Expanding the Disorder-Function Paradigm in the C-Terminal Tails of Erbbs
Abstract
:1. Introduction
2. General Structure and Mechanism of Erbbs
2.1. Similarities and Differences between ErbBs
2.2. The Different Steps of Activation, from Extracellular Ligand Binding to Kinase Activation
2.2.1. Ligand Binding to the ECD and Dimerization
2.2.2. The Transmembrane Helix (TMH) and N-Terminal End of the Juxtamembrane Region (JMA)
2.2.3. The Tyrosine Kinase Domain and Its Activation
2.3. The C-Terminal Tail (CT) and Signal Transduction by Phosphotyrosines
2.3.1. Tyrosine Autophosphorylation Sites
2.3.2. Partners of Phosphorylated Tyrosines of ErbBs
2.3.3. Shortcuts and Pitfall of the Common Signal Transduction Model by CTs
3. CTs of ErbBs, More Than Disordered Ropes with pY Anchors
3.1. Sequence Characteristics of the Four CT-ErbBs
3.2. The Need for Tight Regulation: The Specificity of Tyrosine Phosphorylation Sites
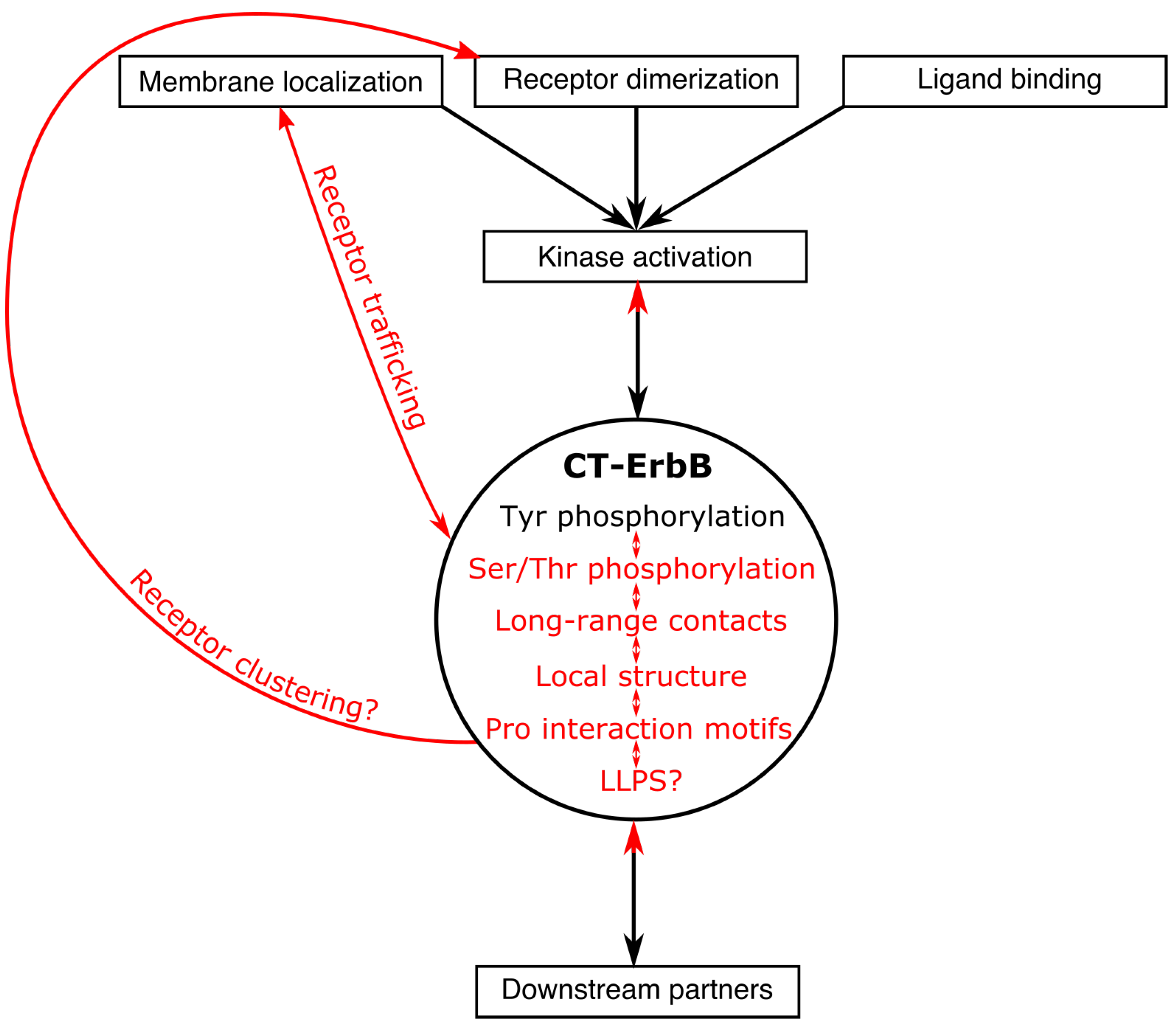
3.3. Regulatory Roles of CT-ErbBs
3.3.1. Interdependence of Phosphorylation Sites
3.3.2. Regulation of Kinase Activity by the CT
3.3.3. The CT Is Involved in ErbB Receptor Trafficking
3.3.4. Possible Regulation of Receptor Clustering by Phase Separation of the CT
3.4. Structural Description Outside of Regions Interacting with the Kinase
3.4.1. Local Structure
3.4.2. Compaction and Long-Range Contacts
3.5. Comparison with the Intracellular Tail of Other RTKs
3.5.1. Inhibition and Activation Mechanisms
3.5.2. An Overlooked Role of Prolines in ErbBs and Other RTKs?
4. Outlook and Outstanding Questions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALK | Anaplastic Lymphoma Kinase receptor |
| CD | Circular Dichroism |
| CT | C-terminal Tail |
| EGFR | Epidermal Growth Factor Receptor |
| ELM | Eukaryotic Linear Motif |
| Erbin | ErbB2 Interacting Protein |
| FGFR | Fibroblast Growth Factor Receptor |
| HER | Human Epidermal growth factor Receptor |
| IDP | Intrinsically Disordered Protein |
| IDR | Intrinsically Disordered Region |
| LLPS | Liquid-liquid phase separation |
| LMTK | Lemur Tyrosine Kinase receptor |
| LTK | Leukocyte Tyrosine Kinase receptor |
| MAPK | Mitogen-Activated Protein Kinases |
| MEMO | Mediator of ErbB2-driven cell MOtility |
| PI3K | Phosphoinositide 3-kinase |
| PLCG1 | Phospholipase C-1 |
| PPII | Polyproline type II |
| PTB domain | phosphotyrosine binding domain |
| PTPN18 | Protein Tyrosine Phosphatase Non-receptor type 18 |
| ROR | Retinoid-related Orphan Receptor |
| RTK | Receptor Tyrosine Kinase |
| SH2 | SRC Homology 2 Domain |
| SH3 | SRC Homology 3 Domain |
| STAT | Signal Transducers and Activators of Transcription |
References
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [Green Version]
- O’Keefe, E.; Hollenberg, M.D.; Cuatrecasas, P. Epidermal Growth Factor. Characteristics of Specific Binding in Membranes from Liver, Placenta, and Other Target Tissues. Arch. Biochem. Biophys. 1974, 164, 518–526. [Google Scholar] [CrossRef]
- Cohen, S.; Fava, R.A.; Sawyer, S.T. Purification and Characterization of Epidermal Growth Factor Receptor/Protein Kinase from Normal Mouse Liver. Proc. Natl. Acad. Sci. USA 1982, 79, 6237–6241. [Google Scholar] [CrossRef] [Green Version]
- Burden, S.; Yarden, Y. Neuregulins and Their Receptors: A Versatile Signaling Module in Organogenesis and Oncogenesis. Neuron 1997, 18, 847–855. [Google Scholar] [CrossRef] [Green Version]
- Chan, R.; Hardy, W.R.; Laing, M.A.; Hardy, S.E.; Muller, W.J. The Catalytic Activity of the ErbB-2 Receptor Tyrosine Kinase Is Essential for Embryonic Development. Mol. Cell. Biol. 2002, 22, 1073–1078. [Google Scholar] [CrossRef] [Green Version]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. The ErbB Signaling Network: Receptor Heterodimerization in Development and Cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef] [Green Version]
- Pinkas-Kramarski, R.; Soussan, L.; Waterman, H.; Levkowitz, G.; Alroy, I.; Klapper, L.; Lavi, S.; Seger, R.; Ratzkin, B.J.; Sela, M.; et al. Diversification of Neu Differentiation Factor and Epidermal Growth Factor Signaling by Combinatorial Receptor Interactions. EMBO J. 1996, 15, 2452–2467. [Google Scholar] [CrossRef]
- Vaskovsky, A.; Lupowitz, Z.; Erlich, S.; Pinkas-Kramarski, R. ErbB-4 Activation Promotes Neurite Outgrowth in PC12 Cells. J. Neurochem. 2000, 74, 979–987. [Google Scholar] [CrossRef]
- Jeon, M.; Lee, J.; Nam, S.; Shin, I.; Lee, J.; Kim, S. Induction of Fibronectin by HER2 Overexpression Triggers Adhesion and Invasion of Breast Cancer Cells. Exp. Cell Res. 2015, 333, 116–126. [Google Scholar] [CrossRef]
- Chausovsky, A.; Waterman, H.; Elbaum, M.; Yarden, Y.; Geiger, B.; Bershadsky, A.D. Molecular Requirements for the Effect of Neuregulin on Cell Spreading, Motility and Colony Organization. Oncogene 2000, 19, 878–888. [Google Scholar] [CrossRef] [Green Version]
- Marone, R.; Hess, D.; Dankort, D.; Muller, W.J.; Hynes, N.E.; Badache, A. Memo Mediates ErbB2-Driven Cell Motility. Nat. Cell Biol. 2004, 6, 515–522. [Google Scholar] [CrossRef]
- Giani, C.; Casalini, P.; Pupa, S.M.; De Vecchi, R.; Ardini, E.; Colnaghi, M.I.; Giordano, A.; Ménard, S. Increased Expression of C-erbB-2 in Hormone-Dependent Breast Cancer Cells Inhibits Cell Growth and Induces Differentiation. Oncogene 1998, 17, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB Signalling Network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Downward, J.; Yarden, Y.; Mayes, E.; Scrace, G.; Totty, N.; Stockwell, P.; Ullrich, A.; Schlessinger, J.; Waterfield, M.D. Close Similarity of Epidermal Growth Factor Receptor and V-Erb-B Oncogene Protein Sequences. Nature 1984, 307, 521–527. [Google Scholar] [CrossRef]
- Schechter, A.L.; Stern, D.F.; Vaidyanathan, L.; Decker, S.J.; Drebin, J.A.; Greene, M.I.; Weinberg, R.A. The Neu Oncogene: An Erb-B-Related Gene Encoding a 185,000-Mr Tumour Antigen. Nature 1984, 312, 513–516. [Google Scholar] [CrossRef]
- Tebbutt, N.; Pedersen, M.W.; Johns, T.G. Targeting the ERBB Family in Cancer: Couples Therapy. Nat. Rev. Cancer 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Chaudhury, A.R.; Gerecke, K.M.; Wyss, J.M.; Morgan, D.G.; Gordon, M.N.; Carroll, S.L. Neuregulin-1 and ErbB4 Immunoreactivity Is Associated with Neuritic Plaques in Alzheimer Disease Brain and in a Transgenic Model of Alzheimer Disease. J. Neuropathol. Exp. Neurol. 2003, 62, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Nakata, A.; Miyagawa, J.I.; Yamashita, S.; Nishida, M.; Tamura, R.; Yamamori, K.; Nakamura, T.; Nozaki, S.; Kameda-Takemura, K.; Kawata, S.; et al. Localization of Heparin-Binding Epidermal Growth Factor-like Growth Factor in Human Coronary Arteries: Possible Roles of HB-EGF in the Formation of Coronary Atherosclerosis. Circulation 1996, 94, 2778–2786. [Google Scholar] [CrossRef]
- Hubbard, S.R. Structural Analysis of Receptor Tyrosine Kinases. Prog. Biophys. Mol. Biol. 1999, 71, 343–358. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The Importance of Intrinsic Disorder for Protein Phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Zhou, H.X.; Pang, X.; Lu, C. Rate Constants and Mechanisms of Intrinsically Disordered Proteins Binding to Structured Targets. Phys. Chem. Chem. Phys. 2012, 14, 10466–10476. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P. Multisteric Regulation by Structural Disorder in Modular Signaling Proteins: An Extension of the Concept of Allostery. Chem. Rev. 2014, 114, 6715–6732. [Google Scholar] [CrossRef] [Green Version]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.; Obradović, Z.; Dunker, A. Intrinsic Disorder in Cell-Signaling and Cancer-Associated Proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing Your ID: Intrinsic Disorder as an ID for Recognition, Regulation and Cell Signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef]
- Keppel, T.R.; Sarpong, K.; Murray, E.M.; Monsey, J.; Zhu, J.; Bose, R. Biophysical Evidence for Intrinsic Disorder in the C-Terminal Tails of the Epidermal Growth Factor Receptor (EGFR) and HER3 Receptor Tyrosine Kinases. J. Biol. Chem. 2017, 292, 597–610. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Pinet, L.; Assrir, N.; Elantak, L.; Guerlesquin, F.; Badache, A.; Lescop, E.; van Heijenoort, C. 1H, 13C and 15N Assignments of the C-Terminal Intrinsically Disordered Cytosolic Fragment of the Receptor Tyrosine Kinase ErbB2. Biomol. NMR Assign 2018, 12, 23–26. [Google Scholar] [CrossRef]
- Okamoto, K.; Sako, Y. Single-Molecule Förster Resonance Energy Transfer Measurement Reveals the Dynamic Partially Ordered Structure of the Epidermal Growth Factor Receptor C-Tail Domain. J. Phys. Chem. B 2019, 123, 571–581. [Google Scholar] [CrossRef]
- Regmi, R.; Srinivasan, S.; Latham, A.P.; Kukshal, V.; Cui, W.; Zhang, B.; Bose, R.; Schlau-Cohen, G.S. Phosphorylation-Dependent Conformations of the Disordered Carboxyl-Terminus Domain in the Epidermal Growth Factor Receptor. J. Phys. Chem. Lett. 2020, 10037–10044. [Google Scholar] [CrossRef]
- Pinet, L.; Wang, Y.H.; Deville, C.; Lescop, E.; Guerlesquin, F.; Badache, A.; Bontems, F.; Morellet, N.; Durand, D.; Assrir, N.; et al. Structural and Dynamic Characterization of the C-Terminal Tail of ErbB2: Disordered but Not Random. Biophys. J. 2021, 120, 1869–1882. [Google Scholar] [CrossRef]
- Jura, N.; Endres, N.F.; Engel, K.; Deindl, S.; Das, R.; Lamers, M.H.; Wemmer, D.E.; Zhang, X.; Kuriyan, J. Mechanism for Activation of the EGF Receptor Catalytic Domain by the Juxtamembrane Segment. Cell 2009, 137, 1293–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirza, A.; Mustafa, M.; Talevich, E.; Kannan, N. Co-Conserved Features Associated with Cis Regulation of ErbB Tyrosine Kinases. PLoS ONE 2010, 5, e14310. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, E.; Das, R.; Wang, Q.; Collier, T.S.; Cantor, A.; Huang, Y.; Wong, K.; Mirza, A.; Barros, T.; Grob, P.; et al. Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor. Mol. Cell Biol. 2015, 35, 3083–3102. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.Y.; Koland, J.G. Conformational Changes Accompany Phosphorylation of the Epidermal Growth Factor Receptor C-Terminal Domain. Protein Sci. A Publ. Protein Soc. 2005, 14, 2793–2803. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.Y.; Hazlett, T.L.; Koland, J.G. Structure and Dynamics of the Epidermal Growth Factor Receptor C-Terminal Phosphorylation Domain. Protein Sci. 2006, 15, 1142–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slamon, D.; Clark, G.; Wong, S.; Levin, W.; Ullrich, A.; McGuire, W. Human Breast Cancer: Correlation of Relapse and Survival with Amplification of the HER-2/Neu Oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, F.; Telesco, S.E.; Liu, Y.; Radhakrishnan, R.; Lemmon, M.A. ErbB3/HER3 Intracellular Domain Is Competent to Bind ATP and Catalyze Autophosphorylation. Proc. Natl. Acad. Sci. USA 2010, 107, 7692–7697. [Google Scholar] [CrossRef] [Green Version]
- Stein, R.A.; Staros, J.V. Evolutionary Analysis of the ErbB Receptor and Ligand Families. J. Mol. Evol. 2000, 50, 397–412. [Google Scholar] [CrossRef]
- Stein, R.A.; Staros, J.V. Insights into the Evolution of the ErbB Receptor Family and Their Ligands from Sequence Analysis. BMC Evol. Biol. 2006, 6, 79. [Google Scholar] [CrossRef] [Green Version]
- Lemmon, M.A.; Schlessinger, J.; Ferguson, K.M. The EGFR Family: Not so Prototypical Receptor Tyrosine Kinases. Cold Spring Harb. Perspect. Biol. 2014, 6, a020768. [Google Scholar] [CrossRef]
- Ogiso, H.; Ishitani, R.; Nureki, O.; Fukai, S.; Yamanaka, M.; Kim, J.H.; Saito, K.; Sakamoto, A.; Inoue, M.; Shirouzu, M.; et al. Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. Cell 2002, 110, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, K.M.; Berger, M.B.; Mendrola, J.M.; Cho, H.s.; Leahy, D.J.; Lemmon, M.A. EGF Activates Its Receptor by Removing Interactions That Autoinhibit Ectodomain Dimerization. Mol. Cell 2003, 11, 507–517. [Google Scholar] [CrossRef]
- Wilson, K.J.; Mill, C.; Lambert, S.; Buchman, J.; Wilson, T.R.; Hernandez-Gordillo, V.; Gallo, R.M.; Ades, L.M.; Settleman, J.; Riese, D.J. EGFR Ligands Exhibit Functional Differences in Models of Paracrine and Autocrine Signaling. Growth Factors 2012, 30, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Macdonald-Obermann, J.L.; Pike, L.J. Different Epidermal Growth Factor (EGF) Receptor Ligands Show Distinct Kinetics and Biased or Partial Agonism for Homodimer and Heterodimer Formation. J. Biol. Chem. 2014, 289, 26178–26188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freed, D.M.; Bessman, N.J.; Kiyatkin, A.; Salazar-Cavazos, E.; Byrne, P.O.; Moore, J.O.; Valley, C.C.; Ferguson, K.M.; Leahy, D.J.; Lidke, D.S.; et al. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell 2017, 171, 683–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriki, T.; Maruyama, H.; Maruyama, I.N. Activation of Preformed EGF Receptor Dimers by Ligand-Induced Rotation of the Transmembrane Domain. J. Mol. Biol. 2001, 311, 1011–1026. [Google Scholar] [CrossRef]
- Tao, R.H.; Maruyama, I.N. All EGF(ErbB) Receptors Have Preformed Homo- and Heterodimeric Structures in Living Cells. J. Cell Sci. 2008, 121, 3207–3217. [Google Scholar] [CrossRef] [Green Version]
- Purba, E.; Saita, E.i.; Maruyama, I. Activation of the EGF Receptor by Ligand Binding and Oncogenic Mutations: The “Rotation Model”. Cells 2017, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Arndt-Jovin, D.J.; Botelho, M.G.; Jovin, T.M. Structure-Function Relationships of ErbB RTKs in the Plasma Membrane of Living Cells. Cold Spring Harb. Perspect. Biol. 2014, 6, a008961. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, K.M. Extracellular Domains Drive Homo- but Not Hetero-Dimerization of erbB Receptors. EMBO J. 2000, 19, 4632–4643. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Schlessinger, J. Self-Phosphorylation of Epidermal Growth Factor Receptor: Evidence for a Model of Intermolecular Allosteric Activation. Biochemistry 1987, 26, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Tzahar, E.; Waterman, H.; Chen, X.; Levkowitz, G.; Karunagaran, D.; Lavi, S.; Ratzkin, B.J.; Yarden, Y. A Hierarchical Network of Interreceptor Interactions Determines Signal Transduction by Neu Differentiation Factor/Neuregulin and Epidermal Growth Factor. Mol. Cell. Biol. 1996, 16, 5276–5287. [Google Scholar] [CrossRef] [Green Version]
- Mendrola, J.M.; Berger, M.B.; King, M.C.; Lemmon, M.A. The Single Transmembrane Domains of ErbB Receptors Self-Associate in Cell Membranes. J. Biol. Chem. 2002, 277, 4704–4712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharpe, S.; Barber, K.R.; Grant, C.W.M. Evidence of a Tendency to Self-Association of the Transmembrane Domain of ErbB-2 in Fluid Phospholipid Bilayers. Biochemistry 2002, 41, 2341–2352. [Google Scholar] [CrossRef] [PubMed]
- Duneau, J.P.; Vegh, A.P.; Sturgis, J.N. A Dimerization Hierarchy in the Transmembrane Domains of the HER Receptor Family. Biochemistry 2007, 46, 2010–2019. [Google Scholar] [CrossRef] [Green Version]
- Mineev, K.S.; Bocharov, E.V.; Pustovalova, Y.E.; Bocharova, O.V.; Chupin, V.V.; Arseniev, A.S. Spatial Structure of the Transmembrane Domain Heterodimer of ErbB1 and ErbB2 Receptor Tyrosine Kinases. J. Mol. Biol. 2010, 400, 231–243. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Weinberg, R.A. Increased Tyrosine Kinase Activity Associated with the Protein Encoded by the Activated Neu Oncogene. Proc. Natl. Acad. Sci. USA 1988, 85, 5394–5398. [Google Scholar] [CrossRef] [Green Version]
- Bell, C.A.; Tynan, J.A.; Hart, K.C.; Meyer, A.N.; Robertson, S.C.; Donoghue, D.J. Rotational Coupling of the Transmembrane and Kinase Domains of the Neu Receptor Tyrosine Kinase. Mol. Biol. Cell 2000, 11, 3589–3599. [Google Scholar] [CrossRef] [Green Version]
- Bragin, P.E.; Mineev, K.S.; Bocharova, O.V.; Volynsky, P.E.; Bocharov, E.V.; Arseniev, A.S. HER2 Transmembrane Domain Dimerization Coupled with Self-Association of Membrane-Embedded Cytoplasmic Juxtamembrane Regions. J. Mol. Biol. 2016, 428, 52–61. [Google Scholar] [CrossRef]
- Red Brewer, M.; Choi, S.H.; Alvarado, D.; Moravcevic, K.; Pozzi, A.; Lemmon, M.A.; Carpenter, G. The Juxtamembrane Region of the EGF Receptor Functions as an Activation Domain. Mol. Cell 2009, 34, 641–651. [Google Scholar] [CrossRef] [Green Version]
- Endres, N.F.; Das, R.; Smith, A.W.; Arkhipov, A.; Kovacs, E.; Huang, Y.; Pelton, J.G.; Shan, Y.; Shaw, D.E.; Wemmer, D.E.; et al. Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell 2013, 152, 543–556. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gureasko, J.; Shen, K.; Cole, P.A.; Kuriyan, J. An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor. Cell 2006, 125, 1137–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkhipov, A.; Shan, Y.; Das, R.; Endres, N.F.N.; Eastwood, M.P.M.; Wemmer, D.E.D.; Kuriyan, J.; Shaw, D.E. Architecture and Membrane Interactions of the EGF Receptor. Cell 2013, 152, 557–569. [Google Scholar] [CrossRef] [Green Version]
- Pawar, A.B.; Sengupta, D. Effect of Membrane Composition on Receptor Association: Implications of Cancer Lipidomics on ErbB Receptors. J. Membr. Biol. 2018, 251, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R. Juxtamembrane Autoinhibition in Receptor Tyrosine Kinases. Nat. Rev. Mol. Cell Biol. 2004, 5, 464–471. [Google Scholar] [CrossRef]
- Gotoh, N.; Tojo, A.; Hino, M.; Yazaki, Y.; Shibuya, M. A Highly Conserved Tyrosine Residue at Codon 845 within the Kinase Domain Is Not Required for the Transforming Activity of Human Epidermal Growth Factor Receptor. Biochem. Biophys. Res. Commun. 1992, 186, 768–774. [Google Scholar] [CrossRef]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef] [Green Version]
- Wood, E.R.; Truesdale, A.T.; McDonald, O.B.; Yuan, D.; Hassell, A.; Dickerson, S.H.; Ellis, B.; Pennisi, C.; Horne, E.; Lackey, K.; et al. A Unique Structure for Epidermal Growth Factor Receptor Bound to GW572016 (Lapatinib). Cancer Res. 2004, 64, 6652–6659. [Google Scholar] [CrossRef] [Green Version]
- Macdonald-Obermann, J.L.; Piwnica-Worms, D.; Pike, L.J. Mechanics of EGF Receptor/ErbB2 Kinase Activation Revealed by Luciferase Fragment Complementation Imaging. Proc. Natl. Acad. Sci. USA 2012, 109, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Ward, M.D.; Leahy, D.J. Kinase Activator-Receiver Preference in ErbB Heterodimers Is Determined by Intracellular Regions and Is Not Coupled to Extracellular Asymmetry. J. Biol. Chem. 2015, 290, 1570–1579. [Google Scholar] [CrossRef] [Green Version]
- Thiel, K.W.; Carpenter, G. Epidermal Growth Factor Receptor Juxtamembrane Region Regulates Allosteric Tyrosine Kinase Activation. Proc. Natl. Acad. Sci. USA 2007, 104, 19238–19243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.; LeVea, C.M.; Freeman, J.K.; Dougall, W.C.; Greene, M.I. Heterodimerization of Epidermal Growth Factor Receptor and Wild-Type or Kinase-Deficient Neu: A Mechanism of Interreceptor Kinase Activation and Transphosphorylation. Proc. Natl. Acad. Sci. USA 1994, 91, 1500–1504. [Google Scholar] [CrossRef] [Green Version]
- Tanos, B.; Pendergast, A.M. Abl Tyrosine Kinase Regulates Endocytosis of the Epidermal Growth Factor Receptor. J. Biol. Chem. 2006, 281, 32714–32723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biscardi, J.S.; Maa, M.C.; Tice, D.A.; Cox, M.E.; Leu, T.H.; Parsons, S.J. C-Src-Mediated Phosphorylation of the Epidermal Growth Factor Receptor on Tyr845 and Tyr1101 Is Associated with Modulation of Receptor Function. J. Biol. Chem. 1999, 274, 8335–8343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stover, D.R.; Becker, M.; Liebetanz, J.; Lydon, N.B. Src Phosphorylation of the Epidermal Growth Factor Receptor at Novel Sites Mediates Receptor Interaction with Src and P85α(*). J. Biol. Chem. 1995, 270, 15591–15597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertics, P.J.; Weber, W.; Cochet, C.; Gill, G.N. Regulation of the Epidermal Growth Factor Receptor by Phosphorylation. J. Cell. Biochem. 1985, 29, 195–208. [Google Scholar] [CrossRef]
- Margolis, B.L.; Lax, I.; Kris, R.; Dombalagian, M.; Honegger, A.M.; Howk, R.; Givol, D.; Ullrich, A.; Schlessinger, J. All Autophosphorylation Sites of Epidermal Growth Factor (EGF) Receptor and HER2/Neu Are Located in Their Carboxyl-Terminal Tails. Identification of a Novel Site in EGF Receptor. J. Biol. Chem. 1989, 264, 10667–10671. [Google Scholar] [CrossRef]
- Walton, G.M.; Chen, W.S.; Rosenfeld, M.G.; Gill, G.N. Analysis of Deletions of the Carboxyl Terminus of the Epidermal Growth Factor Receptor Reveals Self-Phosphorylation at Tyrosine 992 and Enhanced in Vivo Tyrosine Phosphorylation of Cell Substrates. J. Boil. Chem. 1990, 265, 1750–1754. [Google Scholar] [CrossRef]
- Koland, J.G. Coarse-Grained Molecular Simulation of Epidermal Growth Factor Receptor Protein Tyrosine Kinase Multi-Site Self-Phosphorylation. PLoS Comput. Biol. 2014, 10, e1003435. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.L.; Kim, D.; Lee, S.; Kim, S.J.; Noh, J.E.; Kim, J.H.; Chae, Y.C.; Lee, J.B.; Ryu, S.H. Pairwise Detection of Site-Specific Receptor Phosphorylations Using Single-Molecule Blotting. Nat. Commun. 2016, 7, 11107. [Google Scholar] [CrossRef] [Green Version]
- Hazan, R.; Margolis, B.; Dombalagian, M.; Ullrich, A.; Zilberstein, A.; Schlessinger, J. Identification of Autophosphorylation Sites of HER2/Neu. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1990, 1, 3–7. [Google Scholar]
- Segatto, O.; Lonardo, F.; Pierce, J.H.; Bottaro, D.P.; Di Fiore, P.P. The Role of Autophosphorylation in Modulation of erbB-2 Transforming Function. New Biol. 1990, 2, 187–195. [Google Scholar]
- Olayioye, M.A.; Graus-Porta, D.; Beerli, R.R.; Rohrer, J.; Gay, B.; Hynes, N.E. ErbB-1 and ErbB-2 Acquire Distinct Signaling Properties Dependent upon Their Dimerization Partner. Mol. Cell Biol. 1998, 18, 5042–5051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, W.X.; Deng, L.; Mann, M. Phosphotyrosine Interactome of the ErbB-Receptor Kinase Family. Mol. Syst. Biol. 2005, 1, E1–E13. [Google Scholar] [CrossRef] [Green Version]
- Qiu, C.; Lienhard, S.; Hynes, N.E.; Badache, A.; Leahy, D.J. Memo Is Homologous to Nonheme Iron Dioxygenases and Binds an ErbB2-Derived Phosphopeptide in Its Vestigial Active Site. J. Biol. Chem. 2008, 283, 2734–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feracci, M.; Pimentel, C.; Bornet, O.; Roche, P.; Salaun, D.; Badache, A.; Guerlesquin, F. MEMO Associated with an ErbB2 Receptor Phosphopeptide Reveals a New Phosphotyrosine Motif. FEBS Lett. 2011, 585, 2688–2692. [Google Scholar] [CrossRef] [PubMed]
- Zaoui, K.; Honoré, S.; Isnardon, D.; Braguer, D.; Badache, A. Memo-RhoA-mDia1 Signaling Controls Microtubules, the Actin Network, and Adhesion Site Formation in Migrating Cells. J. Cell Biol. 2008, 183, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Rahuel, J.; Gay, B.; Erdmann, D.; Strauss, A.; Garcia-Echeverria, C.; Furet, P.; Caravatti, G.; Fretz, H.; Schoepfer, J.; Grutter, M.G. Structural Basis for Specificity of GRB2-SH2 Revealed by a Novel Ligand Binding Mode. Nat. Struct. Biol. 1996, 3, 586–589. [Google Scholar] [CrossRef]
- Papaioannou, D.; Geibel, S.; Kunze, M.B.; Kay, C.W.; Waksman, G. Structural and Biophysical Investigation of the Interaction of a Mutant Grb2 SH2 Domain (W121G) with Its Cognate Phosphopeptide. Protein Sci. 2016, 25, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Van Der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-Translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef] [Green Version]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an Intrinsically Disordered Protein by Phosphorylation as a Regulatory Switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Bozoky, Z.; Krzeminski, M.; Muhandiram, R.; Birtley, J.R.; Al-Zahrani, A.; Thomas, P.J.; Frizzell, R.A.; Ford, R.C.; Forman-Kay, J.D.; Wright, P.E. Regulatory R Region of the CFTR Chloride Channel Is a Dynamic Integrator of Phospho-Dependent Intra- and Intermolecular Interactions. Proc. Natl. Acad. Sci. USA 2013, 110, E4427–E4436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Dyson, H.J.; Wright, P.E. A Phosphorylation-Dependent Switch in the Disordered P53 Transactivation Domain Regulates DNA Binding. Proc. Natl. Acad. Sci. USA 2021, 118, e2021456118. [Google Scholar] [CrossRef]
- Kathiriya, J.J.; Pathak, R.R.; Clayman, E.; Xue, B.; Uversky, V.N.; Davé, V. Presence and Utility of Intrinsically Disordered Regions in Kinases. Mol. Biosyst. 2014, 10, 2876–2888. [Google Scholar] [CrossRef]
- Niggenaber, J.; Hardick, J.; Lategahn, J.; Rauh, D. Structure Defines Function: Clinically Relevant Mutations in ErbB Kinases. J. Med. Chem. 2020, 63, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Pines, G.; Huang, P.H.; Zwang, Y.; White, F.M.; Yarden, Y. EGFRvIV: A Previously Uncharacterized Oncogenic Mutant Reveals a Kinase Autoinhibitory Mechanism. Oncogene 2010, 29, 5850–5860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [Green Version]
- Mészáros, B.; Erdös, G.; Dosztányi, Z. IUPred2A: Context-Dependent Prediction of Protein Disorder as a Function of Redox State and Protein Binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Uversky, V.N. The Alphabet of Intrinsic Disorder II. Various Roles of Glutamic Acid in Ordered and Intrinsically Disordered Proteins. Intrinsically Disord. Proteins 2013, 1, e24684. [Google Scholar] [CrossRef] [Green Version]
- Hunter, T.; Sefton, B.M. Transforming Gene Product of Rous Sarcoma Virus Phosphorylates Tyrosine. Proc. Natl. Acad. Sci. USA 1980, 77, 1311–1315. [Google Scholar] [CrossRef] [Green Version]
- Hunter, T. Tyrosine Phosphorylation: Thirty Years and Counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, D.R.; Wu, Y.M.; Lin, S.F. The Protein Tyrosine Kinase Family of the Human Genome. Oncogene 2000, 19, 5548–5557. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Liu, Y.; Shaw, S. Protein Kinase Specificity: A Strategic Collaboration between Kinase Peptide Specificity and Substrate Recruitment. Cell Cycle 2005, 4, 52–56. [Google Scholar] [CrossRef] [Green Version]
- Dankort, D.L.; Wang, Z.; Blackmore, V.; Moran, M.F.; Muller, W.J. Distinct Tyrosine Autophosphorylation Sites Negatively and Positively Modulate Neu-Mediated Transformation. Mol. Cell. Boil. 1997, 17, 5410–5425. [Google Scholar] [CrossRef] [Green Version]
- Dankort, D.; Jeyabalan, N.; Jones, N.; Dumont, D.J.; Muller, W.J. Multiple ErbB-2/Neu Phosphorylation Sites Mediate Transformation through Distinct Effector Proteins. J. Biol. Chem. 2001, 276, 38921–38928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dankort, D.; Maslikowski, B.; Warner, N.; Kanno, N.; Kim, H.; Wang, Z.; Moran, M.F.; Oshima, R.G.; Cardiff, R.D.; Muller, W.J. Grb2 and Shc Adapter Proteins Play Distinct Roles in Neu ( ErbB-2)–Induced Mammary Tumorigenesis: Implications for Human Breast Cancer. Mol. Cell. Boil. 2001, 21, 1540–1551. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, N.; Tojo, A.; Muroya, K.; Hashimoto, Y.; Hattori, S.; Nakamura, S.; Takenawa, T.; Yazaki, Y.; Shibuya, M. Epidermal Growth Factor-Receptor Mutant Lacking the Autophosphorylation Sites Induces Phosphorylation of Shc Protein and Shc-Grb2/ASH Association and Retains Mitogenic Activity. Proc. Natl. Acad. Sci. USA 1994, 91, 167–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorkin, A.; Helin, K.; Waters, C.M.; Carpenter, G.; Beguinot, L. Multiple Autophosphorylation Sites of the Epidermal Growth Factor Receptor Are Essential for Receptor Kinase Activity and Internalization. Contrasting Significance of Tyrosine 992 in the Native and Truncated Receptors. J. Boil. Chem. 1992, 267, 8672–8678. [Google Scholar] [CrossRef]
- Alvarez, C.V.; Shon, K.J.; Miloso, M.; Beguinot, L. Structural Requirements of the Epidermal Growth Factor Receptor for Tyrosine Phosphorylation of Eps8 and Eps15, Substrates Lacking Src SH2 Homology Domains. J. Biol. Chem. 1995, 270, 16271–16276. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Koland, J.G. Nucleotide Binding by the Epidermal Growth Factor Receptor Protein-Tyrosine Kinase. J. Biol. Chem. 1996, 271, 311–318. [Google Scholar] [CrossRef] [Green Version]
- Gill, K.; Macdonald-Obermann, J.L.; Pike, L.J. Epidermal Growth Factor Receptors Containing a Single Tyrosine in Their C-Terminal Tail Bind Different Effector Molecules and Are Signaling-Competent. J. Biol. Chem. 2017, 292, 20744–20755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustafa, M.; Mirza, A.; Kannan, N. Conformational Regulation of the EGFR Kinase Core by the Juxtamembrane and C-Terminal Tail: A Molecular Dynamics Study. ProteinsStruct. Funct. Bioinform. 2011, 79, 99–114. [Google Scholar] [CrossRef]
- Bublil, E.M.; Pines, G.; Patel, G.; Fruhwirth, G.; Ng, T.; Yarden, Y. Kinase-Mediated Quasi-Dimers of EGFR. FASEB J. 2010, 24, 4744–4755. [Google Scholar] [CrossRef]
- Lerdrup, M.; Bruun, S.; Grandal, M.V.; Roepstorff, K.; Kristensen, M.M.; Hommelgaard, A.M.; van Deurs, B. Endocytic Down-Regulation of ErbB2 Is Stimulated by Cleavage of Its C-Terminus. Mol. Biol. Cell 2007, 18, 3656–3666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorkin, A.; Di Fiore, P.P.; Carpenter, G. The Carboxyl Terminus of Epidermal Growth Factor Receptor/erbB-2 Chimerae Is Internalization Impaired. Oncogene 1993, 8, 3021–3028. [Google Scholar] [PubMed]
- Baulida, J.; Kraus, M.H.; Alimandi, M.; Di Fiore, P.P.; Carpenter, G. All ErbB Receptors Other than the Epidermal Growth Factor Receptor Are Endocytosis Impaired. J. Biol. Chem. 1996, 271, 5251–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, M.; Malik, M.S.; Skeie, M.; Bertelsen, V.; Stang, E. Protein Kinase C Regulates ErbB3 Turnover. Exp. Cell Res. 2019, 382, 111473. [Google Scholar] [CrossRef]
- Sundvall, M.; Korhonen, A.; Paatero, I.; Gaudio, E.; Melino, G.; Croce, C.M.; Aqeilan, R.I.; Elenius, K. Isoform-Specific Monoubiquitination, Endocytosis, and Degradation of Alternatively Spliced ErbB4 Isoforms. Proc. Natl. Acad. Sci. USA 2008, 105, 4162–4167. [Google Scholar] [CrossRef] [Green Version]
- Bertelsen, V.; Stang, E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes 2014, 4, 424–446. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, M.; Malik, M.S.; Nikolaysen, F.; Skeie, M.; Stang, E. Protein Kinase C Mediated Internalization of ErbB2 Is Independent of Clathrin, Ubiquitination and Hsp90 Dissociation. Exp. Cell Res. 2018, 371, 139–150. [Google Scholar] [CrossRef]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The Eukaryotic Linear Motif Resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [Green Version]
- Sorkin, A.; Mazzotti, M.; Sorkina, T.; Scotto, L.; Beguinot, L. Epidermal Growth Factor Receptor Interaction with Clathrin Adaptors Is Mediated by the Tyr974-Containing Internalization Motif*. J. Biol. Chem. 1996, 271, 13377–13384. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Khvorova, A.; Marshall, W.; Sorkin, A. Analysis of Clathrin-Mediated Endocytosis of Epidermal Growth Factor Receptor by RNA Interference*[Boxs]. J. Biol. Chem. 2004, 279, 16657–16661. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Shen, C.; Luo, S.; Traoré, W.; Marchetto, S.; Santoni, M.J.; Xu, L.; Wu, B.; Shi, C.; Mei, J.; et al. Role of Erbin in ErbB2-Dependent Breast Tumor Growth. Proc. Natl. Acad. Sci. USA 2014, 111, E4429–E4438. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Taylor, P.; Moran, M.F. Proteomic Analysis of the Epidermal Growth Factor Receptor (EGFR) Interactome and Post-Translational Modifications Associated with Receptor Endocytosis in Response to EGF and Stress. Mol. Cell Proteom. 2014, 13, 1644–1658. [Google Scholar] [CrossRef] [Green Version]
- Tong, J.; Taylor, P.; Peterman, S.M.; Prakash, A.; Moran, M.F. Epidermal Growth Factor Receptor Phosphorylation Sites Ser991 and Tyr998 Are Implicated in the Regulation of Receptor Endocytosis and Phosphorylations at Ser1039 and Thr1041. Mol. Cell Proteom. 2009, 8, 2131–2144. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Huang, F.; Marusyk, A.; Sorkin, A. Grb2 Regulates Internalization of EGF Receptors through Clathrin-Coated Pits. MBoC 2003, 14, 858–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699. [Google Scholar] [CrossRef] [Green Version]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The Disordered P Granule Protein LAF-1 Drives Phase Separation into Droplets with Tunable Viscosity and Dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, L.B.; Ditlev, J.A.; Rosen, M.K. Regulation of Transmembrane Signaling by Phase Separation. Annu. Rev. Biophys. 2019, 48, 465–494. [Google Scholar] [CrossRef]
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase Separation of Signaling Molecules Promotes T Cell Receptor Signal Transduction. Science 2016, 352, 595–599. [Google Scholar] [CrossRef] [Green Version]
- Chemes, L.B.; Alonso, L.G.; Noval, M.G.; de Prat-Gay, G. Circular Dichroism Techniques for the Analysis of Intrinsically Disordered Proteins and Domains. In Intrinsically Disordered Protein Analysis: Volume 1, Methods and Experimental Tools; Uversky, V.N., Dunker, A.K., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; pp. 387–404. [Google Scholar] [CrossRef]
- Narwani, T.J.; Santuz, H.; Shinada, N.; Melarkode Vattekatte, A.; Ghouzam, Y.; Srinivasan, N.; Gelly, J.C.; de Brevern, A.G. Recent Advances on Polyproline II. Amino Acids 2017, 49, 705–713. [Google Scholar] [CrossRef] [Green Version]
- Mansiaux, Y.; Joseph, A.P.; Gelly, J.C.; de Brevern, A.G. Assignment of Polyproline Ii Conformation and Analysis of Sequence-Structure Relationship. PLoS ONE 2011, 6, e18401. [Google Scholar] [CrossRef]
- Saksela, K.; Permi, P. SH3 Domain Ligand Binding: What’s the Consensus and Where’s the Specificity? FEBS Lett. 2012, 586, 2609–2614. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Pendergast, A.M.; Hung, M.C. Dominant-Negative Mutants of Grb2 Induced Reversal of the Transformed Phenotypes Caused by the Point Mutation-Activated Rat HER-2/Neu. J. Boil. Chem. 1995, 270, 30717–30724. [Google Scholar] [CrossRef] [Green Version]
- Bornet, O.; Nouailler, M.; Feracci, M.; Sebban-Kreuzer, C.; Byrne, D.; Halimi, H.; Morelli, X.; Badache, A.; Guerlesquin, F. Identification of a Src Kinase SH3 Binding Site in the C-Terminal Domain of the Human ErbB2 Receptor Tyrosine Kinase. FEBS Lett. 2014, 588, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, A.; Beguinot, L.; Bishayee, S. Phosphorylation of Tyrosine 992, 1068, and 1086 Is Required for Conformational Change of the Human Epidermal Growth Factor Receptor C-Terminal Tail. Mol. Biol. Cell 1999, 10, 525–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett, T.P.J.; McKern, N.M.; Lou, M.; Elleman, T.C.; Adams, T.E.; Lovrecz, G.O.; Zhu, H.J.; Walker, F.; Frenkel, M.J.; Hoyne, P.A.; et al. Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor α. Cell 2002, 110, 763–773. [Google Scholar] [CrossRef] [Green Version]
- Wendler, F.; Purice, T.M.; Simon, T.; Stebbing, J.; Giamas, G. The LMTK-Family of Kinases: Emerging Important Players in Cell Physiology and Pathogenesis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2021, 1867, 165372. [Google Scholar] [CrossRef]
- Niu, X.L.; Peters, K.G.; Kontos, C.D. Deletion of the Carboxyl Terminus of Tie2 Enhances Kinase Activity, Signaling, and Function: Evidence for an Autoinhibitory Mechanism. J. Biol. Chem. 2002, 277, 31768–31773. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Z.; George, R.; Lin, C.C.; Suen, K.M.; Levitt, J.A.; Suhling, K.; Ladbury, J.E. Direct Binding of Grb2 SH3 Domain to FGFR2 Regulates SHP2 Function. Cell. Signal. 2010, 22, 23–33. [Google Scholar] [CrossRef]
- Lin, C.C.C.C.; Melo, F.A.; Ghosh, R.; Suen, K.M.; Stagg, L.J.; Kirkpatrick, J.; Arold, S.T.; Ahmed, Z.; Ladbury, J.E. Inhibition of Basal FGF Receptor Signaling by Dimeric Grb2. Cell 2012, 149, 1514–1524. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Z.; Lin, C.C.; Suen, K.M.; Melo, F.A.; Levitt, J.A.; Suhling, K.; Ladbury, J.E. Grb2 Controls Phosphorylation of FGFR2 by Inhibiting Receptor Kinase and Shp2 Phosphatase Activity. J. Cell Biol. 2013, 200, 493–504. [Google Scholar] [CrossRef] [Green Version]
- Timsah, Z.; Ahmed, Z.; Lin, C.C.; Melo, F.A.; Stagg, L.J.; Leonard, P.G.; Jeyabal, P.; Berrout, J.; O’Neil, R.G.; Bogdanov, M.; et al. Competition between Grb2 and Plcγ1 for FGFR2 Regulates Basal Phospholipase Activity and Invasion. Nat. Struct. Mol. Biol. 2014, 21, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Wieteska, L.; Poncet-Montange, G.; Suen, K.M.; Arold, S.T.; Ahmed, Z.; Ladbury, J.E. Regulation of Kinase Activity by Combined Action of Juxtamembrane and C-Terminal Regions of Receptors. BioRxiv 2020. [Google Scholar] [CrossRef]
- Warnet, X.L.; Bakke Krog, H.; Sevillano-Quispe, O.G.; Poulsen, H.; Kjaergaard, M. The C-Terminal Domains of the NMDA Receptor: How Intrinsically Disordered Tails Affect Signalling, Plasticity and Disease. Eur. J. Neurosci. 2021, 54, 6713–6739. [Google Scholar] [CrossRef]
- Seiffert, P.; Bugge, K.; Nygaard, M.; Haxholm, G.W.; Martinsen, J.H.; Pedersen, M.N.; Arleth, L.; Boomsma, W.; Kragelund, B.B. Orchestration of Signaling by Structural Disorder in Class 1 Cytokine Receptors. Cell Commun. Signal. 2020, 18, 132. [Google Scholar] [CrossRef] [PubMed]
- Clemens, L.; Dushek, O.; Allard, J. Intrinsic Disorder in the T Cell Receptor Creates Cooperativity and Controls ZAP70 Binding. Biophys. J. 2021, 120, 379–392. [Google Scholar] [CrossRef]



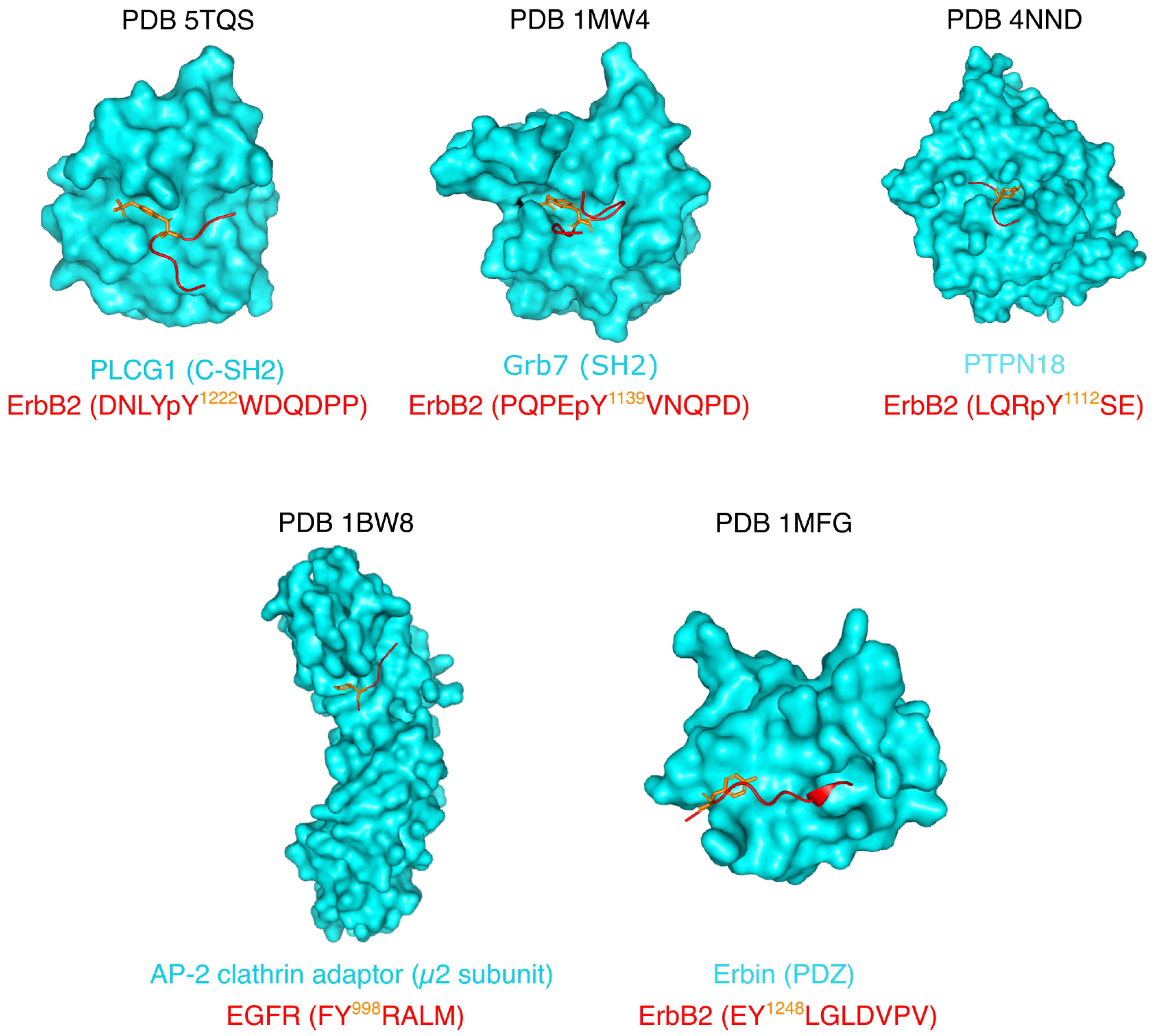
| CT-ErbB1 | CT-ErbB2 | CT-ErbB3 | CT-ErbB4 | Average in Disprot [100] | Average in SwissProt [100] | |
|---|---|---|---|---|---|---|
| Length (residues) | 231 | 268 | 376 | 323 | ND | ND |
| Average IUPred score | 0.58 | 0.75 | 0.72 | 0.59 | ND | ND |
| Number [%] of Tyr | 9 [3.90] | 9 [3.36] | 14 [3.72] | 19 [5.88] | [2.13] | [3.03] |
| Number [%] of Gly | 10 [4.3] | 28 [10.4] | 34 [9.0] | 15 [4.6] | [7.4] | [7.0] |
| Number [%] of Pro | 24 [10.4] | 44 [16.4] | 39 [10.4] | 39 [12.1] | [8.1] | [4.8] |
| Number [%] of Asp | 20 [8.7] | 21 [7.8] | 19 [5.1] | 21 [6.5] | [5.8] | [5.4] |
| Number [%] of Glu | 12 [5.2] | 19 [7.1] | 38 [10.1] | 27 [8.4] | [9.9] | [6.7] |
| Number [%] of Arg | 8 [3.5] | 11 [4.1] | 29 [7.7] | 18 [5.6] | [4.8] | [5.4] |
| Number [%] of Lys | 6 [2.60] | 5 [1.87] | 7 [1.86] | 15 [4.64] | [7.85] | [5.92] |
| Total net charge | −18 | −24 | −21 | −15 | ND | ND |
| pI | 4.43 | 4.29 | 5.05 | 4.91 | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinet, L.; Assrir, N.; van Heijenoort, C. Expanding the Disorder-Function Paradigm in the C-Terminal Tails of Erbbs. Biomolecules 2021, 11, 1690. https://doi.org/10.3390/biom11111690
Pinet L, Assrir N, van Heijenoort C. Expanding the Disorder-Function Paradigm in the C-Terminal Tails of Erbbs. Biomolecules. 2021; 11(11):1690. https://doi.org/10.3390/biom11111690
Chicago/Turabian StylePinet, Louise, Nadine Assrir, and Carine van Heijenoort. 2021. "Expanding the Disorder-Function Paradigm in the C-Terminal Tails of Erbbs" Biomolecules 11, no. 11: 1690. https://doi.org/10.3390/biom11111690






