Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold
Abstract
:1. Introduction
2. Materials and Methods
2.1. G4 Formation of CpG ODNs
2.2. CD Measurement
2.3. Polyacrylamide Gel Electrophoresis (PAGE)
2.4. Stability of ODNs in Serum
2.5. Cell Culture
2.6. Preparation of Mouse Bone Marrow-Derived Macrophage Cells (BMDMs)
2.7. Immunostimulatory Assay
2.8. Statistical Analysis
3. Results
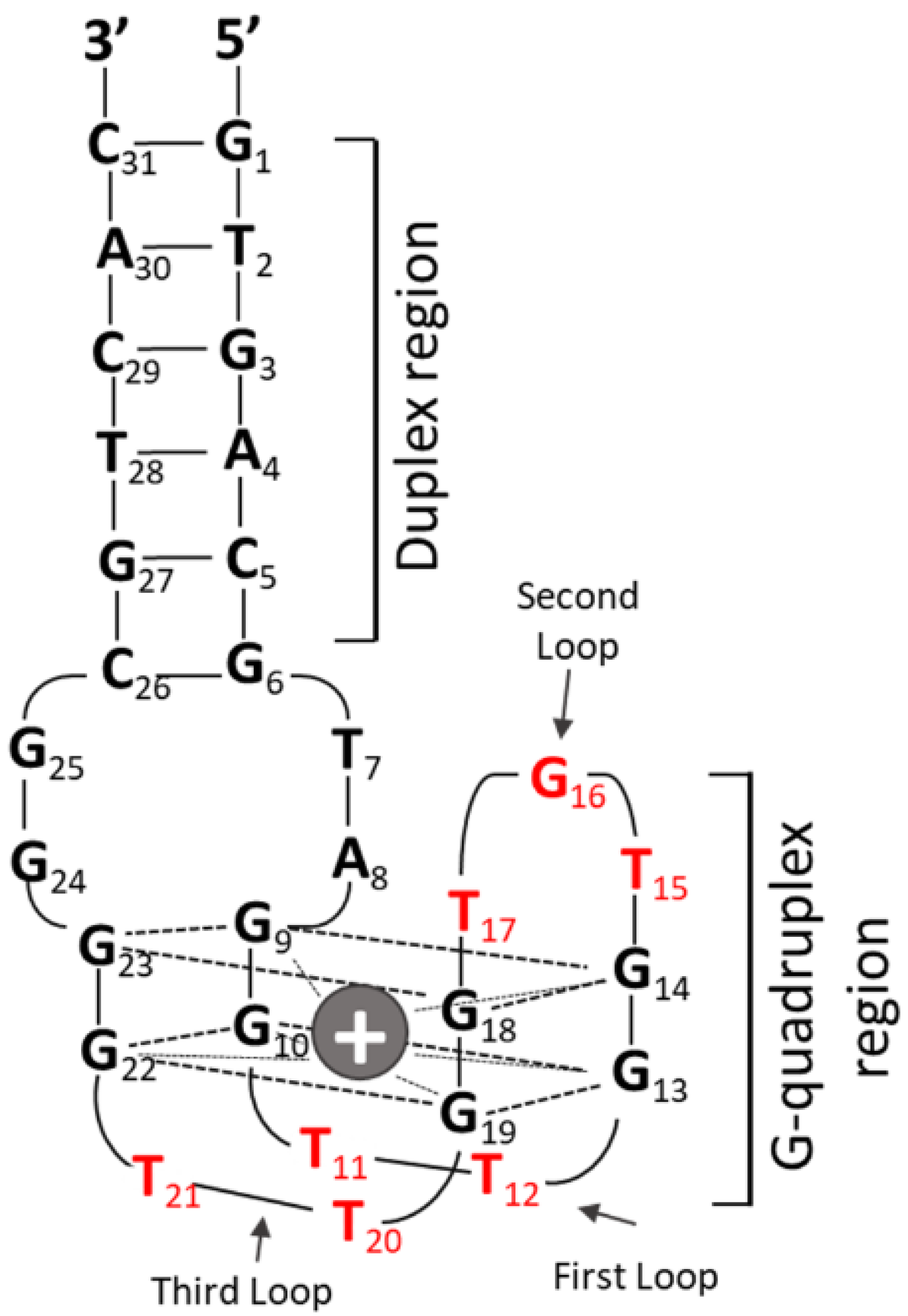
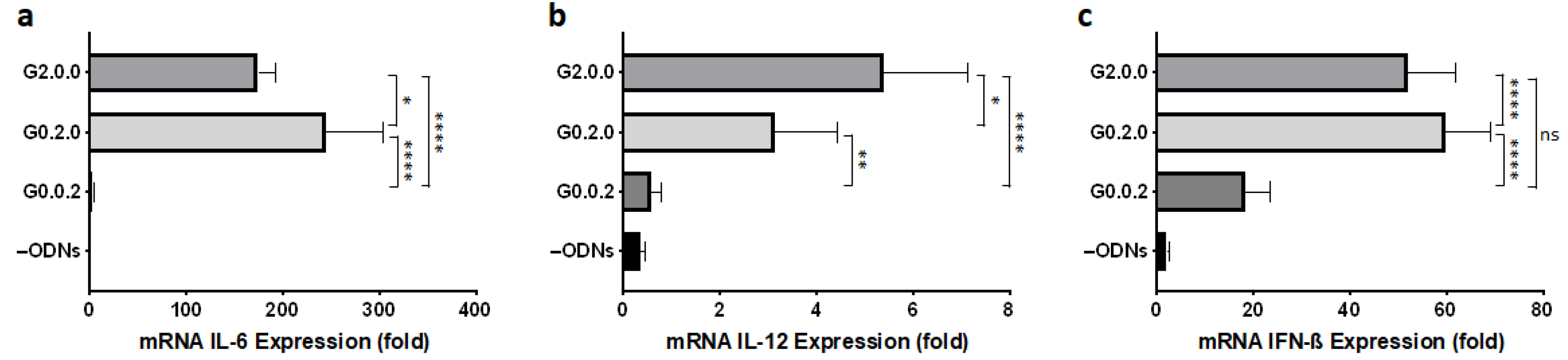
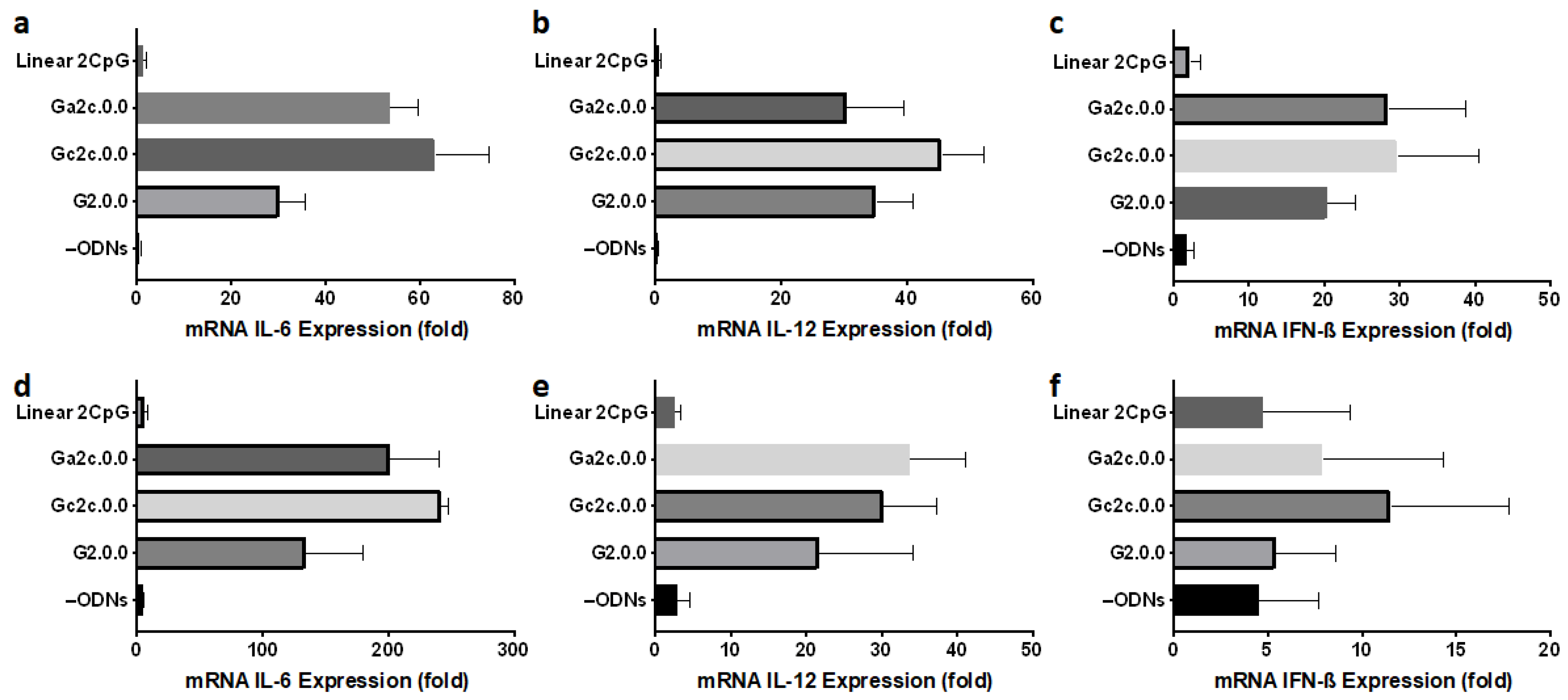
3.1. Effect of CpG Motif Position in Anti-Parallel Duplex/G4 ODNs on G4 Formation and Immunostimulant Function
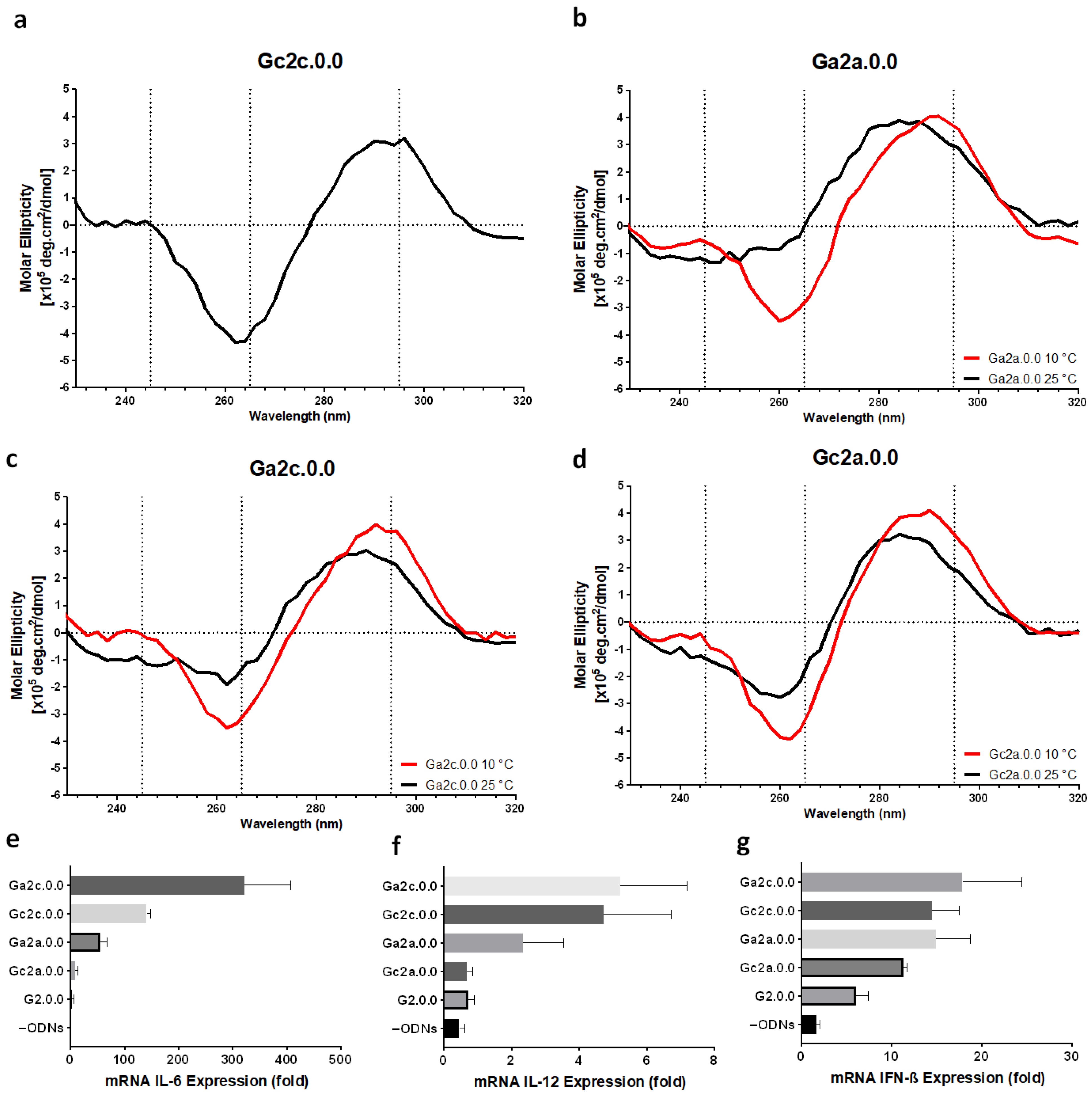
3.2. Influence of Nucleotide Connectors between G-Tract and CpG Loop on G4 Formation and Immunostimulatory Activity
3.3. Electrophoresis Analysis of Anti-Parallel G4 CpG ODNs
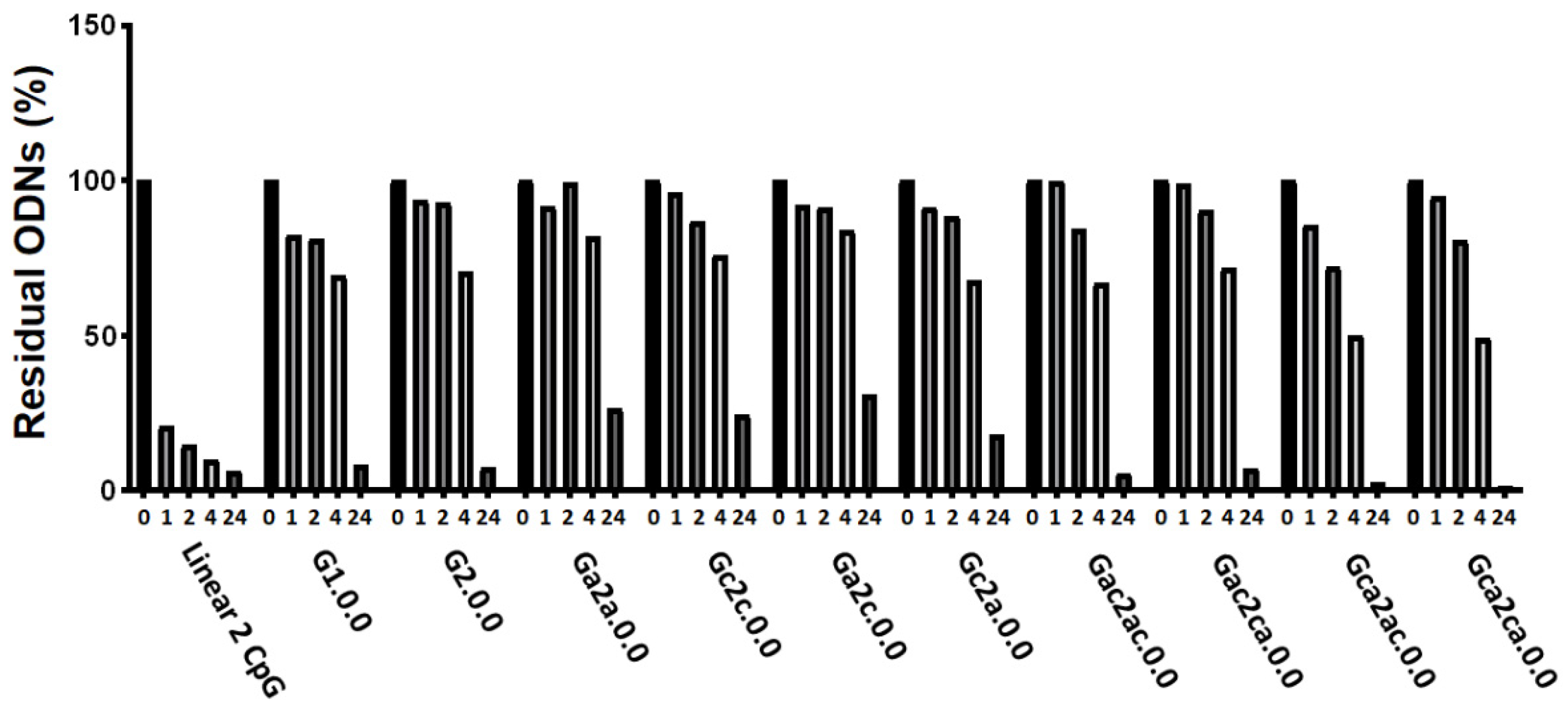
3.4. Serum and Thermal Stability of G4 CpG ODNs
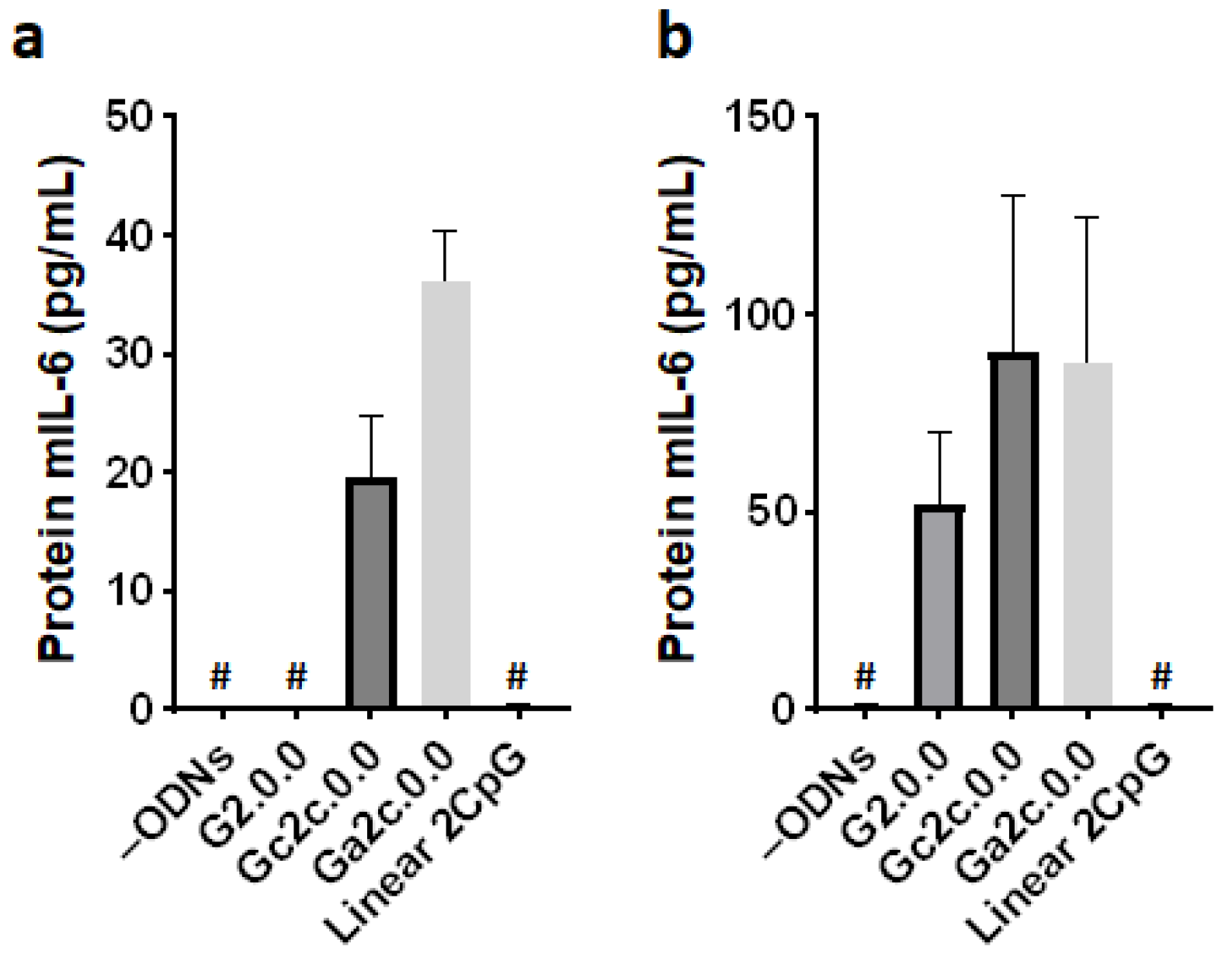
3.5. Immune Responses of Mouse Dendritic Cells and BMDMs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Zhu, H.; Xia, X.; Liang, Z.; Ma, X.; Sun, B. Vaccine adjuvants: Understanding the structure and mechanism of adjuvanticity. Vaccine 2019, 37, 3167–3178. [Google Scholar] [CrossRef] [PubMed]
- Givens, B.E.; Geary, S.M.; Salem, A.K. Nanoparticle-based CpG-oligonucleotide therapy for treating allergic asthma. Immunotherapy 2018, 10, 595–604. [Google Scholar] [CrossRef]
- Kell, S.A.; Kachura, M.A.; Renn, A.; Traquina, P.; Coffman, R.L.; Campbell, J.D. Preclinical development of the TLR9 agonist DV281 as an inhaled aerosolized immunotherapeutic for lung cancer: Pharmacological profile in mice, non-human primates, and human primary cells. Int. Immunopharmacol. 2019, 66, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Kang, S.H.; Gryaznov, S.M.; DeDionisio, L.; Heidenreich, O.; Sullivan, S.; Xu, X.; Nerenberg, M.I. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 1994, 269, 26801–26805. [Google Scholar] [CrossRef]
- Sheehan, J.P.; Lan, H.-C. Phosphorothioate Oligonucleotides Inhibit the Intrinsic Tenase Complex. Blood 1998, 92, 1617–1625. [Google Scholar] [CrossRef]
- Levin, A.A. A review of issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim. Biophys. Acta. 1999, 1489, 69–84. [Google Scholar] [CrossRef]
- Henry, S.P.; Beattie, G.; Yeh, G.; Chappel, A.; Giclas, P.; Mortari, A.; Jagels, M.A.; Kornbrust, D.J.; Levin, A.A. Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int. Immunopharmacol. 2002, 2, 1657–1666. [Google Scholar] [CrossRef]
- Tu, A.T.T.; Hoshi, K.; Ikebukuro, K.; Hanagata, N.; Yamazaki, T. Monomeric G-Quadruplex-Based CpG Oligodeoxynucleotides as Potent Toll-Like Receptor 9 Agonists. Biomacromolecules 2020, 21, 3644–3657. [Google Scholar] [CrossRef]
- Hoshi, K.; Yamazaki, T.; Sugiyama, Y.; Tsukakoshi, K.; Tsugawa, W.; Sode, K.; Ikebukuro, K. G-quadruplex structure improves the immunostimulatory effects of cpg oligonucleotides. Nucleic Acid Ther. 2019, 29, 224–229. [Google Scholar] [CrossRef]
- Liao, W.; Tan, M.; Kusamori, K.; Takakura, Y.; Nishikawa, M. Construction of Monomeric and Dimeric G-Quadruplex-Structured CpG Oligodeoxynucleotides for Enhanced Uptake and Activation in TLR9-Positive Macrophages. Nucleic Acid Ther. 2020, 30, 299–311. [Google Scholar] [CrossRef]
- Hao, F.; Ma, Y.; Guan, Y. Effects of central loop length and metal ions on the thermal stability of G-quadruplexes. Molecules 2019, 24, 1863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Cheng, Y.; Hao, J.; Jia, G.; Zhou, J.; Mergny, J.L.; Li, C. Loop permutation affects the topology and stability of G-quadruplexes. Nucleic Acids Res. 2018, 46, 9264–9275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikebukuro, K.; Okumura, Y.; Sumikura, K.; Karube, I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005, 33, e108. [Google Scholar] [CrossRef] [Green Version]
- Mazurov, A.V.; Titaeva, E.V.; Khaspekova, S.G.; Storojilova, A.N.; Spiridonova, V.A.; Kopylov, A.M.; Dobrovolsky, A.B. Characteristics of a new DNA aptamer, direct inhibitor of thrombin. Bull. Exp. Biol. Med. 2011, 150, 422–425. [Google Scholar] [CrossRef] [PubMed]
- Spiridonova, V.A.; Barinova, K.V.; Glinkina, K.A.; Melnichuk, A.V.; Gainutdynov, A.A.; Safenkova, I.V.; Dzantiev, B.B. A family of DNA aptamers with varied duplex region length that forms complexes with thrombin and prothrombin. FEBS Lett. 2015, 589, 2043–2049. [Google Scholar] [CrossRef] [Green Version]
- Russo Krauss, I.; Spiridonova, V.; Pica, A.; Napolitano, V.; Sica, F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016, 44, 983–991. [Google Scholar] [CrossRef] [Green Version]
- Mergny, J.L.; Lacroix, L. UV melting of G-quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009, 1–15. [Google Scholar] [CrossRef]
- Hanagata, N.; Li, X.; Chen, M.H.; Li, J.; Hattori, S. Double-stranded phosphodiester cytosine-guanine oligodeoxynucleotide complexed with calcium phosphate as a potent vaccine adjuvant for activating cellular and Th1-type humoral immunities. Int. J. Nanomed. 2018, 13, 43–62. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.; Ohba, Y.; Yanai, H.; Negishi, H.; Mizutani, T.; Takaoka, A.; Taya, C.; Taniguchi, T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 2005, 434, 1035–1040. [Google Scholar] [CrossRef]
- Kejnovská, I.; Kypr, J.; Vorlíčková, M. Oligo(dT) is not a correct native PAGE marker for single-stranded DNA. Biochem. Biophys. Res. Commun. 2007, 353, 776–779. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In The Alkali Metal Ions: Their Role for Life, 1st ed.; Sigel, A., Sigel, H., Sigel, R., Eds.; Springer: Cham, Switzerland, 2016; Volume 16, pp. 203–258. [Google Scholar]
- Duque, G.A.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar] [CrossRef] [Green Version]
- Noel, G.; Baetz, N.W.; Staab, J.F.; Donowitz, M.; Kovbasnjuk, O.; Pasetti, M.F.; Zachos, N.C. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 2017, 7, 45270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.Y. M1 and M2 polarization of macrophages: A mini-review. Med. Biol. Sci. Eng. 2019, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Germic, N.; Frangez, Z.; Yousefi, S.; Simon, H.U. Regulation of the innate immune system by autophagy: Monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 2019, 26, 715–727. [Google Scholar] [CrossRef] [PubMed]
- van Dinther, D.; Veninga, H.; Iborra, S.; Borg, E.G.F.; Hoogterp, L.; Olesek, K.; Beijer, M.R.; Schetters, S.T.T.; Kalay, H.; Garcia-Vallejo, J.J.; et al. Functional CD169 on Macrophages Mediates Interaction with Dendritic Cells for CD8 + T Cell Cross-Priming. Cell Rep. 2018, 22, 1484–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Sequence 5′ to 3′ | Length |
|---|---|---|
| G0.0.0 | GTGACGTAGGTTGGTGTGGTTGGGGCGTCAC | 31 |
| G2.0.0 | GTGACGTAGGGTCGTTTTGTCGTTGGTGTGGTTGGGGCGTCAC | 43 |
| G0.2.0 | GTGACGTAGGTTGGGTCGTTTTGTCGTTGGTTGGGGCGTCAC | 42 |
| G0.0.2 | GTGACGTAGGTTGGTGTGGGTCGTTTTGTCGTTGGGGCGTCAC | 43 |
| G1.0.0 | GTGACGTAGGGTCGTTGGTGTGGTTGGGGCGTCAC | 35 |
| Ga2a.0.0 | GTGACGTAGGAGTCGTTTTGTCGTTAGGTGTGGTTGGGGCGTCAC | 45 |
| Gc2c.0.0 | GTGACGTAGGCGTCGTTTTGTCGTTCGGTGTGGTTGGGGCGTCAC | 45 |
| Ga2c.0.0 | GTGACGTAGGAGTCGTTTTGTCGTTCGGTGTGGTTGGGGCGTCAC | 45 |
| Gc2a.0.0 | GTGACGTAGGCGTCGTTTTGTCGTTAGGTGTGGTTGGGGCGTCAC | 45 |
| Gac2ac.0.0 | GTGACGTAGGACGTCGTTTTGTCGTTACGGTGTGGTTGGGGCGTCAC | 47 |
| Gac2ca.0.0 | GTGACGTAGGACGTCGTTTTGTCGTTCAGGTGTGGTTGGGGCGTCAC | 47 |
| Gca2ac.0.0 | GTGACGTAGGCAGTCGTTTTGTCGTTACGGTGTGGTTGGGGCGTCAC | 47 |
| Gca2ca.0.0 | GTGACGTAGGACGTCGTTTTGTCGTTCAGGTGTGGTTGGGGCGTCAC | 47 |
| Linear 2CpG | GTCGTTTTGTCGTT |
| Name | Loop Sequence | Tm at 1st Cooling (℃) | Tm at 2nd Heating (℃) | ||
|---|---|---|---|---|---|
| 1st | 2nd | 3rd | |||
| G0.0.0 | TT | TGT | TT | 41.8 | 42.8 |
| G1.0.0 | GTCGTT | TGT | TT | 40.2 | 41.2 |
| G2.0.0 | GTCGTTTTGTCGTT | TGT | TT | 35.6 | 35.6 |
| G0.0.2 | TT | TGT | GTCGTTTTGTCGTT | 32.9 | 33.4 |
| Ga2a.0.0 | AGTCGTTTTGTCGTTA | TGT | TT | 33.6 | 34.9 |
| Gc2c.0.0 | CGTCGTTTTGTCGTTC | TGT | TT | 32.5 | 32.8 |
| Ga2c.0.0 | AGTCGTTTTGTCGTTC | TGT | TT | 30.2 | 29.5 |
| Gc2a.0.0 | CGTCGTTTTGTCGTTA | TGT | TT | 25.9 | 25.2 |
| Gac2ac.0.0 | ACGTCGTTTTGTCGTTAC | TGT | TT | 33.6 | 34.6 |
| Gca2ac.0.0 | CAGTCGTTTTGTCGTTAC | TGT | TT | 32.5 | 34.7 |
| Gca2ca.0.0 | CAGTCGTTTTGTCGTTCA | TGT | TT | 34.9 | 30.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safitri, F.A.; Tu, A.T.T.; Hoshi, K.; Shobo, M.; Zhao, D.; Witarto, A.B.; Sumarsono, S.H.; Giri-Rachman, E.A.; Tsukakoshi, K.; Ikebukuro, K.; et al. Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold. Biomolecules 2021, 11, 1617. https://doi.org/10.3390/biom11111617
Safitri FA, Tu ATT, Hoshi K, Shobo M, Zhao D, Witarto AB, Sumarsono SH, Giri-Rachman EA, Tsukakoshi K, Ikebukuro K, et al. Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold. Biomolecules. 2021; 11(11):1617. https://doi.org/10.3390/biom11111617
Chicago/Turabian StyleSafitri, Fika Ayu, Anh Thi Tram Tu, Kazuaki Hoshi, Miwako Shobo, Dandan Zhao, Arief Budi Witarto, Sony Heru Sumarsono, Ernawati Arifin Giri-Rachman, Kaori Tsukakoshi, Kazunori Ikebukuro, and et al. 2021. "Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold" Biomolecules 11, no. 11: 1617. https://doi.org/10.3390/biom11111617
APA StyleSafitri, F. A., Tu, A. T. T., Hoshi, K., Shobo, M., Zhao, D., Witarto, A. B., Sumarsono, S. H., Giri-Rachman, E. A., Tsukakoshi, K., Ikebukuro, K., & Yamazaki, T. (2021). Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold. Biomolecules, 11(11), 1617. https://doi.org/10.3390/biom11111617







