and stress responses. J. Exp. Bot. 2019, 70, 4069–4073. [Google Scholar] [CrossRef]
- Vicente, J.G.; Holub, E.B. Xanthomonas campestrispv.campestris(cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 2013, 14, 2–18. [Google Scholar] [CrossRef]
- de Souza, J.T.; de Boer, M.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 2003, 69, 7161–7172. [Google Scholar] [CrossRef] [Green Version]
- Tikunov, Y.M.; Laptenok, S.; Hall, R.D.; Bovy, A.; De Vos, R.C.H. MSClust: A tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 2012, 8, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.; Bino, R.J.; Hall, R. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Lommen, A. MetAlign: Interface-Driven, Versatile Metabolomics Tool for Hyphenated Full-Scan Mass Spectrometry Data Preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 25 February 2021).
- Metlin. Available online: https://metlin.scripps.edu/ (accessed on 25 February 2021).
- Jeon, J.-S.; Carreno-Quintero, N.; van Eekelen, H.D.L.M.; De Vos, R.C.H.; Raaijmakers, J.M.; Etalo, D.W. Impact of root-associated strains of three Paraburkholderia species on primary and secondary metabolism of Brassica oleracea. Sci. Rep. 2021, 11, 2781. [Google Scholar] [CrossRef] [PubMed]
- Cribari-Neto, F.; Zeileis, A. Beta Regression in R. J. Stat. Softw. 2010, 34, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Meister, A. Glutathione biosynthesis and its inhibition. Methods Enzymol. 1995, 252, 26–30. [Google Scholar] [CrossRef]
- Geu-Flores, F.; Moldrup, M.E.; Bottcher, C.; Olsen, C.E.; Scheel, D.; Halkier, B.A. Cytosolic gamma-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant. Cell. 2011, 23, 2456–2469. [Google Scholar] [CrossRef] [Green Version]
- Textor, S.; Bartram, S.; Falk, K.L.; Hick, A.; Pickett, J.A.; Gershenzon, J.; Kroymann, J. Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 2004, 218, 1026–1035. [Google Scholar] [CrossRef]
- Textor, S.; de Kraker, J.-W.; Hause, B.; Gershenzon, J.; Tokuhisa, J. MAM3 Catalyzes the Formation of All Aliphatic Glucosinolate Chain Lengths in Arabidopsis. Plant Physiol. 2007, 144, 60–71. [Google Scholar] [CrossRef] [Green Version]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid Accumulation in Arabidopsis Repressed in Lignin Synthesis Affects Auxin Transport and Plant Growth. Plant Cell 2007, 19, 148–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, B.M.; Errafi, S.; Bucher, R.; Dobrev, P.; Geisler, M.; Bigler, L.; Zazimalova, E.; Ringli, C. 7-Rhamnosylated Flavonols Modulate Homeostasis of the Plant Hormone Auxin and Affect Plant Development. J. Biol. Chem. 2016, 291, 5385–5395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddox, C.E.; Laur, L.M.; Tian, L. Antibacterial Activity of Phenolic Compounds Against the Phytopathogen Xylella fastidiosa. Curr. Microbiol. 2010, 60, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Yang, W.; Tang, F.; Chen, X.; Ren, L. Antibacterial Activities of Flavonoids: Structure-Activity Relationship and Mechanism. Curr. Med. Chem. 2014, 22, 132–149. [Google Scholar] [CrossRef]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Reimers, P.; Leach, J. Race-specific resistance to Xanthomonas oryzae pv. oryzae conferred by bacterial blight resistance gene Xa-10 in rice (Oryza sativa) involves accumulation of a lignin-like substance in host tissues. Physiol. Mol. Plant Pathol. 1991, 38, 39–55. [Google Scholar] [CrossRef]
- Madloo, P.; Lema, M.; Francisco, M.; Soengas, P. Role of Major Glucosinolates in the Defense of Kale Against Sclerotinia sclerotiorum and Xanthomonas campestris pv. campestris. Phytopathol. 2019, 109, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.; Piślewska-Bednarek, M.; Svatoš, A.; Schneider, B.; Doubský, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A Glucosinolate Metabolism Pathway in Living Plant Cells Mediates Broad-Spectrum Antifungal Defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef]
- Foyer, C.H. Ascorbic acid. In Antioxidants in Higher Plants; CRC Press: Boca Raton, FL, USA, 2017; pp. 39–66. [Google Scholar]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free. Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, J.-S.; Etalo, D.W.; Carreno-Quintero, N.; de Vos, R.C.H.; Raaijmakers, J.M. Effects of Sulfur Assimilation in Pseudomonas fluorescens SS101 on Growth, Defense, and Metabolome of Different Brassicaceae. Biomolecules 2021, 11, 1704. https://doi.org/10.3390/biom11111704
Jeon J-S, Etalo DW, Carreno-Quintero N, de Vos RCH, Raaijmakers JM. Effects of Sulfur Assimilation in Pseudomonas fluorescens SS101 on Growth, Defense, and Metabolome of Different Brassicaceae. Biomolecules. 2021; 11(11):1704. https://doi.org/10.3390/biom11111704
Chicago/Turabian StyleJeon, Je-Seung, Desalegn W. Etalo, Natalia Carreno-Quintero, Ric C. H. de Vos, and Jos M. Raaijmakers. 2021. "Effects of Sulfur Assimilation in Pseudomonas fluorescens SS101 on Growth, Defense, and Metabolome of Different Brassicaceae" Biomolecules 11, no. 11: 1704. https://doi.org/10.3390/biom11111704
APA StyleJeon, J.-S., Etalo, D. W., Carreno-Quintero, N., de Vos, R. C. H., & Raaijmakers, J. M. (2021). Effects of Sulfur Assimilation in Pseudomonas fluorescens SS101 on Growth, Defense, and Metabolome of Different Brassicaceae. Biomolecules, 11(11), 1704. https://doi.org/10.3390/biom11111704





