Vascular Tissue Specific miRNA Profiles Reveal Novel Correlations with Risk Factors in Coronary Artery Disease
Abstract
:1. Introduction
2. Materials and Methods
3. Results
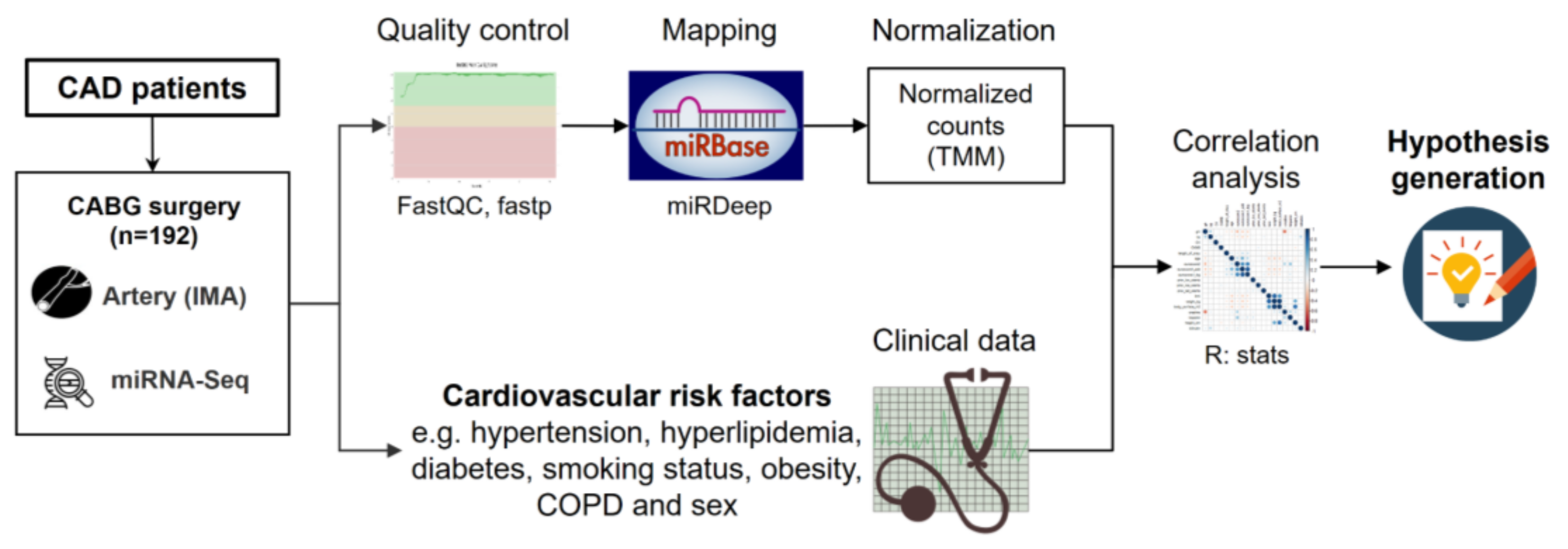
3.1. Study Workflow
3.2. Study Cohort
3.3. miRNAs in IMA Samples—A Novel Atlas
3.4. miRNAs and Cardiovascular Risk Factors
3.5. Clinical Relevance and Potential Confounders
4. Discussion
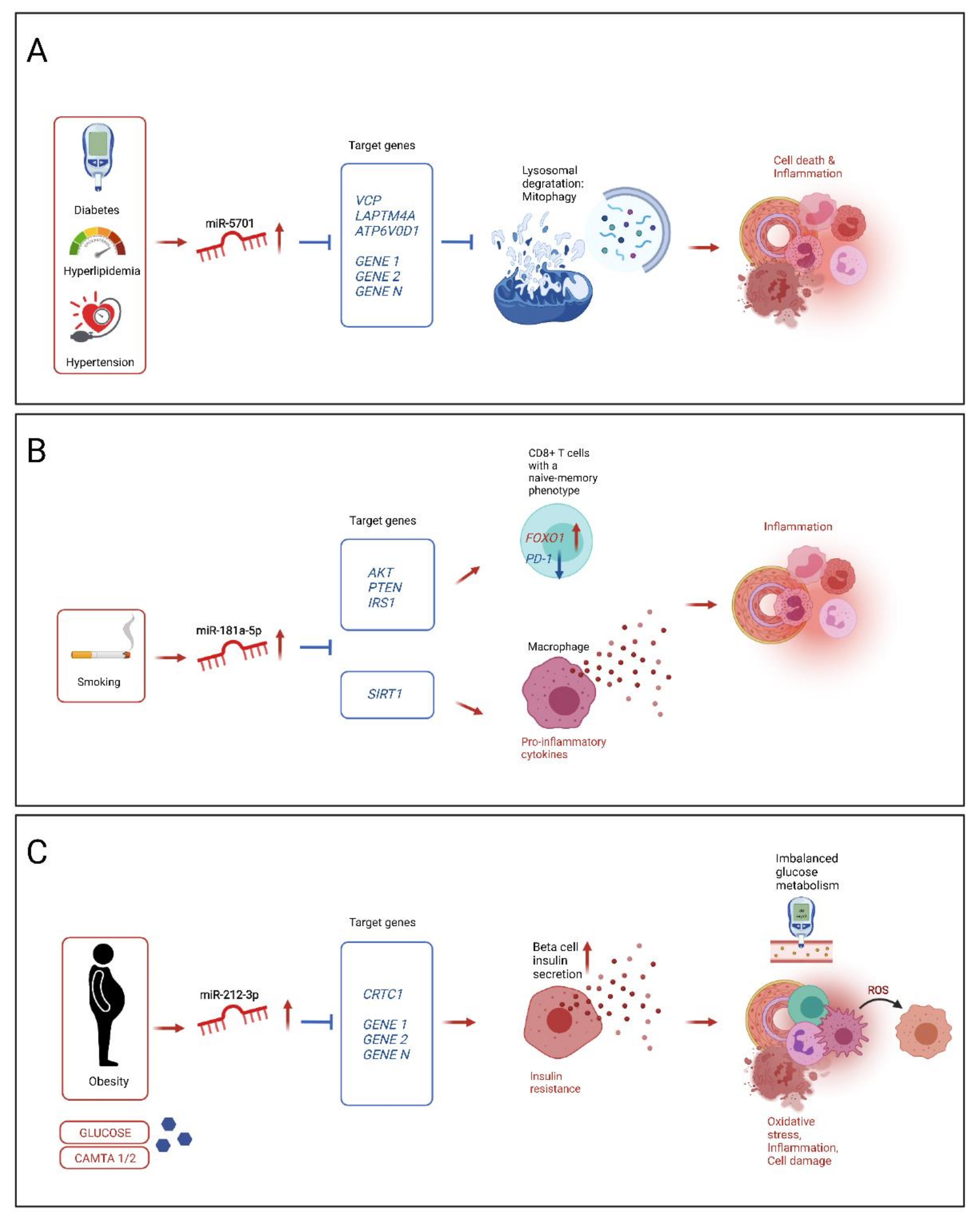
4.1. miRNA Found to Play a Role in CAD/MI, Associations with Risk Factors
4.2. Potential Mechanisms of Selected Novel Mirnas Influencing Atherosclerosis Progression on Response to Risk Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez, A.D.; Mathers, C.D.; Ezzati, M.; Jamison, D.T.; Murray, C.J. Global and regional burden of disease and risk factors, 2001: Systematic analysis of population health data. Lancet 2006, 367, 1747–1757. [Google Scholar] [CrossRef]
- Collaborators, G.B.D.R.F. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef]
- Davey Smith, G.; Ebrahim, S.; Lewis, S.; Hansell, A.L.; Palmer, L.J.; Burton, P.R. Genetic epidemiology and public health: Hope, hype, and future prospects. Lancet 2005, 366, 1484–1498. [Google Scholar] [CrossRef]
- Erdmann, J.; Kessler, T.; Munoz Venegas, L.; Schunkert, H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc. Res. 2018, 114, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Vilne, B.; Schunkert, H. Integrating Genes Affecting Coronary Artery Disease in Functional Networks by Multi-OMICs Approach. Front. Cardiovasc. Med. 2018, 5, 89. [Google Scholar] [CrossRef]
- Samani, N.J.; Erdmann, J.; Hall, A.S.; Hengstenberg, C.; Mangino, M.; Mayer, B.; Dixon, R.J.; Meitinger, T.; Braund, P.; Wichmann, H.E.; et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007, 357, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Erdmann, J.; Grosshennig, A.; Braund, P.S.; Konig, I.R.; Hengstenberg, C.; Hall, A.S.; Linsel-Nitschke, P.; Kathiresan, S.; Wright, B.; Tregouet, D.A.; et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 2009, 41, 280–282. [Google Scholar] [CrossRef]
- Tregouet, D.A.; Konig, I.R.; Erdmann, J.; Munteanu, A.; Braund, P.S.; Hall, A.S.; Grosshennig, A.; Linsel-Nitschke, P.; Perret, C.; DeSuremain, M.; et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat. Genet. 2009, 41, 283–285. [Google Scholar] [CrossRef]
- Schunkert, H.; Konig, I.R.; Kathiresan, S.; Reilly, M.P.; Assimes, T.L.; Holm, H.; Preuss, M.; Stewart, A.F.; Barbalic, M.; Gieger, C.; et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011, 43, 333–338. [Google Scholar] [CrossRef]
- The CARDIoGRAMplusC4D Consortium; Deloukas, P.; Kanoni, S.; Willenborg, C.; Farrall, M.; Assimes, T.L.; Thompson, J.R.; Ingelsson, E.; Saleheen, D.; Erdmann, J.; et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013, 45, 25–33. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; Konig, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated With Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.P.; Goel, A.; Butterworth, A.S.; Kanoni, S.; Webb, T.R.; Marouli, E.; Zeng, L.; Ntalla, I.; Lai, F.Y.; Hopewell, J.C.; et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017, 49, 1385–1391. [Google Scholar] [CrossRef]
- Howson, J.M.M.; Zhao, W.; Barnes, D.R.; Ho, W.K.; Young, R.; Paul, D.S.; Waite, L.L.; Freitag, D.F.; Fauman, E.B.; Salfati, E.L.; et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017, 49, 1113–1119. [Google Scholar] [CrossRef] [Green Version]
- van der Harst, P.; Verweij, N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Aragam, K.G.; Jiang, T.; Goel, A.; Kanoni, S.; Wolford, B.N.; Weeks, E.M.; Wang, M.; Hindy, G.; Zhou, W.; Grace, C.; et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. medRxiv 2021. [Google Scholar] [CrossRef]
- von Scheidt, M.; Zhao, Y.; de Aguiar Vallim, T.Q.; Che, N.; Wierer, M.; Seldin, M.M.; Franzen, O.; Kurt, Z.; Pang, S.; Bongiovanni, D.; et al. Transcription Factor MAFF (MAF Basic Leucine Zipper Transcription Factor F) Regulates an Atherosclerosis Relevant Network Connecting Inflammation and Cholesterol Metabolism. Circulation 2021, 143, 1809–1823. [Google Scholar] [CrossRef]
- Braenne, I.; Civelek, M.; Vilne, B.; Di Narzo, A.; Johnson, A.D.; Zhao, Y.; Reiz, B.; Codoni, V.; Webb, T.R.; Foroughi Asl, H.; et al. Prediction of Causal Candidate Genes in Coronary Artery Disease Loci. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2207–2217. [Google Scholar] [CrossRef] [Green Version]
- Kessler, T.; Vilne, B.; Schunkert, H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol. Med. 2016, 8, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, J.; Freudenberg, J.M.; Meng, Q.; Rajpal, D.K.; Yang, X. Network-Based Identification and Prioritization of Key Regulators of Coronary Artery Disease Loci. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 928–941. [Google Scholar] [CrossRef] [Green Version]
- Vilne, B.; Skogsberg, J.; Foroughi Asl, H.; Talukdar, H.A.; Kessler, T.; Bjorkegren, J.L.M.; Schunkert, H. Network analysis reveals a causal role of mitochondrial gene activity in atherosclerotic lesion formation. Atherosclerosis 2017, 267, 39–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schunkert, H.; von Scheidt, M.; Kessler, T.; Stiller, B.; Zeng, L.; Vilne, B. Genetics of coronary artery disease in the light of genome-wide association studies. Clin. Res. Cardiol. 2018, 107, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Lempiainen, H.; Braenne, I.; Michoel, T.; Tragante, V.; Vilne, B.; Webb, T.R.; Kyriakou, T.; Eichner, J.; Zeng, L.; Willenborg, C.; et al. Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci. Rep. 2018, 8, 3434. [Google Scholar] [CrossRef] [Green Version]
- Kessler, T.; Zhang, L.; Liu, Z.; Yin, X.; Huang, Y.; Wang, Y.; Fu, Y.; Mayr, M.; Ge, Q.; Xu, Q.; et al. ADAMTS-7 inhibits re-endothelialization of injured arteries and promotes vascular remodeling through cleavage of thrombospondin-1. Circulation 2015, 131, 1191–1201. [Google Scholar] [CrossRef] [Green Version]
- Kessler, T.; Wobst, J.; Wolf, B.; Eckhold, J.; Vilne, B.; Hollstein, R.; von Ameln, S.; Dang, T.A.; Sager, H.B.; Moritz Rumpf, P.; et al. Functional Characterization of the GUCY1A3 Coronary Artery Disease Risk Locus. Circulation 2017, 136, 476–489. [Google Scholar] [CrossRef]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic variants in microRNA genes: Impact on microRNA expression, function, and disease. Front. Genet. 2015, 6, 186. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, X.Q.; Ge, P.C.; Liu, Z.; Jia, H.; Chen, X.; An, F.H.; Li, L.H.; Chen, Z.H.; Mao, H.W.; Li, Z.Y.; et al. Interaction between microRNA expression and classical risk factors in the risk of coronary heart disease. Sci. Rep. 2015, 5, 14925. [Google Scholar] [CrossRef] [Green Version]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Cordes, K.R.; Srivastava, D. MicroRNA regulation of cardiovascular development. Circ. Res. 2009, 104, 724–732. [Google Scholar] [CrossRef]
- Kessler, T.; Erdmann, J.; Vilne, B.; Bruse, P.; Kurowski, V.; Diemert, P.; Schunkert, H.; Sager, H.B. Serum microRNA-1233 is a specific biomarker for diagnosing acute pulmonary embolism. J. Transl. Med. 2016, 14, 120. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zhang, M.; Sanagawa, A.; Mori, C.; Ito, S.; Iwaki, S.; Satoh, H.; Fujii, S. Circulating microRNA-126 in patients with coronary artery disease: Correlation with LDL cholesterol. Thromb. J. 2012, 10, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, M.A.; Fareed, M.T.; Argenio, S.L.; Agunwamba, A.O.; Hanson, T.R. Coronary artery disease. Prim. Care 2013, 40, 1–16. [Google Scholar] [CrossRef]
- Nabel, E.G.; Braunwald, E. A tale of coronary artery disease and myocardial infarction. N. Engl. J. Med. 2012, 366, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Libby, P. Inflammation in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2045–2051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libby, P.; Theroux, P. Pathophysiology of coronary artery disease. Circulation 2005, 111, 3481–3488. [Google Scholar] [CrossRef] [Green Version]
- Diseases, G.B.D.; Injuries, C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Sanchis-Gomar, F.; Perez-Quilis, C.; Leischik, R.; Lucia, A. Epidemiology of coronary heart disease and acute coronary syndrome. Ann. Transl. Med. 2016, 4, 256. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.S.; Lai, E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, K.A.; Xiang, J. Mechanisms of cellular communication through intercellular protein transfer. J. Cell Mol. Med. 2011, 15, 1458–1473. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, S.; Ding, J.; Zhao, Y.; Liang, L.; Liu, T.; Zhan, R.; He, X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3’ untranslated region. Oncogene 2010, 29, 2302–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visseren, F.L.J.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Back, M.; Benetos, A.; Biffi, A.; Boavida, J.M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 September 2021).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J.R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Caputo, M.; Saif, J.; Rajakaruna, C.; Brooks, M.; Angelini, G.D.; Emanueli, C. MicroRNAs in vascular tissue engineering and post-ischemic neovascularization. Adv. Drug. Deliv. Rev. 2015, 88, 78–91. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Li, Z.; Shen, D.; Zhu, D.; Huang, K.; Su, T.; Dinh, P.U.; Cores, J.; Cheng, K. Publisher Correction: Exosome-eluting stents for vascular healing after ischaemic injury. Nat. Biomed. Eng. 2021, 1–15. [Google Scholar] [CrossRef]
- Polyakova, E.A.; Zaraiskii, M.I.; Mikhaylov, E.N.; Baranova, E.I.; Galagudza, M.M.; Shlyakhto, E.V. Association of myocardial and serum miRNA expression patterns with the presence and extent of coronary artery disease: A cross-sectional study. Int. J. Cardiol. 2021, 322, 9–15. [Google Scholar] [CrossRef]
- Siasos, G.; Bletsa, E.; Stampouloglou, P.K.; Oikonomou, E.; Tsigkou, V.; Paschou, S.A.; Vlasis, K.; Marinos, G.; Vavuranakis, M.; Stefanadis, C.; et al. MicroRNAs in cardiovascular disease. Hellenic J. Cardiol. 2020, 61, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Peters, L.J.F.; Biessen, E.A.L.; Hohl, M.; Weber, C.; van der Vorst, E.P.C.; Santovito, D. Small Things Matter: Relevance of MicroRNAs in Cardiovascular Disease. Front. Physiol. 2020, 11, 793. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Wu, L.J.; Li, J.J.; Xiao, H.B.; He, Y.; Yan, Y.X. A meta-analysis of dysregulated miRNAs in coronary heart disease. Life Sci. 2018, 215, 170–181. [Google Scholar] [CrossRef]
- Zhao, Y.; Ponnusamy, M.; Zhang, L.; Zhang, Y.; Liu, C.; Yu, W.; Wang, K.; Li, P. The role of miR-214 in cardiovascular diseases. Eur. J. Pharmacol. 2017, 816, 138–145. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, J.; Gong, Y.; Chen, M.; Chen, J.; Zhao, W.; Tan, S. Hsa-miR-140-5p down-regulates LDL receptor and attenuates LDL-C uptake in human hepatocytes. Atherosclerosis 2020, 297, 111–119. [Google Scholar] [CrossRef]
- Yan, Z.; Zang, B.; Gong, X.; Ren, J.; Wang, R. MiR-214-3p exacerbates kidney damages and inflammation induced by hyperlipidemic pancreatitis complicated with acute renal injury. Life Sci. 2020, 241, 117118. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.C.; Liu, P.Y.; Lin, H.F.; Lin, W.Y.; Liao, J.K.; Juo, S.H. Two functional polymorphisms of ROCK2 enhance arterial stiffening through inhibiting its activity and expression. J. Mol. Cell Cardiol. 2015, 79, 180–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; He, H.W.; Wang, Z.M.; Zhao, H.; Lian, X.Q.; Wang, Y.S.; Zhu, J.; Yan, J.J.; Zhang, D.G.; Yang, Z.J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzimato, V.; Chen, P.; Barreby, E.; Morgantini, C.; Levi, L.; Vankova, A.; Jager, J.; Sulen, A.; Diotallevi, M.; Shen, J.X.; et al. Hepatic miR-144 drives fumarase activity preventing NRF2 activation during obesity. Gastroenterology 2021. [Google Scholar] [CrossRef]
- Hanouskova, B.; Vavrova, G.; Ambroz, M.; Bousova, I.; Karlsen, T.A.; Skalova, L.; Matouskova, P. MicroRNAs mediated regulation of glutathione peroxidase 7 expression and its changes during adipogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194734. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, Z.; Guo, X.; Kong, X. The roles of microRNAs in the pathogenesis of chronic obstructive pulmonary disease. Int. Immunopharmacol. 2019, 67, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Lareyre, F.; Clement, M.; Moratal, C.; Loyer, X.; Jean-Baptiste, E.; Hassen-Khodja, R.; Chinetti, G.; Mallat, Z.; Raffort, J. Differential micro-RNA expression in diabetic patients with abdominal aortic aneurysm. Biochimie 2019, 162, 1–7. [Google Scholar] [CrossRef]
- Pofi, R.; Giannetta, E.; Galea, N.; Francone, M.; Campolo, F.; Barbagallo, F.; Gianfrilli, D.; Venneri, M.A.; Filardi, T.; Cristini, C.; et al. Diabetic Cardiomiopathy Progression is Triggered by miR122-5p and Involves Extracellular Matrix: A 5-Year Prospective Study. JACC Cardiovasc. Imaging 2021, 14, 1130–1142. [Google Scholar] [CrossRef]
- Burke, G.M.; Genuardi, M.; Shappell, H.; D’Agostino, R.B., Sr.; Magnani, J.W. Temporal Associations Between Smoking and Cardiovascular Disease, 1971 to 2006 (from the Framingham Heart Study). Am. J. Cardiol. 2017, 120, 1787–1791. [Google Scholar] [CrossRef]
- Kannel, W.B.; Dawber, T.R.; Kagan, A.; Revotskie, N.; Stokes, J., 3rd. Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann. Intern. Med. 1961, 55, 33–50. [Google Scholar] [CrossRef]
- Katta, N.; Loethen, T.; Lavie, C.J.; Alpert, M.A. Obesity and Coronary Heart Disease: Epidemiology, Pathology, and Coronary Artery Imaging. Curr. Probl. Cardiol. 2021, 46, 100655. [Google Scholar] [CrossRef]
- Onuma, O.K.; Pencina, K.; Qazi, S.; Massaro, J.M.; D’Agostino, R.B., Sr.; Chuang, M.L.; Fox, C.S.; Hoffmann, U.; O’Donnell, C.J. Relation of Risk Factors and Abdominal Aortic Calcium to Progression of Coronary Artery Calcium (from the Framingham Heart Study). Am. J. Cardiol. 2017, 119, 1584–1589. [Google Scholar] [CrossRef] [Green Version]
- Tsao, C.W.; Preis, S.R.; Peloso, G.M.; Hwang, S.J.; Kathiresan, S.; Fox, C.S.; Cupples, L.A.; Hoffmann, U.; O’Donnell, C.J. Relations of long-term and contemporary lipid levels and lipid genetic risk scores with coronary artery calcium in the framingham heart study. J. Am. Coll. Cardiol. 2012, 60, 2364–2371. [Google Scholar] [CrossRef]
- Wong, N.D.; Levy, D. Legacy of the framingham heart study: Rationale, design, initial findings, and implications. Glob. Heart 2013, 8, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Guddeti, R.R.; Matsuzawa, Y.; Liu, L.P.; Su, L.X.; Guo, D.; Nie, S.P.; Du, J.; Zhang, M. Plasma Levels of microRNA-145 Are Associated with Severity of Coronary Artery Disease. PLoS ONE 2015, 10, e0123477. [Google Scholar] [CrossRef] [Green Version]
- Jusic, A.; Devaux, Y.; Action, E.U.-C.C. Noncoding RNAs in Hypertension. Hypertension 2019, 74, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, P.; Tin, A.; Grove, M.L.; Ma, J.; Boerwinkle, E.; Coresh, J.; Chakravarti, A. MicroRNAs in the miR-17 and miR-15 families are downregulated in chronic kidney disease with hypertension. PLoS ONE 2017, 12, e0176734. [Google Scholar] [CrossRef] [Green Version]
- Sundar, I.K.; Li, D.; Rahman, I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J. Extracell Vesicles 2019, 8, 1684816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asulin, N.; Volinsky, N.; Grosman-Rimon, L.; Kachel, E.; Sternik, L.; Raanani, E.; Altshuler, R.; Magen, I.; Ben-Zvi, I.; Margalit, N.; et al. Differential microRNAs expression in calcified versus rheumatic aortic valve disease. J. Card. Surg 2020, 35, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Mo, L.; Lei, T.; Yan, Y.; Han, S.; Miao, J.; Gao, Y.; Wang, X.; Zhao, W.; Huang, C. miR-5701 promoted apoptosis of clear cell renal cell carcinoma cells by targeting phosphodiesterase-1B. Anticancer Drugs 2021, 32, 855–863. [Google Scholar] [CrossRef]
- Prajapati, P.; Sripada, L.; Singh, K.; Roy, M.; Bhatelia, K.; Dalwadi, P.; Singh, R. Systemic Analysis of miRNAs in PD Stress Condition: miR-5701 Modulates Mitochondrial-Lysosomal Cross Talk to Regulate Neuronal Death. Mol. Neurobiol. 2018, 55, 4689–4701. [Google Scholar] [CrossRef]
- Wasen, C.; Ospelt, C.; Camponeschi, A.; Erlandsson, M.C.; Andersson, K.M.E.; Silfversward, S.T.; Gay, S.; Bokarewa, M.I. Nicotine Changes the microRNA Profile to Regulate the FOXO Memory Program of CD8(+) T Cells in Rheumatoid Arthritis. Front. Immunol. 2020, 11, 1474. [Google Scholar] [CrossRef]
- Zidar, D.A.; Mudd, J.C.; Juchnowski, S.; Lopes, J.P.; Sparks, S.; Park, S.S.; Ishikawa, M.; Osborne, R.; Washam, J.B.; Chan, C.; et al. Altered Maturation Status and Possible Immune Exhaustion of CD8 T Lymphocytes in the Peripheral Blood of Patients Presenting With Acute Coronary Syndromes. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Kolbus, D.; Ljungcrantz, I.; Andersson, L.; Hedblad, B.; Fredrikson, G.N.; Bjorkbacka, H.; Nilsson, J. Association between CD8+ T-cell subsets and cardiovascular disease. J. Intern. Med. 2013, 274, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, H. MicroRNA-181a-5p Regulates Inflammatory Response of Macrophages in Sepsis. Open Med. 2019, 14, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Mollet, I.G.; Malm, H.A.; Wendt, A.; Orho-Melander, M.; Eliasson, L. Integrator of Stress Responses Calmodulin Binding Transcription Activator 1 (Camta1) Regulates miR-212/miR-132 Expression and Insulin Secretion. J. Biol. Chem. 2016, 291, 18440–18452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malm, H.A.; Mollet, I.G.; Berggreen, C.; Orho-Melander, M.; Esguerra, J.L.; Goransson, O.; Eliasson, L. Transcriptional regulation of the miR-212/miR-132 cluster in insulin-secreting beta-cells by cAMP-regulated transcriptional co-activator 1 and salt-inducible kinases. Mol. Cell Endocrinol. 2016, 424, 23–33. [Google Scholar] [CrossRef]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuniga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Cao, Y.; Dai, S.; Luo, S.; Guo, Q.; Wang, E. Plasma MIR-212-3p as a biomarker for acute right heart failure with pulmonary artery hypertension. Ann. Transl. Med. 2020, 8, 1571. [Google Scholar] [CrossRef]

| Characteristics | Female (n = 28) | Male (n = 160) | p-Value |
|---|---|---|---|
| Age, years–mean ± SD | 68.6 ± 7.0 | 70.6 ± 8.2 | 0.18 |
| BMI *–mean ± SD | 28.2 ± 6.1 | 28.2 ± 3.8 | 0.97 |
| Rhythm–no. (%) | |||
| Sinus rhythm at first presentation | 26 (92.9) | 127 (79.4) | 0.12 |
| Prior atrial fibrillation | 5 (17.9) | 34 (21.2) | 0.80 |
| CV * risk factors–no. (%) | |||
| Arterial Hypertension | 23 (82.1) | 144 (90.0) | 0.21 |
| Hyperlipidemia | 22 (78.6) | 132 (82.5) | 0.60 |
| Diabetes mellitus | 10 (35.7) | 57 (35.6) | 1.0 |
| Use of insulin | 2 (7.1) | 18 (11.3) | 0.74 |
| Current smoker | 9 (32.1) | 27 (16.9) | 0.07 |
| Familial disposition | 11 (39.3) | 61 (38.1) | 1.0 |
| Obesity | 9 (32.1) | 63 (40.0) | 0.53 |
| Chronic renal disorder | 1 (3.6) | 19 (11.9) | 0.32 |
| Chronic lung disease | 1 (3.6) | 16 (10.0) | 0.48 |
| Clinical presentation–no. (%) | |||
| Unstable angina | 6 (21.4) | 20 (12.5) | 0.23 |
| NSTEMI | 8 (28.6) | 38 (23.8) | 0.63 |
| Medical history–no. (%) | |||
| Myocardial infarction | 9 (32.1) | 41 (25.6) | 0.49 |
| PCI * | 7 (25.0) | 42 (26.3) | 1.0 |
| CABG * | 0 (0.0) | 1 (0.6) | 1.0 |
| Stroke | 2 (7.1) | 14 (8.8) | 1.0 |
| Peripheral vascular disease | 0 (0.0) | 22 (13.8) | 0.05 |
| LV-EF * ≤50%–no./total no. (%) | 4/21 (19.0) | 33/131 (33.6) | 0.22 |
| Three vessel disease–no. (%) | 19 (67.9) | 143 (89.4) | 0.006 |
| EuroSCORE *–mean ± SD | 6.46 ± 2.28 | 5.46 ± 3.13 | 0.05 |
| Preoperative medication-no./total no. (%) | |||
| Oral anticoagulant | 6/20 (30.0) | 35/120 (29.2) | 1.0 |
| Aspirin | 19/22 (86.4) | 121/138 (87.7) | 0.74 |
| P2Y12-inhibitor (Clopidogrel) | 1/22 (4.5) | 6/138 (4.3) | 1.0 |
| Betablocker | 9/27 (33.3) | 81/153 (52.9) | 0.09 |
| ACE-inhibitor7 | 11/27 (40.7) | 71/153 (46.4) | 0.68 |
| Calcium channel inhibitor | 4/20 (20.0) | 29/117 (24.8) | 0.78 |
| Diuretic | 7/27 (25.9) | 56/153 (36.6) | 0.38 |
| Statin | 22/28 (78.6) | 126/160 (78.8) | 1.0 |
| Laboratory values at admission | |||
| Hemoglobin, mg/dl | 12.0 ± 3.5 | 14.0 ± 3.4 | 0.04 |
| eGFR *, ml/min | 73.0 ± 32.8 | 71.6 ± 24.0 | 0.82 |
| miRNA | Clinical Trait | Correlation Coefficient | p-Value | FDR |
|---|---|---|---|---|
| miR-10b-3p | Sex | 0.21 | 0.004 | <0.05 |
| miR-10b-5p | Sex | 0.29 | <0.001 | <0.01 |
| miR-17-3p | Hypertension | −0.20 | 0.006 | <0.05 |
| miR-21-5p | Sex | −0.21 | 0.004 | <0.05 |
| COPD | 0.20 | 0.005 | <0.05 | |
| miR-151a-5p | COPD | −0.22 | 0.002 | <0.05 |
| miR-181a-5p | Smoking | 0.22 | 0.003 | <0.05 |
| miR-185-5p | Sex | 0.20 | 0.006 | <0.05 |
| Hypertension | −0.19 | 0.009 | 0.06 | |
| miR-194-5p | Hypertension | −0.21 | 0.004 | <0.05 |
| miR-199a-3p | Sex | −0.22 | 0.002 | <0.05 |
| miR-199b-3p | Sex | −0.22 | 0.002 | <0.05 |
| miR-212-3p | Obesity | 0.21 | 0.003 | <0.05 |
| miR-363-3p | Hypertension | −0.20 | 0.005 | <0.05 |
| Sex | 0.20 | 0.007 | 0.05 | |
| miR-548d-5p | Sex | 0.21 | 0.004 | <0.05 |
| miR-744-5p | Sex | −0.20 | 0.006 | <0.05 |
| miR-3117-3p | Sex | −0.22 | 0.003 | <0.05 |
| miR-5683 | Hyperlipidemia | 0.23 | 0.002 | <0.05 |
| Smoking | 0.16 | 0.024 | 0.16 | |
| miR-5701 | Hyperlipidemia | 0.22 | 0.002 | <0.05 |
| Diabetes | 0.20 | 0.005 | <0.05 | |
| Hypertension | 0.20 | 0.007 | 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neiburga, K.D.; Vilne, B.; Bauer, S.; Bongiovanni, D.; Ziegler, T.; Lachmann, M.; Wengert, S.; Hawe, J.S.; Güldener, U.; Westerlund, A.M.; et al. Vascular Tissue Specific miRNA Profiles Reveal Novel Correlations with Risk Factors in Coronary Artery Disease. Biomolecules 2021, 11, 1683. https://doi.org/10.3390/biom11111683
Neiburga KD, Vilne B, Bauer S, Bongiovanni D, Ziegler T, Lachmann M, Wengert S, Hawe JS, Güldener U, Westerlund AM, et al. Vascular Tissue Specific miRNA Profiles Reveal Novel Correlations with Risk Factors in Coronary Artery Disease. Biomolecules. 2021; 11(11):1683. https://doi.org/10.3390/biom11111683
Chicago/Turabian StyleNeiburga, Katrīna D., Baiba Vilne, Sabine Bauer, Dario Bongiovanni, Tilman Ziegler, Mark Lachmann, Simon Wengert, Johann S. Hawe, Ulrich Güldener, Annie M. Westerlund, and et al. 2021. "Vascular Tissue Specific miRNA Profiles Reveal Novel Correlations with Risk Factors in Coronary Artery Disease" Biomolecules 11, no. 11: 1683. https://doi.org/10.3390/biom11111683
APA StyleNeiburga, K. D., Vilne, B., Bauer, S., Bongiovanni, D., Ziegler, T., Lachmann, M., Wengert, S., Hawe, J. S., Güldener, U., Westerlund, A. M., Li, L., Pang, S., Yang, C., Saar, K., Huebner, N., Maegdefessel, L., DigiMed Bayern Consortium, Lange, R., Krane, M., ... von Scheidt, M. (2021). Vascular Tissue Specific miRNA Profiles Reveal Novel Correlations with Risk Factors in Coronary Artery Disease. Biomolecules, 11(11), 1683. https://doi.org/10.3390/biom11111683








