Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Culture Enrichment
2.2. Microbial Community Analysis
2.3. Isolation of Pure Strains and Screening of Isolates
2.4. Phylogenetic Characterization of the Isolated Bacteria
2.5. Optimization of the Parameters for PCL Degradation
2.6. Esterase Assay
2.7. Estimation of the Bacterial Biomass
2.8. Estimation of the Gravimetric Weight
2.9. Gel Permeation Chromatography
2.10. Scanning Electron Microscopy (SEM)
3. Results
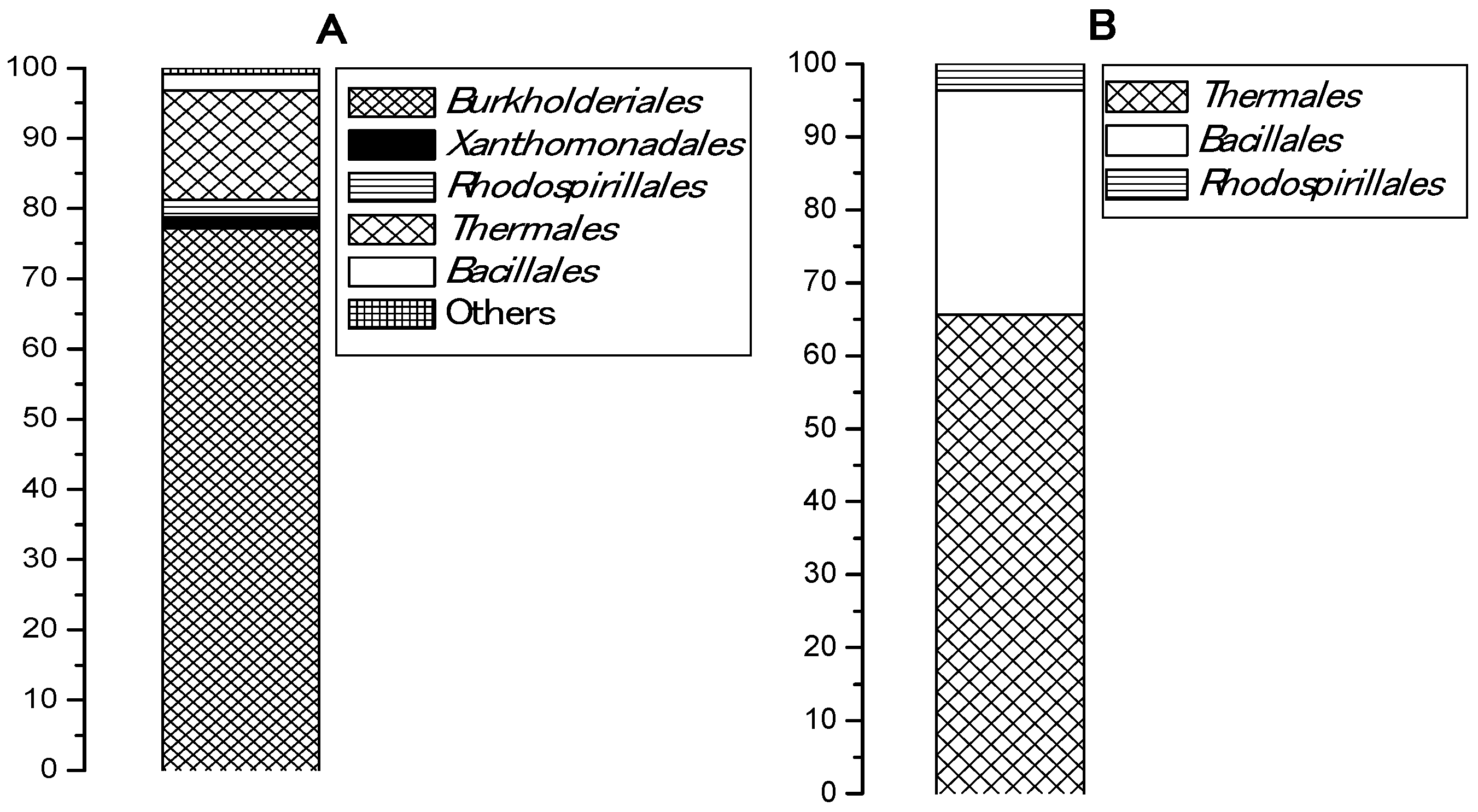
3.1. Phylogenetic Diversity in Samples Cultivated with and without Plastic
3.2. Isolation of PCL Degrading Bacteria
3.3. Optimization of Physico-Chemical Parameters for PCL Degradation
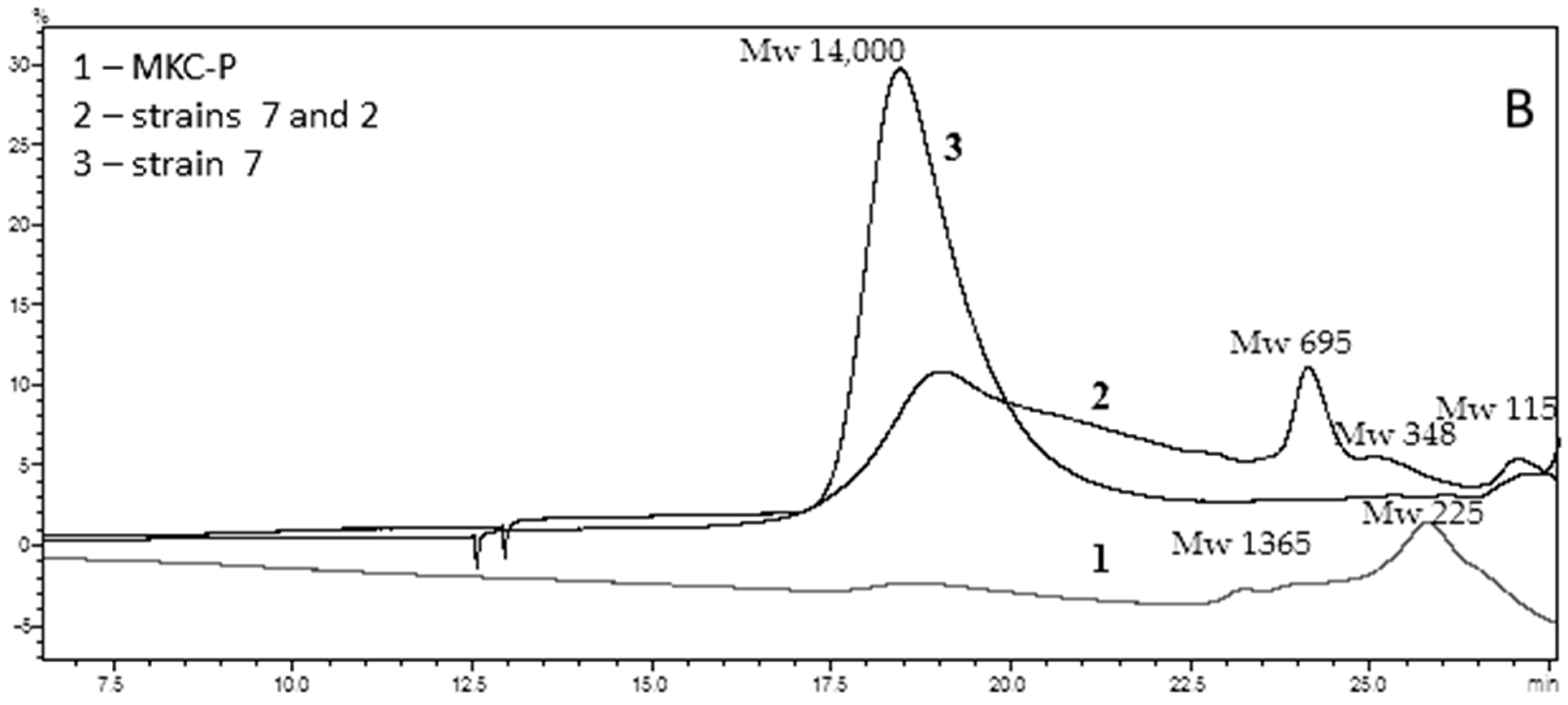
3.4. Characteristics of PCL Biodegradation Process
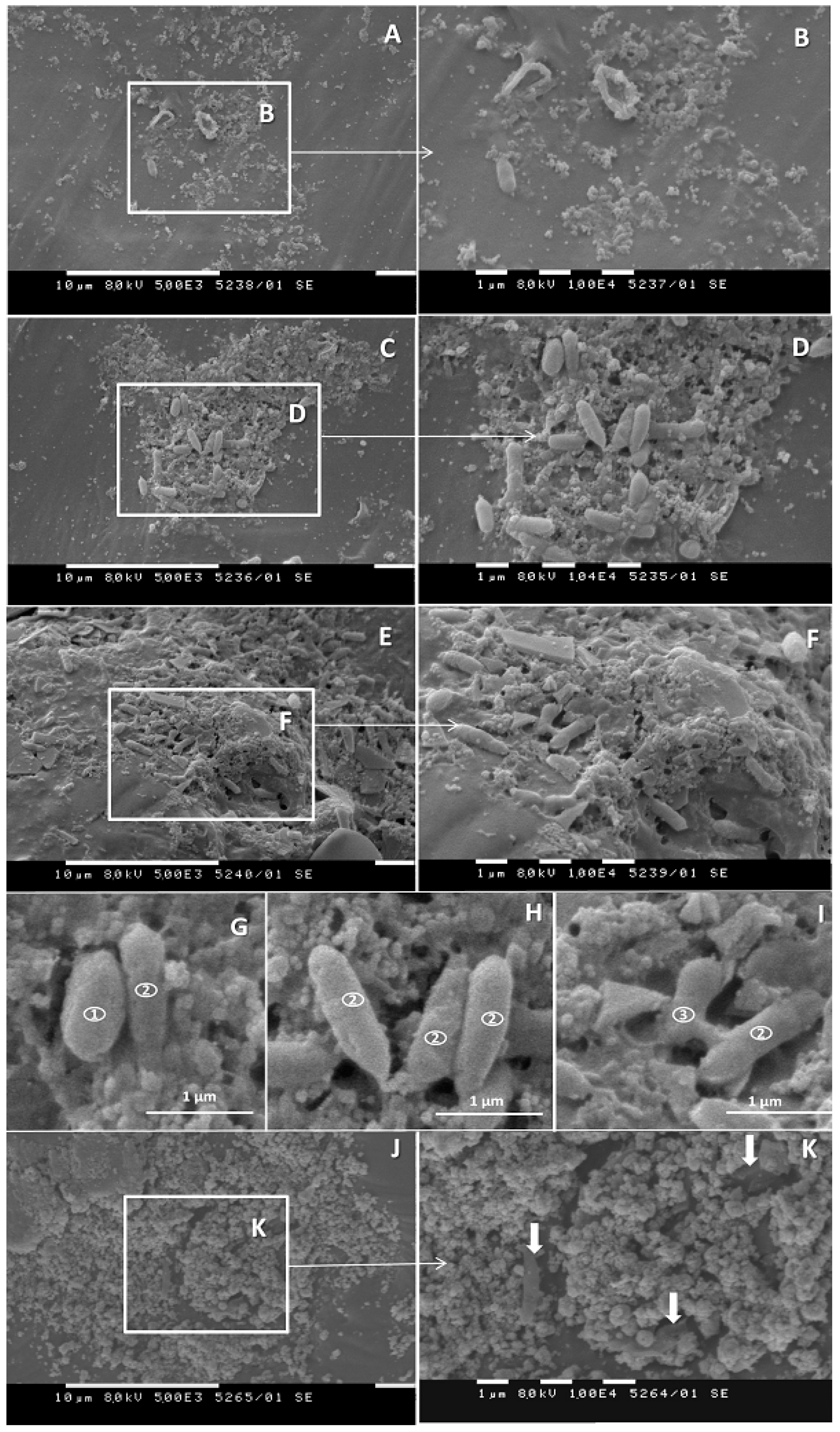
3.5. SEM Investigations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Plastics Europe. Plastics—The Facts 2020: An Analysis of European Plastics Production, Demand and Waste Data. 2020. Available online: https://www.plasticseurope.org/application/files/3416/2270/7211/Plastics_the_facts-WEB-2020_versionJun21_final.pdf (accessed on 27 September 2021).
- Polycaprolactone Market Analysis. Coherent Market Insights. 2020. Available online: https://www.coherentmarketinsights.com/market-insight/polycaprolactone-market-3553 (accessed on 28 April 2021).
- Global Market Study on Polycaprolactone: Adoption of Biodegradable Polymers Enabling Increasing Usage of Polycaprolactone. Persistence Market Research. Available online: https://www.persistencemarketresearch.com/market-research/polycaprolactone-market.asp (accessed on 28 April 2021).
- Pathak, V.M. Review on the current status of polymer degradation: A microbial approach. Bioresour. Bioprocess. 2017, 4, 15. [Google Scholar] [CrossRef]
- Gajanand, E.; Soni, L.K.; Dixit, V.K. Biodegradable polymers: A smart strategy for today’s crucial needs. Crit. Rev. Pharm. Sci. 2014, 3, 1–70. [Google Scholar]
- Debroas, D.; Mone, A.; Ter Halle, A. Plastics in the North Atlantic garbage patch: A boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 2017, 599–600, 1222–1232. [Google Scholar] [CrossRef]
- Sivan, A. New perspectives in plastic biodegradation. Curr. Opin. Biotechnol. 2011, 22, 422–426. [Google Scholar] [CrossRef]
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742. [Google Scholar] [CrossRef]
- Sekiguchi, T.; Saika, A.; Nomura, K.; Watanabe, T.; Fujimoto, Y.; Enoki, M.; Sato, T.; Kato, C.; Kanehiro, H. Biodegradation of aliphatic polyesters soaked in deep seawaters and isolation of poly (ɛ-caprolactone)-degrading bacteria. Polym. Degrad. Stab. 2011, 96, 1397–1403. [Google Scholar] [CrossRef]
- Oda, Y.; Oida, N.; Urakami, T.; Tonomura, K. Polycaprolactone depolymerase produced by the bacterium Alcaligenes faecalis. FEMS Microbiol. Lett. 1997, 152, 339–343. [Google Scholar] [CrossRef]
- Motiwalla, M.J.; Punyarthi, P.P.; Mehta, M.K.; D’Souza, J.S.; Kelkar-Mane, V. Studies on degradation efficiency of polycaprolactone by a naturally occurring bacterium. J. Environ. Biol. 2013, 34, 43–49. [Google Scholar]
- Abou-Zeid, D.M.; Muller, R.J.; Deckwer, W.D. Degradation of natural and synthetic polyesters under anaerobic conditions. J. Biotechnol. 2001, 86, 113–126. [Google Scholar] [CrossRef]
- Pinto, M.; Langer, T.M.; Hüffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, T.; Shahid, M.; Azeem, F.; Rasul, I.; Shah, A.A.; Noman, M.; Hameed, A.; Manzoor, N.; Manzoor, I.; Muhammad, S. Biodegradation of plastics: Current scenario and future prospects for environmental safety. Environ. Sci. Pollut. Res. 2018, 25, 7287–7298. [Google Scholar] [CrossRef]
- Balkova, R.; Hermanova, S.; Voberkova, S.; Damborsky, P.; Richtera, L.; Omelkova, J.; Jancar, J. Structure and morphology of microbial degraded poly (ε-caprolactone)/graphite oxide composite. J. Polym. Environ. 2014, 22, 190–199. [Google Scholar] [CrossRef]
- Balani, K.; Verma, V.; Agarwal, A.; Narayan, R. Physical, thermal, and mechanical properties of polymers. In Biosurfaces: A Materials Science and Engineering Perspective; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 329–344. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, T.C.H.; Dang, D.T.; Thai, H.; Nguyen, T.C.; Tran, T.T.H.; Le, V.H.; Nguyen, V.H.; Tran, H.B.; Pham, T.P.T.; Nguyen, T.G.; et al. Plastic degradation by thermophilic Bacillus sp. BCBT21 isolated from composting agricultural residual in Vietnam. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 015014. [Google Scholar] [CrossRef]
- Hadad, D.; Geresh, S.; Sivan, A. Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J. Appl. Microbiol. 2005, 98, 1093–1100. [Google Scholar] [CrossRef]
- Mahdi, M.S.; Ameen, R.S.; Ibrahim, H.K. Study on degradation of nylon 6 by thermophilic bacteria Anoxybacillus rupiensis Ir3 (JQ912241). Int. J. Adv. Res. Biol. Sci. 2016, 3, 200–209. [Google Scholar] [CrossRef]
- Tomita, K.; Hayashi, N.; Ikeda, N.; Kikuchi, Y. Isolation of a thermophilic bacterium degrading some nylons. Polym. Degrad. Stab. 2003, 81, 511–514. [Google Scholar] [CrossRef]
- Nakasaki, K.; Matsuura, H.; Tanaka, H.; Sakai, T. Synergy of two thermophiles enables decomposition of poly-ɛ-caprolactone under composting conditions. FEMS Microbiol. Ecol. 2006, 58, 373–383. [Google Scholar] [CrossRef]
- Chua, T.K.; Tseng, M.; Yang, M.K. Degradation of Poly (ε-caprolactone) by thermophilic Streptomyces thermoviolaceus subsp. thermoviolaceus 76T-2. AMB Express 2013, 3, e8. [Google Scholar] [CrossRef] [Green Version]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic degradation by extremophilic bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef]
- Almeida, E.L.; Carrillo Rincón, A.F.; Jackson, S.A.; Dobson, A.D. In silico screening and heterologous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone (PCL)-degrading activity, from the marine sponge-derived strain Streptomyces sp. SM14. Front. Microbiol. 2019, 10, 2187. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Rathi, P.; Gupta, R. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 2002, 311, 98–99. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef] [PubMed]
- Dussud, C.; Meistertzheim, A.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M.; et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef]
- Tourova, T.; Sokolova, D.; Nazina, T.; Grouzdev, D.; Kurshev, E.; Laptev, A. Biodiversity of microorganisms colonizing the surface of polystyrene samples exposed to different aqueous environments. Sustainability 2020, 12, 3624. [Google Scholar] [CrossRef]
- Panda, A.K.; Bisht, S.S.; DeMondal, S.; Kumar, N.S.; Gurusubramanian, G.; Panigrahi, A.K. Brevibacillus as a biological tool: A short review. Antonie Van Leeuwenhoek 2014, 105, 623–639. [Google Scholar] [CrossRef]
- Pramila, R.; Kesavaram, P.; Vijaya, R.; Krishnan, M. Brevibacillus parabrevis, Acinetobacter baumannii and Pseudomonas citronellolis-potential candidates for biodegradation of low density polyethylene (LDPE). J. Bacteriol. Res. 2012, 4, 9–14. [Google Scholar] [CrossRef]
- Yan, F.; Wei, R.; Cui, Q.; Bornscheuer, U.T.; Liu, Y.J. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2021, 14, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.-J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.-D. Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Comm. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Urbanek, A.K.; Rymowicz, W.; Mirończuk, A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 2018, 102, 7669–7678. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Qureshi, A.; Purohit, H. Microbial degradation of plastics: Biofilms and degradation pathways. In Contaminants in Agriculture and Environment: Health Risks and Remediation; Agro Environ Media: Haridwar, India, 2019; Volume 1, pp. 184–199. [Google Scholar] [CrossRef]
- Skariyachan, S.; Manjunatha, V.; Sultana, S.; Jois, C.; Bai, V.; Vasist, K.S. Novel bacterial consortia isolated from plastic garbage processing areas demonstrated enhanced degradation for low density polyethylene. Environ. Sci. Pollut. Res. Int. 2016, 23, 18307–18319. [Google Scholar] [CrossRef]
- Yu, Y.; Li, H.; Zeng, Y.; Chen, B. Extracellular enzymes of cold-adapted bacteria from Arctic sea ice, Canada Basin. Polar Biol. 2009, 32, 1539–1547. [Google Scholar] [CrossRef]
- Singh, P.; Singh, S.M.; Dhakephalkar, P. Diversity, cold active enzymes and adaptation strategies of bacteria inhabiting glacier cryoconite holes of High Arctic. Extremophiles 2014, 18, 229–242. [Google Scholar] [CrossRef]
- Castonguay, M.H.; van der Schaaf, S.; Koester, W.; Krooneman, J.; van der Meer, W.; Harmsen, H.; Landini, P. Biofilm formation by Escherichia coli is stimulated by synergistic interactions and co-adhesion mechanisms with adherence-proficient bacteria. Res. Microbiol. 2006, 157, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Madsen, J.S.; Sørensen, S.J.; Burmølle, M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015, 9, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Zupančič, J.; Raghupathi, P.K.; Houf, K.; Burmølle, M.; Sørensen, S.J.; Gunde-Cimerman, N. Synergistic Interactions in microbial biofilms facilitate the establishment of opportunistic pathogenic fungi in household dishwashers. Front. Microbiol. 2018, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Uhlich, G.A.; Rogers, D.P.; Mosier, D.A. Escherichia coli serotype O157:H7 retention on solid surfaces and peroxide resistance is enhanced by dual-strain biofilm formation. Foodborne Pathog. 2010, 7, 935–943. [Google Scholar] [CrossRef]
- Vacheva, A.; Ivanova, R.; Stoitsova, S. Stimulation of sessile growth of a biofilm-deficient strain Escherichia coli K-12 by protein(s) secreted by Enterobacteriaceae species. Biotechnol. Biotechnol. Eq. 2011, 25, 88–91. [Google Scholar] [CrossRef]
- Vacheva, A.; Ivanova, R.; Paunova-Krasteva, T.; Stoitsova, S. Released products of pathogenic bacteria stimulate biofilm formation by Escherichia coli K-12 strains. Antonie Van Leeuwenhoek 2012, 102, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.A.; Anjana, K.; Hinduja, M.; Sujitha, K.; Dharani, G. Review on plastic wastes in marine environment-Biodegradation and biotechnological solutions. Mar. Pollut. Bull. 2020, 150, 110733. [Google Scholar] [CrossRef]
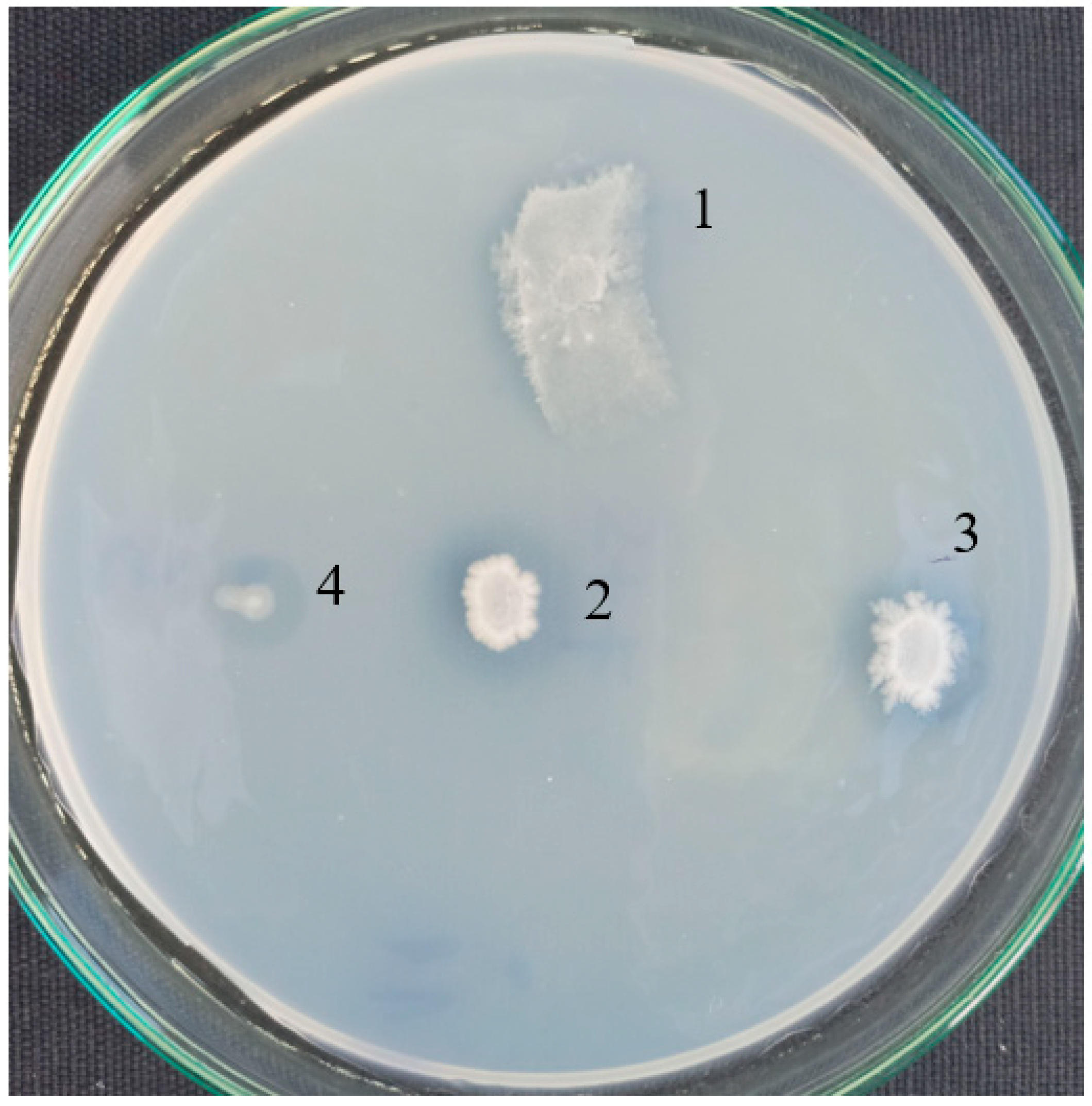
 , MKC-P;
, MKC-P;  , B. thermoruber strain 7;
, B. thermoruber strain 7;  , co-culture of strain 7 and A. thermoaerophilus strain 2.
, co-culture of strain 7 and A. thermoaerophilus strain 2.
 , MKC-P;
, MKC-P;  , B. thermoruber strain 7;
, B. thermoruber strain 7;  , co-culture of strain 7 and A. thermoaerophilus strain 2.
, co-culture of strain 7 and A. thermoaerophilus strain 2.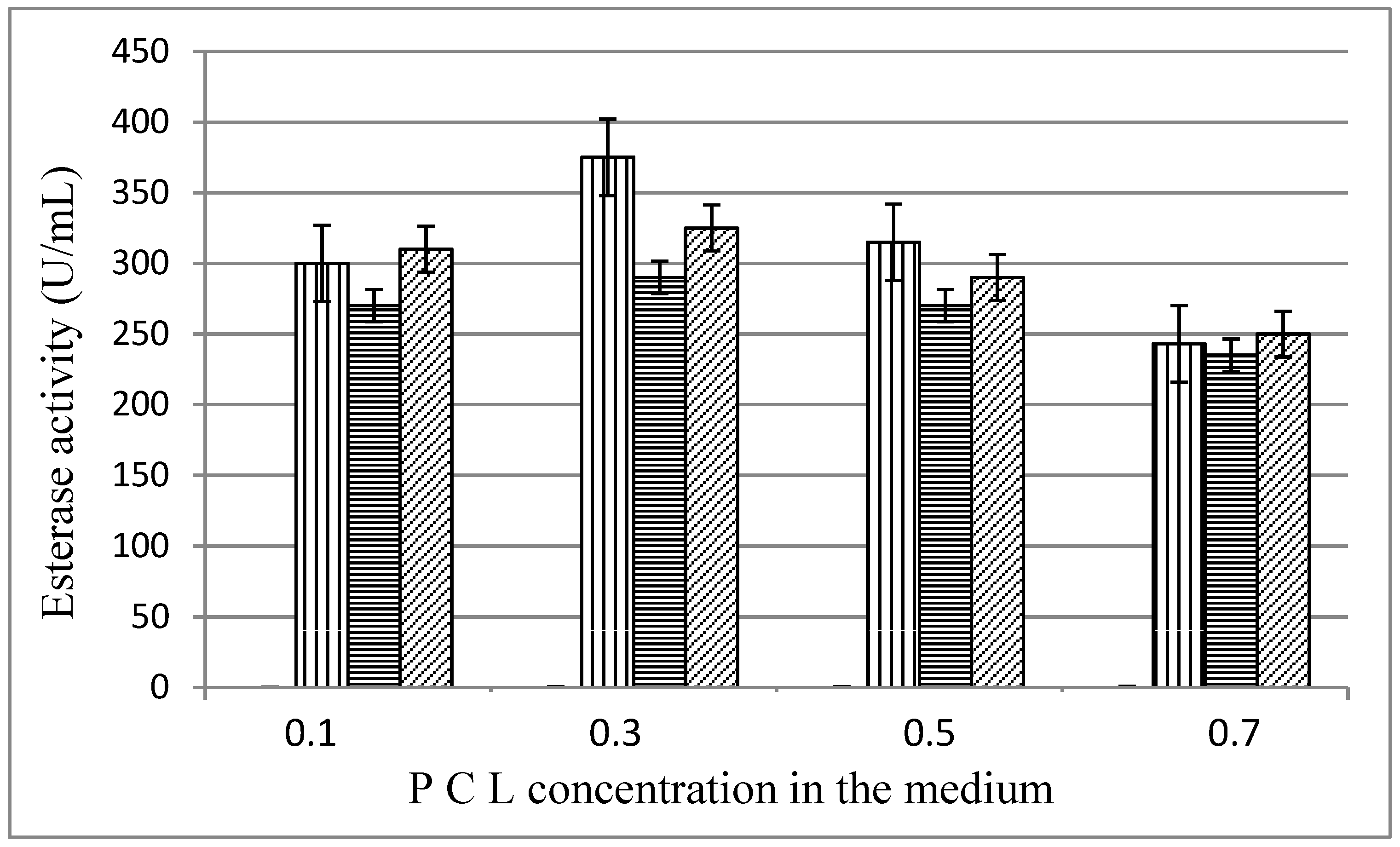


| Identified Group | Phylogenetic Level | Reads, % |
|---|---|---|
| Caldimonas sp. | Phylum Proteobacteria, Order Burkholderiales | 42.2 |
| Tepidimonas ignava | Phylum Proteobacteria, Order Burkholderiales | 21.0 |
| Meiothermus sp. | Phylum Deinococcus-Thermus, Order Thermales | 14.5 |
| Tepidimonas sp. | Phylum Proteobacteria Order Burkholderiales | 13.7 |
| Bacillales | Phylum Firmicutes, Order Bacillales | 2.3 |
| Thermomonas sp. | Phylum Proteobacteria, Order Xanthomonadales | 1.5 |
| Elioraea tepidiphila | Phylum Proteobacteria Order Rhodospirillales | 1.1 |
| Thermaceae | Phylum Deinococcus-Thermus, Order Thermales | 1.1 |
| Roseomonas | Class Alphaproteobacteria Order Rhodospirillales Family Acetobacteraceae | 0.9 |
| Acetobacteraceae | Class Alphaproteobacteria Order: Rhodospirillales | 0.5 |
| Burkholderiales | Phylum Proteobacteria Order: Burkholderiales | 0.3 |
| Identified Group | Phylogenetic Affiliation | Reads (%) |
|---|---|---|
| Meiothermus | Phylum Deinococcus-Thermus, Order Thermales | 58.1 |
| Brevibacillus | Phylum Firmicutes Order Bacillales Family Paenibacillaceae | 18.5 |
| Bacillales | Phylum Firmicutes, Order Bacillales | 11.6 |
| Thermaceae | Phylum Deinococcus-Thermus, Order Thermales | 7.5 |
| Elioraea tepidiphila | Phylum Proteobacteria Order Rhodospirillales | 3.7 |
| Paenibacillus | Phylum Firmicutes Order Bacillales | 0.4 |
| Bacillaceae | Phylum Firmicutes | 0.2 |
| Highest-Homology Organism (Maximum % Identity of 16S rRNA Gene Sequence) | Sequence Number | Esterase Activity of Pure Strains (U/mL) | Esterase Activity in Co-Cultures (U/mL) with B. thermoruber |
|---|---|---|---|
| Aneurinibacillus thermoaerophilus strain 2 | MW927323 | 115 | 325 |
| Brevibacillus thermoruber strain 7 | MW541896 | 290 | - |
| Meiothermus cateniformans strain 12 | MW927332 | 42 | 70 |
| MKC-P | PRJNA766622 | 375 |
| Microorganisms | Week | Final Weight | Weight Loss per 7 Days (mg) | PCL Degradation Rate (mg/d) | Weight Loss per 7 Days (%) | Protein Contents (mg/mL) | Esterase Assay U/mL | ||
|---|---|---|---|---|---|---|---|---|---|
| Protein in Free Cells | Protein in Biofilms | Total Protein | |||||||
| Control | 4 | 150 ± 9 | 0 | 0 | 0 | 0 | - | 0 | 0 |
| Community | 1 | 88.2 | 61.8 | 8.83 | 41.2 | 0.23 | 1.31 | 1.54 | 375 |
| 2 | 48.8 | 39.4 | 5.63 | 26.3 | 0.45 | 1.56 | 2.01 | 375 | |
| 3 | 20.1 | 28.7 | 4.1 | 19.1 | 0.63 | 1.39 | 2.02 | 333.8 | |
| 4 | 0 | 20.1 | 2.87 | 13.4 | 0.17 | 0.84 | 1.01 | 291.6 | |
| B. thermoruber strain 7 | 1 | 134.7 | 15.3 | 2.18 | 10.2 | 0.75 | 0.05 | 0.80 | 290 |
| 2 | 115.2 | 19.5 | 2.78 | 13 | 0.78 | 0.15 | 0.93 | 290 | |
| 3 | 84.2 | 31.0 | 4.42 | 20.6 | 0.84 | 0.19 | 1.03 | 258 | |
| 4 | 54.5 | 29.7 | 4.24 | 19.8 | 0.11 | 0.14 | 0.25 | 125 | |
| B. thermoruber strain 7 + A. thermoaerophilus strain 2 | 1 | 125 | 25 | 3.57 | 16.7 | 0.82 | 0.13 | 0.95 | 325 |
| 2 | 96.8 | 28.2 | 4.02 | 18.8 | 1.06 | 0.19 | 1.25 | 308 | |
| 3 | 67.6 | 29.2 | 4.17 | 19.5 | 1.18 | 0.19 | 1.37 | 280 | |
| 4 | 37.7 | 29.9 | 4.27 | 19.9 | 0.72 | 0.17 | 0.89 | 248 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atanasova, N.; Paunova-Krasteva, T.; Stoitsova, S.; Radchenkova, N.; Boyadzhieva, I.; Petrov, K.; Kambourova, M. Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring. Biomolecules 2021, 11, 1488. https://doi.org/10.3390/biom11101488
Atanasova N, Paunova-Krasteva T, Stoitsova S, Radchenkova N, Boyadzhieva I, Petrov K, Kambourova M. Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring. Biomolecules. 2021; 11(10):1488. https://doi.org/10.3390/biom11101488
Chicago/Turabian StyleAtanasova, Nikolina, Tsvetelina Paunova-Krasteva, Stoyanka Stoitsova, Nadja Radchenkova, Ivanka Boyadzhieva, Kaloyan Petrov, and Margarita Kambourova. 2021. "Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring" Biomolecules 11, no. 10: 1488. https://doi.org/10.3390/biom11101488
APA StyleAtanasova, N., Paunova-Krasteva, T., Stoitsova, S., Radchenkova, N., Boyadzhieva, I., Petrov, K., & Kambourova, M. (2021). Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring. Biomolecules, 11(10), 1488. https://doi.org/10.3390/biom11101488







