Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review
Abstract
1. Introduction
2. Choice of Metabolomics Techniques, Study Design and Bio-Sample Handling to Conduct Metabolomics Study to Understand Metabolic Alterations by NSAIDs
2.1. Metabolomics and Metabolic Fingerprinting Techniques
2.2. Study Design of the Metabolomics Experiment in NSAIDs Based Toxicology Study
2.3. Sample Preparations and Storage for Metabolomics Experiment
3. Metabolomics of Murine Response toward NSAIDs Administration
3.1. Lipidomics Signature in NSAIDs Administration
3.2. Metabolic Fingerprint in Urine by NSAIDs Administration
3.3. Metabolic Signatures in Serum/Plasma Induced by NSAIDs
3.4. Metabolic Signatures in Stomach Induced by NSAIDs
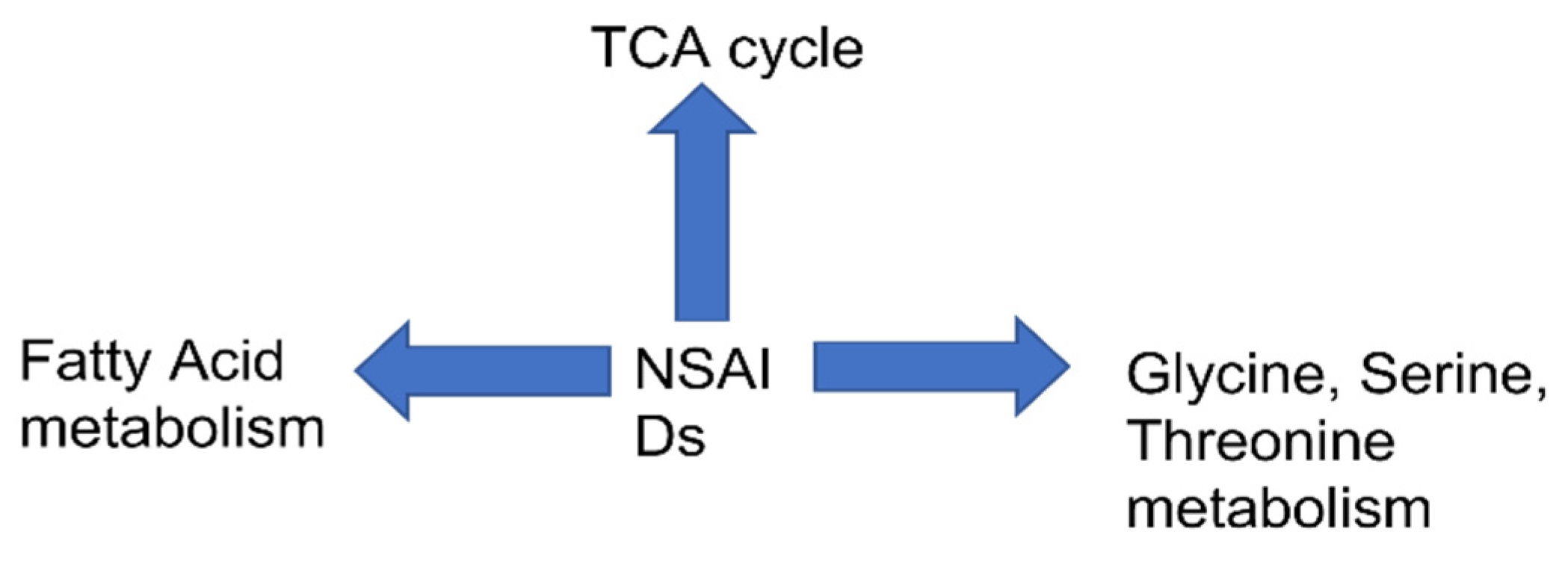
3.5. Pathway Analysis
4. Discussion and Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larive, C.K.; Gregory, A.; Barding, J.; Dinges, M.M. NMR Spectroscopy for Metabolomics and Metabolic Profiling. Anal. Chem. 2014, 87, 133–146. [Google Scholar] [CrossRef]
- Liu, X.; Locasale, J.W. Metabolomics—A primer. Trends Biochem. Sci. 2017, 42, 274. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, I.; Scarpignato, C.; Holmgren, E.; Olszewiski, M.; Raindford, K.D.; Lanas, A. Mechanisms of Damage to the Gastrointestinal Tract From Nonsteroidal Anti-Inflammatory Drugs. Gastroenterology 2018, 154, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Bombardier, C.; Laine, L.; Reicin, A.; Shapiro, D.; Vargas-Burgos, R.; Davis, B.; Day, R.; Ferraz, M.B.; Hawkey, C.J.; Hochberg, M.C.; et al. Comparison of upper gastrointestinal toxicity of rofecoxib and naproxen in patients with rheumatoid arthritis. N. Engl. J. Med. 2000, 343, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Grosser, T.; Ricciotti, E.; FitzGerald, G.A. The Cardiovascular P–harmacology of Nonsteroidal Anti-Inflammatory Drugs. Trends Pharm. Sci. 2017, 38, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Larkai, E.N.; Smith, J.L.; Lidsky, M.D.; Graham, D.Y. Gastroduodenal mucosa and dyspeptic symptoms in arthritic patients during chronic nonsteroidal anti-inflammatory drug use. Am. J. Gastroenterol. 1987, 82, 1153–1158. [Google Scholar]
- Sostres, C.; Gargallo, C.J.; Lanas, A. Nonsteroidal anti-inflammatory drugs and upper and lower gastrointestinal mucosal damage. Arthritis Res. 2013, 15, S3. [Google Scholar] [CrossRef] [PubMed]
- Tai, F.W.D.; McAlindon, M.E. NSAIDs and the small bowel. Curr. Opin. Gastroenterol. 2018, 34, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.G. NSAIDs and increased blood pressure. What is the clinical significance? Drug Saf. 1997, 17, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Li, N.; Yang, J.; Li, N.; Qiu, H.; Ai, D.; Chiamvimonvat, N.; Zhu, Y.; Hammock, B.D. Metabolic profiling of murine plasma reveals an unexpected biomarker in rofecoxib-mediated cardiovascular events. Proc. Natl. Acad. Sci. USA 2010, 107, 17017–17022. [Google Scholar] [CrossRef]
- Meek, I.L.; van de Laar, M.A.; Vonkeman, H.E. Non-Steroidal Anti-Inflammatory Drugs: An Overview of Cardiovascular Risks. Pharmaceuticals 2010, 3, 2146. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic Phenotyping in Health and Disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Ellis, D.I. Metabolomics: Current analytical platforms and methodologies. TrAC Trends Anal. Chem. 2005, 24, 285–294. [Google Scholar] [CrossRef]
- Sengupta, A.; Ghosh, S.; Sharma, S.; Sonawat, H.M. 1H NMR metabonomics indicates continued metabolic changes and sexual dimorphism post-parasite clearance in self-limiting murine malaria model. PLoS ONE 2013, 8, e66954. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robinson, J.I.; Weir, W.H.; Crowley, J.R.; Hink, T.; Reske, K.A.; Kwon, J.H.; Burnham, C.D.; Dubberke, E.R.; Mucha, P.J.; Henderson, J.P. Metabolomic networks connect host-microbiome processes to human Clostridioides difficile infections. J. Clin. Investig. 2019, 129, 3792–3806. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.N.; Raftery, D. Can NMR solve some significant challenges in metabolomics? J. Magn. Reson. 2015, 260, 144–160. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sengupta, A.; Chandra, K. Quantitative metabolic profiling of NMR spectral signatures of branched chain amino acids in blood serum. Amin. Acids 2015, 47, 2229–2236. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; González-Mariscal, I.; Egan, J.M.; Moaddel, R. Targeted proteomics of cannabinoid receptor CB1 and the CB1b isoform. J. Pharm. Biomed. Anal. 2017, 144, 154–158. [Google Scholar] [CrossRef]
- Merrell, K.; Southwick, K.; Graves, S.W.; Esplin, M.S.; Lewis, N.E.; Thulin, C.D. Analysis of Low-Abundance, Low-Molecular-Weight Serum Proteins Using Mass Spectrometry. J. Biomol. Tech. 2004, 15, 238. [Google Scholar]
- Du, X.; Zeisel, S.H. Spectral deconvolution for gas chromatography mass spectrometry-based metabolomics: Current status and future perspectives. Comput. Struct. Biotechnol. J. 2013, 4, e201301013. [Google Scholar] [CrossRef] [PubMed]
- Ravanbakhsh, S.; Liu, P.; Bjordahl, T.C.; Mandal, R.; Grant, J.R.; Wilson, M.; Eisner, R.; Sinelnikov, I.; Hu, X.; Luchinat, C.; et al. Accurate, Fully-Automated NMR Spectral Profiling for Metabolomics. PLoS ONE 2015, 10, e0124219. [Google Scholar] [CrossRef] [PubMed]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Catchpole, G.S.; Beckmann, M.; Enot, D.P.; Mondhe, M.; Zywicki, B.; Taylor, J.; Hardy, N.; Smith, A.; King, R.D.; Kell, D.B.; et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA 2005, 102, 14458–14462. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sengupta, A.; Sharma, S.; Sonawat, H.M. Metabolic fingerprints of serum, brain, and liver are distinct for mice with cerebral and noncerebral malaria: A 1H NMR spectroscopy-based metabonomic study. J. Proteome Res. 2012, 11, 4992–5004. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sengupta, A.; Sharma, S.; Sonawat, H.M. Metabolic Perturbations of Kidney and Spleen in Murine Cerebral Malaria: 1H NMR-Based Metabolomic Study. PLoS ONE 2013, 8, e73113. [Google Scholar] [CrossRef][Green Version]
- Sengupta, A.; Ghosh, S.; Pathak, S.; Gogtay, N.; Thatte, U.; Doshi, M.; Sharma, S.; Sonawat, H.M. Metabolomic analysis of urine samples of vivax malaria in-patients for biomarker identification. Metabolomics 2015, 11, 1351–1362. [Google Scholar] [CrossRef]
- Ghosh, S.; Sengupta, A.; Sharma, S.; Sonawat, H.M. Early prediction of cerebral malaria by (1)H NMR based metabolomics. Malar. J. 2016, 15, 198. [Google Scholar] [CrossRef] [PubMed]
- Pemmari, A.; Tuure, L.; Hämäläinen, M.; Leppänen, T.; Vuolteenaho, K.; Moilanen, T.; Moilanen, E. Comprehensive effects of ibuprofen on gene expression in chondrocytes as determined by RNA-Seq. Osteoarthr. Cart. 2019, 27, S378. [Google Scholar] [CrossRef]
- Gonçalves, V.; Henriques, A.F.; Matos, P.; Jordan, P. Ibuprofen disrupts a WNK1/GSK3β/SRPK1 protein complex required for expression of tumor-related splicing variant RAC1B in colorectal cells. Oncotarget 2020, 11, 4421–4437. [Google Scholar] [CrossRef]
- So, Y.U.; Myeon, W.C.; Kim, K.B.; Seon, H.K.; Ji, S.O.; Hye, Y.O.; Hwa, J.L.; Ki, H.C. Pattern recognition analysis for the prediction of adverse effects by nonsteroidal anti-inflammatory drugs using 1H NMR-based metabolomics in rats. Anal. Chem. 2009, 81, 4734–4741. [Google Scholar]
- Smith, L.; Villaret-Cazadamont, J.; Claus, S.P.; Canlet, C.; Guillou, H.; Cabaton, N.J.; Ellero-Simatos, S. Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites 2020, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Golbs, A.; Neuber, S.; Kamlage, B.; Christiansen, N.; Bethan, B.; Rennefahrt, U.; Schatz, P.; Lars, L. Effects of Long-Term Storage at -80 °C on the Human Plasma Metabolome. Metabolites 2019, 9, 99. [Google Scholar] [CrossRef]
- Hyötyläinen, T. Critical evaluation of sample pretreatment techniques. Anal. Bioanal. Chem. 2009, 394, 743–758. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Thongboonkerd, V.; Saetun, P. Bacterial Overgrowth Affects Urinary Proteome Analysis: Recommendation for Centrifugation, Temperature, Duration, and the Use of Preservatives during Sample Collection. J. Proteome Res. 2007, 6, 4173–4181. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.-Y.; Timofeyev, V.; Qiu, H.; Hwang, S.H.; Tuteja, D.; Lu, L.; Yang, J.; Mochida, H.; Low, R.; et al. Beneficial Effects of Soluble Epoxide Hydrolase Inhibitors in Myocardial Infarction Model: Insight Gained Using Metabolomic Approaches. J. Mol. Cell. Cardiol. 2009, 47, 835. [Google Scholar] [CrossRef] [PubMed]
- <monospace>Rivera-Velez, S.M.; Broughton-Neiswanger, L.E.; Suarez, M.; Piñeyro, P.; Navas, J.; Chen, S.; Hwang, J.; Villarino, N.F. Repeated administration of the NSAID meloxicam alters the plasma and urine lipidome. Sci. Rep. 2019, 9, 4303. [Google Scholar]
- Um, S.Y.; Chung, M.W.; Kim, K.-B.; Kim, S.H.; Oh, J.S.; Oh, H.Y.; Lee, H.J.; Choi, K.H. Pattern recognition analysis for the prediction of adverse effects by nonsteroidal anti-inflammatory drugs using 1H NMR-based metabolomics in rats. Anal. Chem. 2009, 81, 4734–4741. [Google Scholar] [CrossRef]
- Lv, H.; Liu, L.; Palacios, G.; Chen, X. Metabolomic analysis characterizes tissue specific indomethacin-induced metabolic perturbations of rats. Analyst 2011, 136, 2260–2269. [Google Scholar] [CrossRef]
- Jung, J.; Park, M.; Park, H.J.; Shim, S.B.; Cho, Y.H.; Kim, J.; Lee, H.S.; Ryu, D.H.; Choi, D.; Hwang, G.S. 1H NMR-based metabolic profiling of naproxen-induced toxicity in rats. Toxicol. Lett. 2011, 200, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Ohishi, M.; Ota, S.; Suzumura, K.; Naraoka, H.; Ohata, T.; Seki, J.; Miyamae, Y.; Honma, M.; Soga, T. Metabolic profiling to identify potential serum biomarkers for gastric ulceration induced by nonsteroid anti-inflammatory drugs. J. Proteome Res. 2013, 12, 1399–1407. [Google Scholar] [CrossRef] [PubMed]
- Yekta, R.F.; Amiri-Dashatan, N.; Koushki, M.; Dadpay, M.; Goshadrou, F. A Metabolomic Study to Identify Potential Tissue Biomarkers for Indomethacin-Induced Gastric Ulcer in Rats. Avicenna J. Med. Biotechnol. 2019, 11, 299. [Google Scholar]
- Zhang, J.; Song, H.; Jiang, S.; Chen, Z.; Tong, S.; Lin, F.; Wen, C.; Zang, X.; Hu, L. Fisher Discrimination of Metabolic Changes in Rats Treated with Aspirin and Ibuprofen. Pharmacology 2017, 100, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.Y.; Hong, Q.; Harris, T.R.; Sirish, P.; Hammack, B.D.; Chiamvimonvat, N. Use of metabolomic profiling in the study of arachidonic acid metabolism in cardiovascular disease. Congest. Heart Fail. 2011, 17, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Lanza, F.L.; Royer, G.L., Jr.; Nelson, R.S.; Chen, T.T.; Seckman, C.E.; Rack, R.M. The effects of ibuprofen, indomethacin, aspirin, naproxen, and placebo on the gastric mucosa of normal volunteers: A gastroscopic and photographic study. Dig. Dis. Sci. 1979, 24, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.G.A.; Purmalis, A.; Vandermeer, D.A.; Denlinger, R.H. The propionic acids. Gastrointestinal toxicity in various species. Toxicol. Pathol. 1988, 16, 245–250. [Google Scholar] [CrossRef]
- Narabayashi, K.; Ito, Y.; Eid, N.; Maemura, K.; Inoue, T.; Takeuchi, T.; Otsuki, Y.; Higuchi, K. Indomethacin suppresses LAMP-2 expression and induces lipophagy and lipoapoptosis in rat enterocytes via the ER stress pathway. J. Gastroenterol. 2015, 50, 541–554. [Google Scholar] [CrossRef]
- Franceschelli, S.; Moltedo, O.; Amodio, G.; Tajana, G.; Romomdelli, P. In the Huh7 Hepatoma Cells Diclofenac and Indomethacin Activate Differently the Unfolded Protein Response and Induce ER Stress Apoptosis. Open Biochem. J. 2011, 5, 45–51. [Google Scholar] [CrossRef]
- Petrescu, I.; Tarba, C. Uncoupling effects of diclofenac and aspirin in the perfused liver and isolated hepatic mitochondria of rat. Biochim. Biophys. Acta 1997, 1318, 385–394. [Google Scholar] [CrossRef]
- Moreno-Sánchez, R.; Bravo, C.; Vásquez, C.; Ayala, G.; Silveira, L.H.; Martínez-Lavín, M. Inhibition and uncoupling of oxidative phosphorylation by nonsteroidal anti-inflammatory drugs: Study in mitochondria, submitochondrial particles, cells, and whole heart. Biochem. Pharm. 1999, 57, 743–752. [Google Scholar] [CrossRef]
- Sanabria, E.-H.; Heiremans, E.; Arroyo, M.C.; Props, R.; Leclercq, L.; Snoeys, J.; de Wiele, T.V. Short-term supplementation of celecoxib-shifted butyrate production on a simulated model of the gut microbial ecosystem and ameliorated in vitro inflammation. NPJ Biofilms Microbiomes 2020, 6. [Google Scholar] [CrossRef]
- Rogers, M.A.; Aronoff, D.M. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin. Microbiol. Infect. 2016, 22, 178–e1. [Google Scholar] [CrossRef]

| Drugs | Biofluids/ Tissues | Dose | Timepoint | Species | Metabolites | Analytical Platform | Publications |
|---|---|---|---|---|---|---|---|
| Celecoxib | Urine | 133 mg/kg | 5 h | Rats | Citrate(dn), 2-oxoglutarate(dn), Acetate(dn), Hippurate(dn) | NMR | [31] |
| Indomethacin | Urine | 25 mg/kg | 5 h | Rats | Allantoin(up), Citrate(dn), 2-Oxoglutarate(dn), Taurine(dn), Hippurate(dn), Dimethylamine(up) | NMR | [31] |
| Ibuprofen | Urine | 800 mg/kg | 5 h | Rats | Allantoin(up), Citrate(dn), 2-Oxoglutarate(dn), Taurine(dn), Hippurate(dn), Dimethylamine(up) | NMR | [31] |
| Naproxen | Urine | 10 mg/kg-100 mg/kg | 7 h | Rats | Kynurenate (up), pantothenate(dn), citrate (up), creatine(up), creatine(up) phosphate (up), cis-aconitate (up), choline (dn), and betaine(up) | NMR | [41] |
| Indomethacin | Urine | 25 mg/kg | 0–24 h, 24–48 h, 48–72 h | Rats | Prostaglandin E2(dn), 2-methylcitric acid(dn), putreanine(up), (10E,12E)-9-hydroxyoctadeca-10, famotidine(dn), docosanamide(up), creatinine(up), pregnenolone(dn), palmitoleic acid(up), l-carnitine(dn), guanosine(dn), thiamine monophosphate(dn), d-ribulose 5-phosphate(up), nervonic acid(up), proline betaine(dn), spermine(up), 3 chlorotyrosine(up), 5-hydroxy-l-tryptophan(up), 15-keto-prostaglandin F2a(dn) and N1-acetylspermidine(dn). | LC/MS | [40] |
| Aspirin | Stomach | 300 mg/kg | 1 h | Rats | Citrate(dn), O-Acetylcarnitine(dn), 3hydroxybutanoic acid(dn), Proline(dn), Hydroxyproline(dn) | CE-TOF-MS | [42] |
| Aspirin | Stomach | 300 mg/kg | 5 h | Rats | Citrate(dn), cis-Aconitate(dn), succinate(dn) O-Acetylcarnitine(dn), 3hydroxybutanoic acid(dn), Hydroxyproline(dn) | CE-TOF-MS | [42] |
| Ibuprofen | Stomach | 800 mg/kg | 1 h | Rats | Citrate(dn), succinate(dn) O-Acetylcarnitine(dn), 3hydroxybutanoic acid(dn), Hydroxyproline(dn) | CE-TOF-MS | [42] |
| Ibuprofen | Stomach | 800 mg/kg | 5 h | Rats | Citrate(dn), cis-Aconitate(dn), succinate(dn) O-Acetylcarnitine(dn), proline(dn), Hydroxyproline(dn) | CE-TOF-MS | [42] |
| Aspirin | Serum | 300 mg/kg | 1 h | Rats | 3-Hydroxy butanoic acid(dn), proline(dn), hydroxyproline(dn) | CE-TOF-MS | [42] |
| Aspirin | Serum | 300 mg/kg | 5 h | Rats | Cis-Aconitate(dn), o acetyl carnitine(dn), 3-Hydroxy butanoic acid(dn), proline(dn), hydroxyproline(dn) | CE-TOF-MS | [42] |
| Ibuprofen | Serum | 800 mg/kg | 1 h | Rats | 3-Hydroxy butanoic acid(dn), | CE-TOF-MS | [42] |
| Ibuprofen | Serum | 800 mg/kg | 5 h | Rats | Succinate(dn), o-acetyl carnitine(dn), 3-Hydroxy butanoic acid(dn), proline(dn), hydroxyproline(dn) | CE-TOF-MS | [42] |
| Ibuprofen | Serum | 15 mg/kg | 1–3 week | Rats | Acetoacetic acid(nm), l-alanine(nm), Trihydroxybutyric acid(nm), Galacturonic acid(nm), Propanedioic acid(nm), Acetic acid(nm), Ethanedioic acid(nm), Galacturonic acid(nm), L-valine(nm), Mannonic acid(nm), Urea(nm), D-galactose(nm), Ethanedioic acid(nm), l-isoleucine(nm), Propanoic acid(nm), Butenoic acid(nm), d-glucose(nm), L-norvaline(nm), Hexanoic acid(nm), Acetamide(nm). | GC-MS | [44] |
| Aspirin | Serum | 15 mg/kg | 1–3 week | Rats | Trihydroxybutyric acid(nm), l-alanine(nm), Galacturonic acid(nm), D-galactose(nm), l-alanine(nm), Acetamide(nm), Propanedioic acid Acetoacetic acid(nm), Butanoic acid(nm), Arachidonic acid(nm), Ethanedioic acid(nm), L-tyrosine(nm), d-glucose(nm), Propanoic acid(nm), Hexanoic acid(nm). | GC-MS | [44] |
| Indomethacin | stomach | 45 mg/kg | 6 h | Rats | Choline(dn), Cis-aconitate(dn), Tryptophan(dn), Spermidine(dn), Trimethylamine(up), N,N-Dimethylglycine(dn), Acetylcarnitine(dn) Creatinine(dn), Pantothenate(dn), Betaine(dn), Carnitine(dn), Isoleucine(dn), Glucose(dn), Kynurenine(dn), Methionine(dn), Acetylcholine(up) | NMR | [43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, S. Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review. Biomolecules 2021, 11, 1456. https://doi.org/10.3390/biom11101456
Ghosh S. Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review. Biomolecules. 2021; 11(10):1456. https://doi.org/10.3390/biom11101456
Chicago/Turabian StyleGhosh, Soumita. 2021. "Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review" Biomolecules 11, no. 10: 1456. https://doi.org/10.3390/biom11101456
APA StyleGhosh, S. (2021). Metabolomic Studies for Metabolic Alterations Induced by Non-Steroidal Anti-Inflammatory Drugs: Mini Review. Biomolecules, 11(10), 1456. https://doi.org/10.3390/biom11101456





