DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences
Abstract
1. Introduction
1.1. Ensemble-Based Modeling of Protein Internal Dynamics
1.2. Neighbor-Dependent Ramachandran Preferences for Amino Acids
1.3. Benchmark Test Systems
1.3.1. Cd3 Cytoplasmic Domain
1.3.2. The Helical Medial Tail Domain of the Porcine Myosin VI Protein
2. Materials and Methods
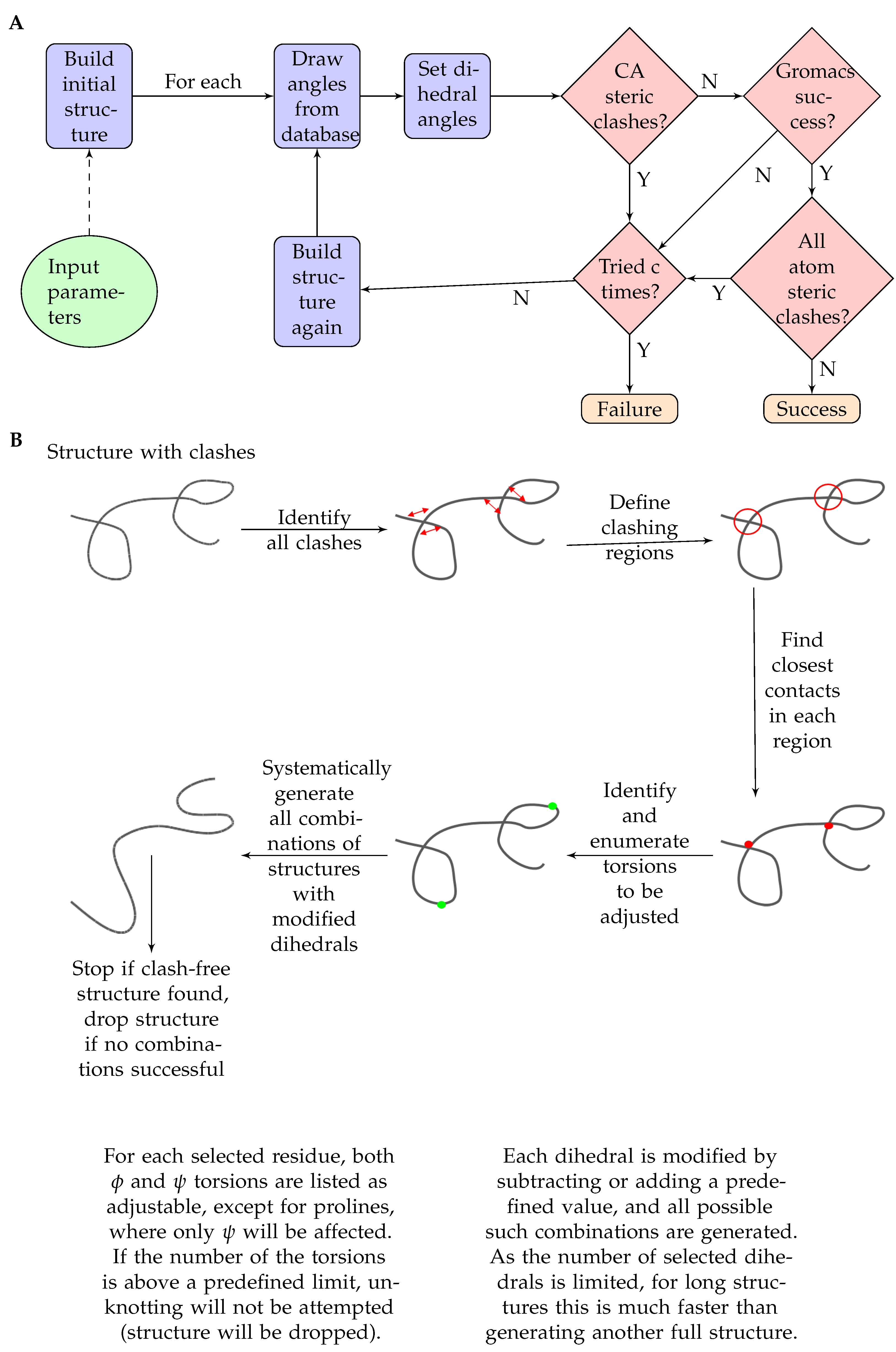
2.1. A Pipeline to Build Three-Dimensional Ensemble Models
- At first, ChimeraX [18] is invoked to build up a very long beta strand based on the given sequence.
- Next, Ramachandran angles for each residue are set based on the neighbor-dependent probabilities reported in [21]. The probabilities are defined for bins of 5x5 degrees and are stored as binary files to optimize speed and storage. Probabilities based on the left (N terminal side sequential) neighbor and right (C terminal side sequential) neighbor, as well as a combined one, derived as described in [21], are available and are denoted LEFT, RIGHT and TRIPLET. A roulette-wheel selection approach is used where a generated random number between 0 and 1 is used to select a bin according to the cumulative probabilities.
- ChimeraX [18] is again used to set the dihedral angles to the previously calculated values.
- For the obtained conformation, a quick check is performed to filter out unrealistic structures based on CA-CA distances below 4 Ångströms. This additional program is written in C++.
- If there are no CA steric clashes, the program Scwrl4 [19] is invoked to optimize the sidechains.
- If there are CA steric clashes, the program attempts to unknot them. If two residues clash, the program tries to perturb the dihedral angles of the residue halfway between them. The perturbation means to add or subtract a given value to the selected dihedral angles. For each angle, these perturbations are combinatorially applied, and the perturbed structures undergo the above clash check again.
- After that, the program runs GROMACS [27] to perform a short energy minimization in vacuum to optimize the structure. The generated log file is checked for success, as it is expected that structures with serious steric problems will fail this step.
- If the optimization is successful, ChimeraX [18] is invoked again to check for all atom steric clashes with its command clashes using default parameters. If there are no steric clashes, the structure is accepted successfully.
- The above steps are performed for each structure to be generated. For unsuccessful trials, the program will try and perform structure generation again until the user-defined limit for trials is reached or an accepted structure is generated.
- The input parameters of the program are:
- -
- The sequence (the only required input parameter);
- -
- The number of structures to be generated;
- -
- The building mode;
- *
- LEFT: considering only the left (N terminal) sequential neighbor in choosing dihedral angles;
- *
- RIGHT: considering only the right (C terminal) sequential neighbor in choosing dihedral angles;
- *
- TRIPLET: considers both sequential neighbors using derived cumulative probabilities;
- *
- WEIGHTED_LEFT: for each residue, a user defined Gaussian distribution can be combined with the Dunbrack distribution;
- *
- WEIGHTED_RIGHT: for each residue, a user defined Gaussian distribution can be combined with the Dunbrack distribution;
- *
- WEIGHTED_TRIPLET: for each residue, a user defined Gaussian distribution can be combined with the derived distribution.
- -
- a filename base for the generated structures (for example „bar_” as a base will result in bar_min_1.pdb, bar_min_2.pdb, etc.);
- -
- The dataset used (TCBIG or Coil only, see [21]);
- -
- The number of trials for a structure;
- -
- Whether a Gromacs optimization step should be performed for each structure;
- -
- Whether temporary files are kept after the run;
- -
- The angle to add or subtract from the dihedral angles at the unknotting steps;
- -
- The maximum number of torsions to be adjusted during unknotting, zero meaning no unknotting.
2.1.1. The Initial Structure
2.1.2. Different Approaches to Handle Sequence Neighborhood
2.1.3. Steric Clashes and Repetition
2.1.4. Selecting and Evaluating Subensembles
2.2. Molecular Dynamics Simulations
3. Results
3.1. Implementation of the DIPEND Pipeline
- Build an initial extended structure (invokes ChimeraX);
- Select the dihedral angles of each residue according to the distribution settings (see Methods);
- Set the dihedral angles to the selected values (invokes ChimeraX);
- Preliminary check for CA-CA clashes to filter out largely unrealistic structures (invokes an in-house C++ program);
- For clashing structures, an “unknotting” attempt can be performed if chosen with the options;
- Optimize side chains (invokes Scwrl4);
- Short energy minimization (invokes GROMACS);
- All-atom clash check (invokes ChimeraX).
3.2. Addition of Secondary Chemical Shift Analysis to the CoNSEnsX Server
3.3. Overview of the DIPEND-Generated Ensembles
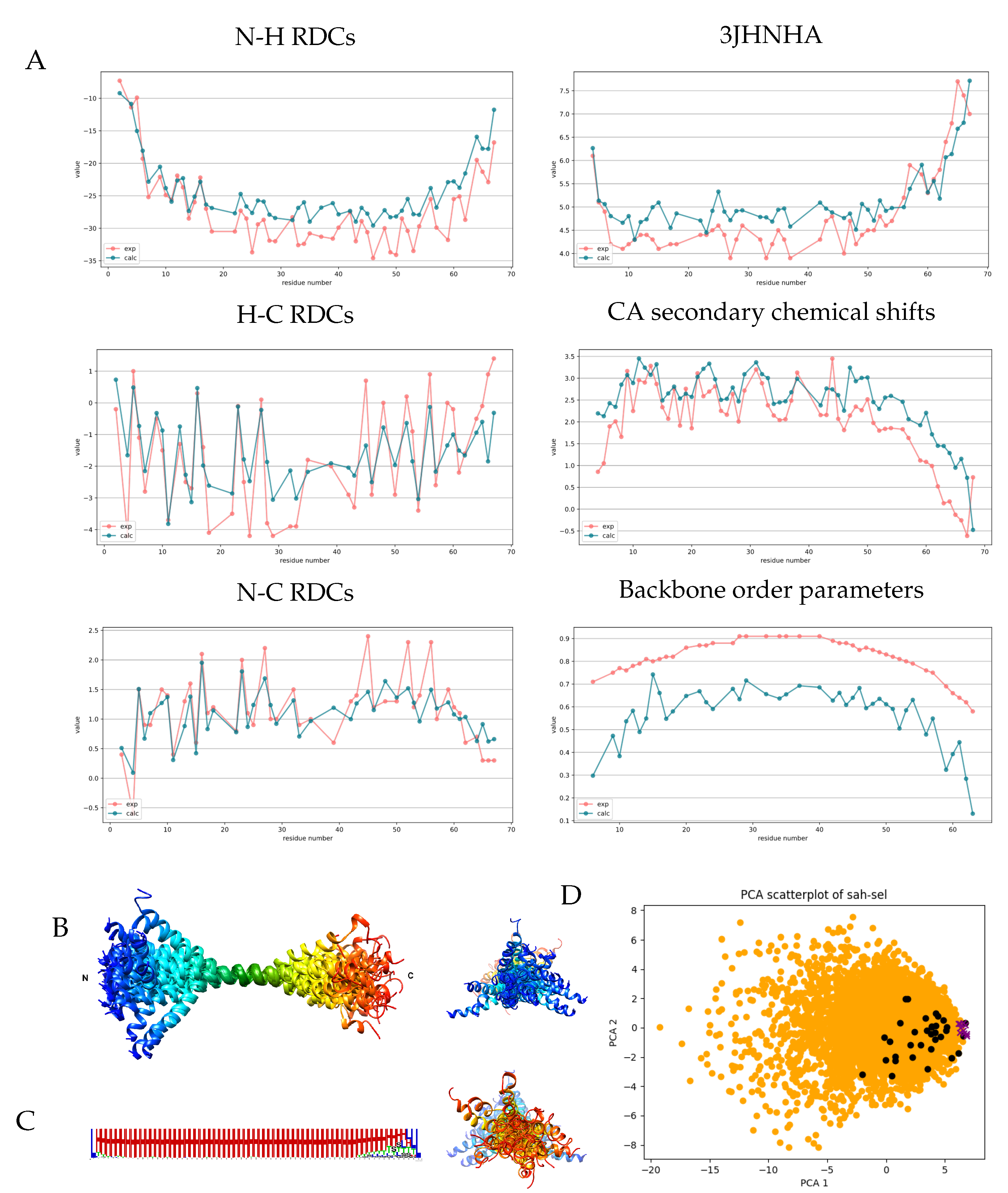
3.4. Analysis of the Cd3 Disordered Cytoplasmic Segment
3.5. The Single -Helical Segment of Myosin VI
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMRB | Biological Magnetic Resonance Data Bank |
| GUI | Graphical User Interface |
| IDP | Intrinsically Disordered Protein |
| NMR | Nuclear Magnetic Resonance |
| NOE | Nuclear Overhauser Effect |
| MD | Molecular Dynamics |
| PDB | Protein Databank |
| PCA | Principal Component Analysis |
| RDC | Residual Dipolar Coupling |
| RMSD | Root Mean Square Deviation |
References
- Pakhrin, S.C.; Shrestha, B.; Adhikari, B.; Kc, D.B. Deep Learning-Based Advances in Protein Structure Prediction. Int. J. Mol. Sci. 2021, 22, 5553. [Google Scholar] [CrossRef] [PubMed]
- Ángyán, A.F.; Gáspári, Z. Ensemble-Based Interpretations of NMR Structural Data to Describe Protein Internal Dynamics. Molecules 2013, 18, 10548–10567. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Heller, G.T.; Camilloni, C.; Vendruscolo, M. Principles of protein structural ensemble determination. Curr. Opin. Struct Biol. 2017, 42, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Lazar, T.; Martínez-Pérez, E.; Quaglia, F.; Hatos, A.; Chemes, L.B.; Iserte, J.A.; Méndez, N.A.; Garrone, N.A.; Saldaño, T.E.; Marchetti, J.; et al. PED in 2021: A major update of the protein ensemble database for intrinsically disordered proteins. Nucleic Acids Res. 2021, 8, D404–D41. [Google Scholar] [CrossRef]
- Quaglia, F.; Lazar, T.; Hatos, A.; Tompa, P.; Piovesan, P.D.; Tosatto, S.C. Exploring curated conformational ensembles of intrinsically disordered proteins in the protein ensemble database. Curr. Protoc. 2021, 1, e192. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Best, R.B.; Depristo, M.A.; Dobson, C.M.; Vendruscolo, M. Simultaneous determination of protein structure and dynamics. Nature 2005, 433, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Lange, O.F.; Lakomek, N.A.; Farès, C.; Schröder, G.F.; Walter, K.F.; Becker, S.; Meiler, J.; Grubmüller, H.; Griesinger, C.; de Groot, B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 2008, 320, 1471–1475. [Google Scholar] [CrossRef]
- Marsh, J.A.; Baker, J.M.R.; Tollinger, M.; Forman-Kay, J.D. Calculation of Residual Dipolar Couplings from Disordered State Ensembles Using Local Alignment. J. Am. Chem. Soc. 2008, 130, 7804–7805. [Google Scholar] [CrossRef]
- Nodet, G.; Salmon, L.; Ozenne, V.; Meier, S.; Jensen, M.R.; Blackledge, M. Quantitative Description of Backbone Conformational Sampling of Unfolded Proteins at Amino Acid Resolution from NMR Residual Dipolar Couplings. J. Am. Chem. Soc. 2009, 131, 17908–17918. [Google Scholar] [CrossRef]
- Krzeminski, M.; Marsh, J.A.; Neale, C.; Choy, W.Y.; Forman-Kay, J.D. Characterization of disordered proteins with ENSEMBLE. Bioinformatics 2013, 29, 398–399. [Google Scholar] [CrossRef]
- Bernadó, P.; Blanchard, L.; Timmins, P.; Marion, D.; Ruigrok, R.W.H.; Blackledge, M. A structural model for unfolded proteins from residual dipolar couplings and small-angle X-ray scattering. Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [Google Scholar] [CrossRef] [PubMed]
- Ozenne, V.; Bauer, F.; Salmon, L.; Huang, J.R.; Jensen, M.R.; Segard, S.; Bernadó, P.; Charavay, C.; Blackledge, M. Flexible-meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 2012, 28, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Estaña, A.; Sibille, N.; Delaforge, E.; Vaisset, M.; Cortés, J.; Bernadó, P. Realistic Ensemble Models of Intrinsically Disordered Proteins Using a Structure-Encoding Coil Database. Structure 2019, 27, 381–391.e2. [Google Scholar] [CrossRef]
- Feldman, H.J.; Hogue, C.V.W. A fast method to sample real protein conformational space. Proteins 2000, 39, 112–131. [Google Scholar] [CrossRef]
- Feldman, H.J.; Hogue, C.W. Probabilistic sampling of protein conformations: New hope for brute force? Proteins Struct. Funct. Bioinform. 2002, 46, 8–23. [Google Scholar] [CrossRef]
- Pietrek, L.M.; Stelzl, L.S.; Hummer, G. Hierarchical Ensembles of Intrinsically Disordered Proteins at Atomic Resolution in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2020, 16, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Senicourt, L.; le Maire, A.; Allemand, F.; Carvalho, J.E.; Guee, L.; Germain, P.; Schubert, M.; Bernadó, P.; Bourguet, W.; Sibille, N. Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR). J. Mol. Biol. 2021, 433, 166899. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Krivov, G.G.; Shapovalov, M.V.; Dunbrack, R.L. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 2009, 77, 778–795. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Ting, D.; Wang, G.; Shapovalov, M.; Mitra, R.; Jordan, M.I.; Dunbrack, R.L., Jr. Neighbor-Dependent Ramachandran Probability Distributions of Amino Acids Developed from a Hierarchical Dirichlet Process Model. PLoS Comput. Biol. 2010, 6, 1–21. [Google Scholar] [CrossRef]
- Dudola, D.; Kovács, B.; Gáspári, Z. CoNSEnsX+ Webserver for the Analysis of Protein Structural Ensembles Reflecting Experimentally Determined Internal Dynamics. J. Chem. Inf. Model. 2017, 57, 1728–1734. [Google Scholar] [CrossRef]
- Huang, Y.; Wange, R.L. T Cell Receptor Signaling: Beyond Complex Complexes. J. Biol. Chem. 2004, 279, 28827–28830. [Google Scholar] [CrossRef]
- Isaksson, L.; Mayzel, M.; Saline, M.; Pedersen, A.; Rosenlöw, J.; Brutscher, B.; Karlsson, B.G.; Orekhov, V.Y. Highly Efficient NMR Assignment of Intrinsically Disordered Proteins: Application to B- and T Cell Receptor Domains. PLoS ONE 2013, 8, e62947. [Google Scholar] [CrossRef]
- Isakov, N. Immunoreceptor tyrosine-based activation motif (ITAM), a unique module linking antigen and Fc receptors to their signaling cascades. J. Leukoc. Biol. 1997, 61, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.A.; Shen, Y.; Ying, J.; Takagi, Y.; Torchia, D.A.; Sellers, J.R.; Bax, A. Remarkable Rigidity of the Single α-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy. J. Am. Chem. Soc. 2019, 141, 9004–9017. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Neal, S.; Nip, A.M.; Zhang, H.; Wishart, D.S. Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR 2003, 26, 215–240. [Google Scholar] [CrossRef] [PubMed]
- Zweckstetter, M.; Bax, A. Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. J. Am. Chem. Soc. 2000, 122, 3791–3792. [Google Scholar] [CrossRef]
- Tamiola, K.; Acar, B.; Mulder, F.A.A. Sequence-Specific Random Coil Chemical Shifts of Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010, 132, 18000–18003. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.C.; Bax, A. Determination of the Backbone Dihedral Angles ϕ in Human Ubiquitin from Reparametrized Empirical Karplus Equations. J. Am. Chem. Soc. 1996, 118, 2483–2494. [Google Scholar] [CrossRef]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Grigera, J.R.; Straatsma, T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Best, R.B.; Vendruscolo, M. Determination of protein structures consistent with NMR order parameters. J. Am. Chem. Soc. 2004, 126, 8090–8091. [Google Scholar] [CrossRef] [PubMed]
- Bottaro, S.; Bengtsen, T.; Lindorff-Larsen, K. Integrating Molecular Simulation and Experimental Data: A Bayesian/Maximum Entropy Reweighting Approach. Methods Mol. Biol. 2020, 2112, 219–240. [Google Scholar] [PubMed]
- Ytreberg, F.M.; Borcherds, W.; Wu, H.; Daughdrill, G.W. Using chemical shifts to generate structural ensembles for intrinsically disordered proteins with converged distributions of secondary structure. Intrinsically Disord Proteins 2015, 3, e984565. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Generated 5000 | CA+CB rmsd | CA rmsd | Selected from MD | |||||
|---|---|---|---|---|---|---|---|---|
| Selected (84 Models) | Selected (45 Models) | (CA rmsd, 29 Models) | ||||||
| rmsd | corr. | rmsd | corr. | rmsd | corr. | rmsd | corr. | |
| CA full | 0.458 | 0.996 | 0.211 | 0.999 | 0.073 | 1.000 | 0.400 | 0.997 |
| CA secondary | 0.296 | 0.696 | 0.953 | 0.451 | ||||
| CB full | 0.550 | 0.999 | 0.137 | 1.000 | 0.601 | 0.998 | 0.831 | 0.997 |
| CB secondary | 0.233 | 0.764 | 0.322 | 0.301 | ||||
| 6OBI (10 Models) | Generated 5000 | Selected (37 Models) | ||||
|---|---|---|---|---|---|---|
| rmsd | corr. | rmsd | corr. | rmsd | corr. | |
| N-H RDC | 4.021 | 0.746 | 7.304 | 0.756 | 3.676 | 0.917 |
| H-C RDC | 1.709 | 0.456 | 1.440 | 0.671 | 1.058 | 0.807 |
| N-C RDC | 0.822 | 0.118 | 0.541 | 0.576 | 0.351 | 0.838 |
| 3JHNHA | 0.602 | 0.789 | 0.765 | 0.625 | 0.518 | 0.903 |
| CA secondary | 0.969 | 0.712 | 0.866 | 0.705 | 0.688 | 0.903 |
| CB secondary | 0.915 | 0.389 | 0.967 | 0.561 | 0.970 | 0.587 |
| N-H S2 | 0.201 | 0.488 | 0.258 | 0.871 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harmat, Z.; Dudola, D.; Gáspári, Z. DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences. Biomolecules 2021, 11, 1505. https://doi.org/10.3390/biom11101505
Harmat Z, Dudola D, Gáspári Z. DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences. Biomolecules. 2021; 11(10):1505. https://doi.org/10.3390/biom11101505
Chicago/Turabian StyleHarmat, Zita, Dániel Dudola, and Zoltán Gáspári. 2021. "DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences" Biomolecules 11, no. 10: 1505. https://doi.org/10.3390/biom11101505
APA StyleHarmat, Z., Dudola, D., & Gáspári, Z. (2021). DIPEND: An Open-Source Pipeline to Generate Ensembles of Disordered Segments Using Neighbor-Dependent Backbone Preferences. Biomolecules, 11(10), 1505. https://doi.org/10.3390/biom11101505






