Immunoprofiles and DNA Methylation of Inflammatory Marker Genes in Ulcerative Colitis-Associated Colorectal Tumorigenesis
Abstract
:1. Introduction
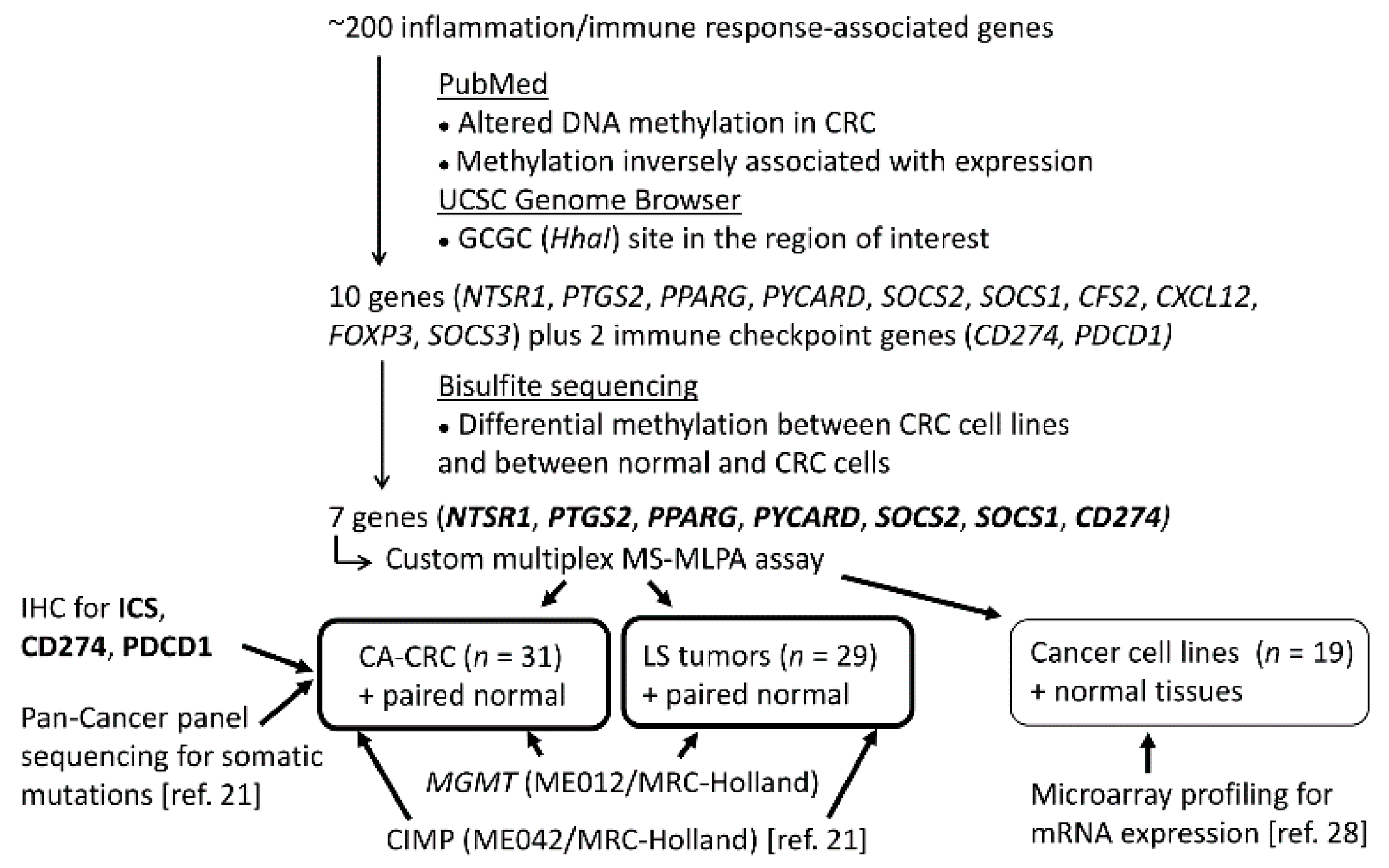
2. Materials and Methods
2.1. Material
2.2. MSI Analysis
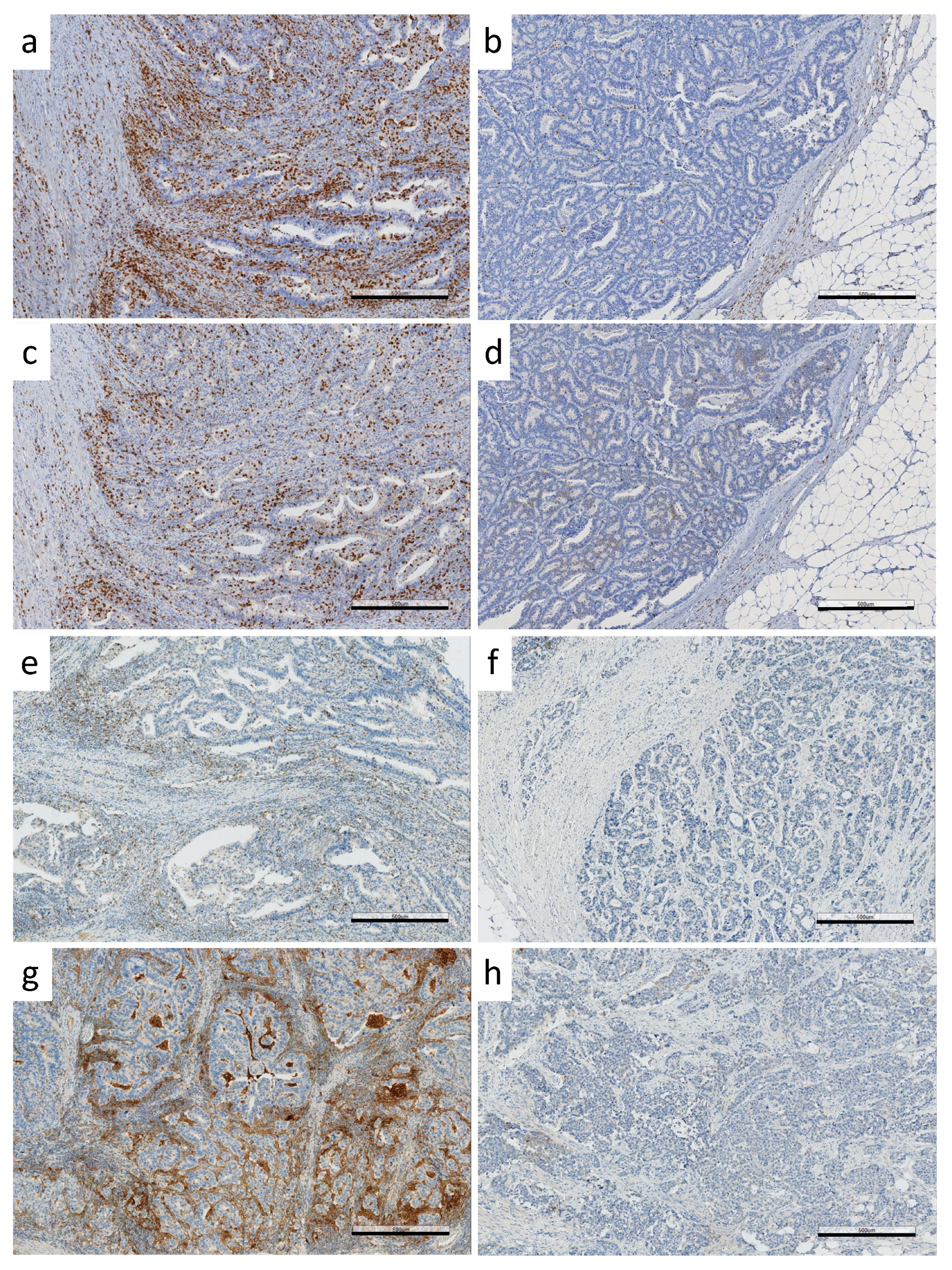
2.3. Immunohistochemical Analysis of PDCD1 and CD274, and Immune Cell Scoring (ICS)
2.4. CpG Island Methylator Phenotype (CIMP) Analysis
2.5. O-6-Methylguanine-DNA-Methyltransferase (MGMT) Methylation Analysis
2.6. Bisulfite Modification and Sequencing
2.7. Custom-Made Methylation Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA) Assay for Inflammation-Related Genes
2.8. mRNA Expression Analysis
2.9. Statistical Analysis
3. Results
3.1. Immunoprofiles of CA-CRC
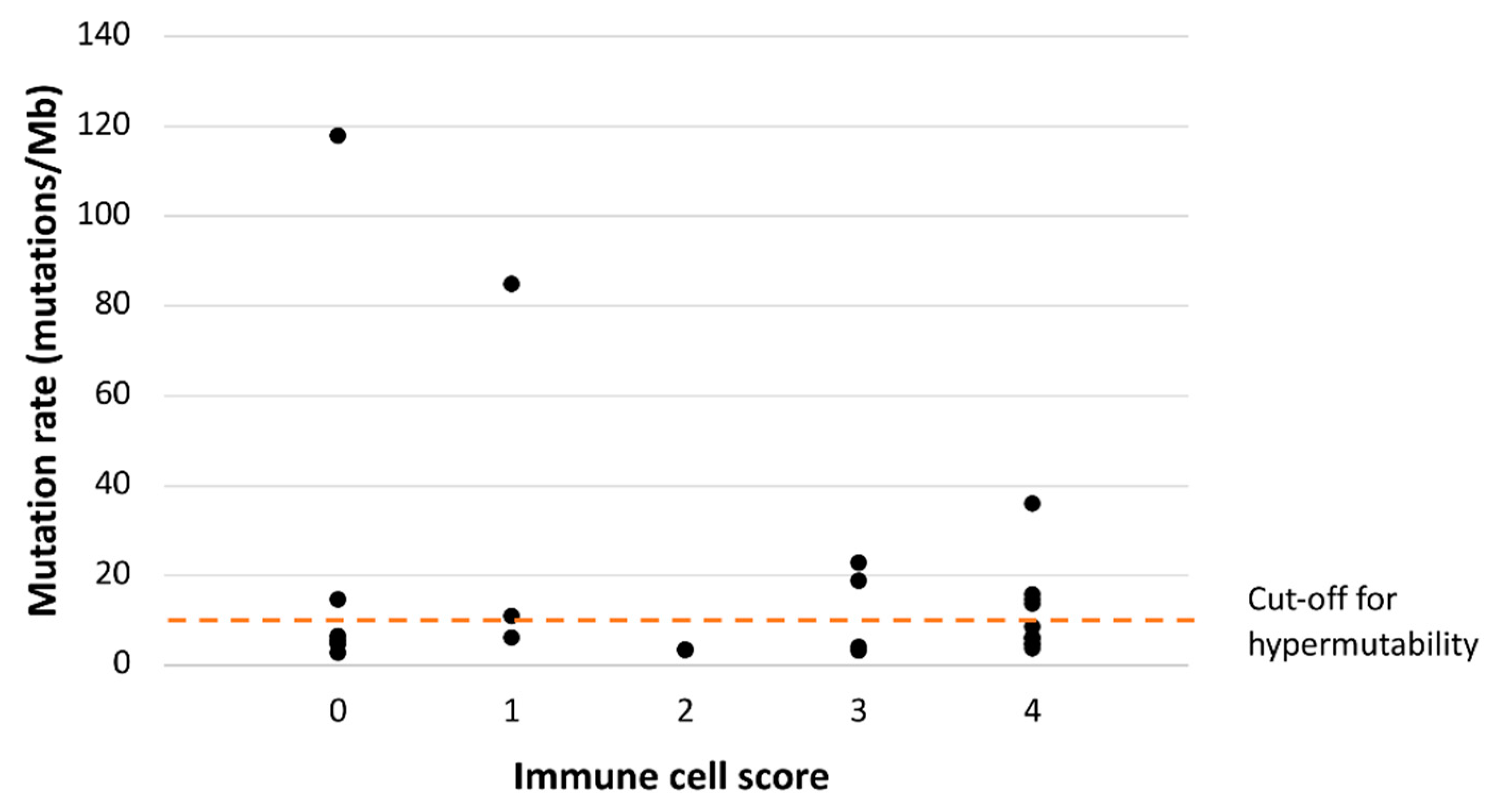
3.2. Immunoprofile vs. Somatic Mutational Status and CIMP
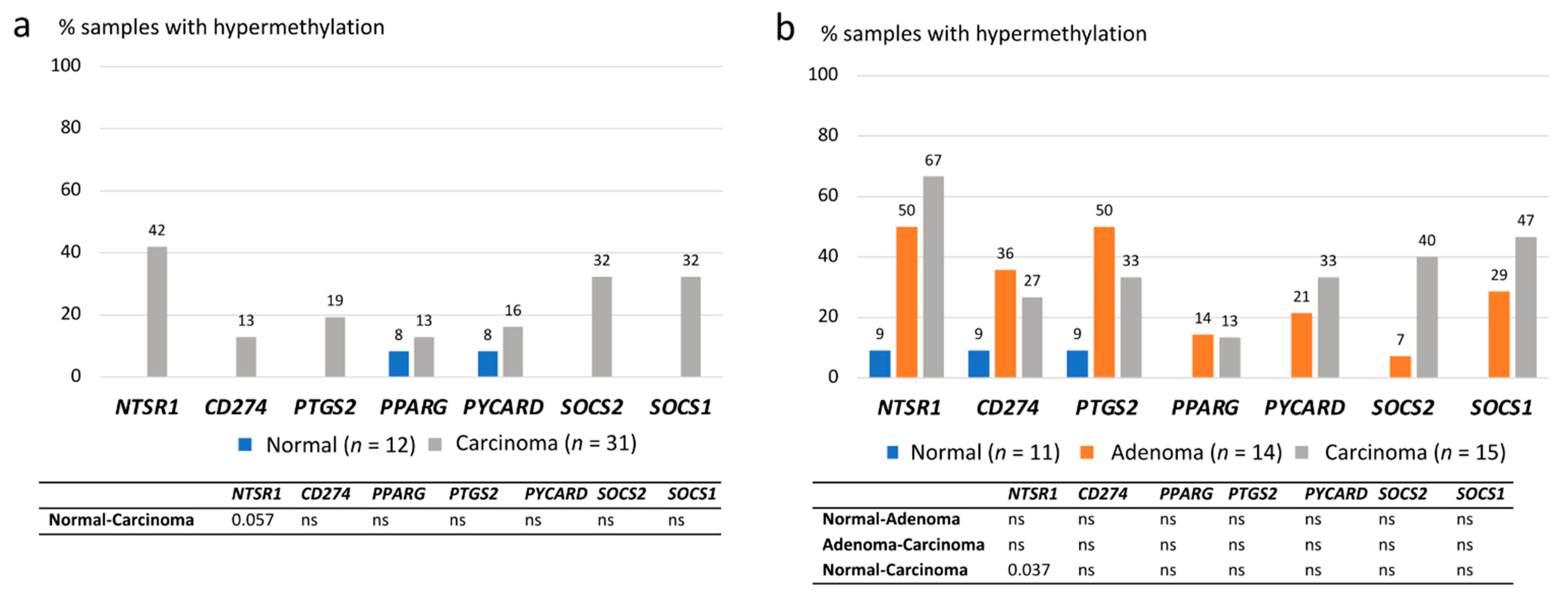
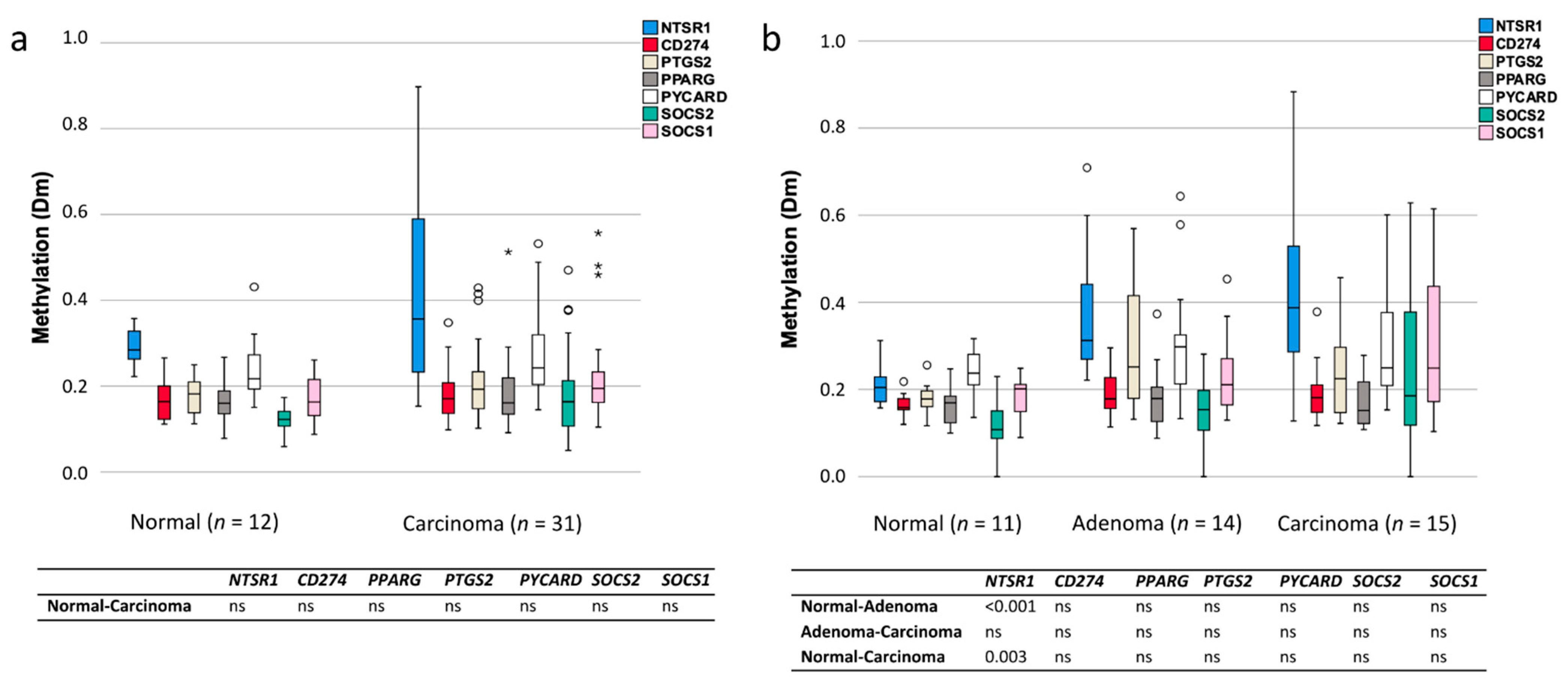
3.3. Methylation of Inflammation-Related Genes in CA-CRC and LS Tumors
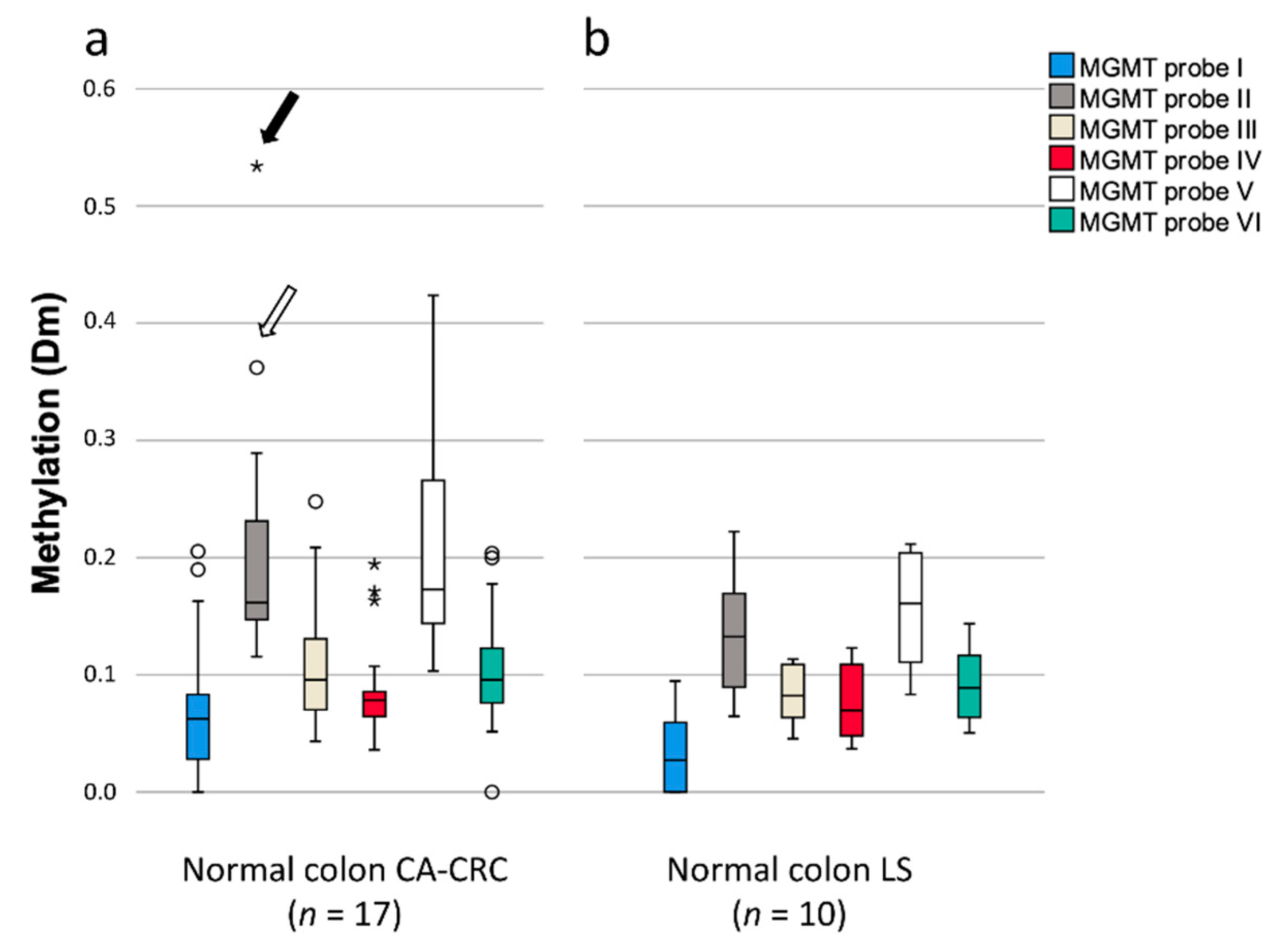
3.4. Methylation of MGMT
3.5. Methylation of Inflammation-Related Genes in Cancer Cell Lines and Association with Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef]
- Al Bakir, I.; Curtius, K.; Graham, T.A. From Colitis to Cancer: An Evolutionary Trajectory That Merges Maths and Biology. Front. Immunol. 2018, 9, 2368. [Google Scholar] [CrossRef]
- Rogler, G. Chronic ulcerative colitis and colorectal cancer. Cancer Lett. 2014, 345, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Hartnett, L.; Egan, L.J. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis 2012, 33, 723–731. [Google Scholar] [CrossRef]
- Watanabe, T.; Konishi, T.; Kishimoto, J.; Kotake, K.; Muto, T.; Sugihara, K.; the Japanese Society for Cancer of the Colon and Rectum. Ulcerative colitis-associated colorectal cancer shows a poorer survival than sporadic colorectal cancer: A nationwide Japanese study. Inflamm. Bowel Dis. 2011, 17, 802–808. [Google Scholar] [CrossRef]
- Drescher, K.M.; Sharma, P.; Lynch, H.T. Current hypotheses on how microsatellite instability leads to enhanced survival of Lynch Syndrome patients. Clin. Dev. Immunol. 2010, 2010, 170432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, P.; Seppala, T.T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Gareth Evans, D.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.H.; et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut 2018, 67, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Thompson, B.A.; Spurdle, A.B.; Plazzer, J.P.; Greenblatt, M.S.; Akagi, K.; Al-Mulla, F.; Bapat, B.; Bernstein, I.; Capella, G.; den Dunnen, J.T.; et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014, 46, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligtenberg, M.J.; Kuiper, R.P.; Chan, T.L.; Goossens, M.; Hebeda, K.M.; Voorendt, M.; Lee, T.Y.; Bodmer, D.; Hoenselaar, E.; Hendriks-Cornelissen, S.J.; et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009, 41, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Cejka, P.; Stojic, L.; Mojas, N.; Russell, A.M.; Heinimann, K.; Cannavo, E.; di Pietro, M.; Marra, G.; Jiricny, J. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003, 22, 2245–2254. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Tsutsumi, S.; Kawaguchi, T.; Nagasaki, K.; Tatsuno, K.; Yamamoto, S.; Sang, F.; Sonoda, K.; Sugawara, M.; Saiura, A.; et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012, 22, 208–219. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Valentin, M.; Seppala, T.T.; Sampson, J.R.; Macrae, F.; Winship, I.; Evans, D.G.; Scott, R.J.; Burn, J.; Moslein, G.; Bernstein, I.; et al. Survival by colon cancer stage and screening interval in Lynch syndrome: A prospective Lynch syndrome database report. Hered. Cancer Clin. Pract. 2019, 17, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biller, L.H.; Syngal, S.; Yurgelun, M.B. Recent advances in Lynch syndrome. Fam. Cancer 2019, 18, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Pages, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Ahtiainen, M.; Wirta, E.V.; Kuopio, T.; Seppala, T.; Rantala, J.; Mecklin, J.P.; Bohm, J. Combined prognostic value of CD274 (PD-L1)/PDCDI (PD-1) expression and immune cell infiltration in colorectal cancer as per mismatch repair status. Mod. Pathol. 2019, 32, 866–883. [Google Scholar] [CrossRef]
- Wirta, E.V.; Seppala, T.; Friman, M.; Vayrynen, J.; Ahtiainen, M.; Kautiainen, H.; Kuopio, T.; Kellokumpu, I.; Mecklin, J.P.; Bohm, J. Immunoscore in mismatch repair-proficient and -deficient colon cancer. J. Pathol. Clin. Res. 2017, 3, 203–213. [Google Scholar] [CrossRef]
- Michael-Robinson, J.M.; Pandeya, N.; Walsh, M.D.; Biemer-Huttmann, A.E.; Eri, R.D.; Buttenshaw, R.L.; Lincoln, D.; Clouston, A.D.; Jass, J.R.; Radford-Smith, G.L. Characterization of tumour-infiltrating lymphocytes and apoptosis in colitis-associated neoplasia: Comparison with sporadic colorectal cancer. J. Pathol. 2006, 208, 381–387. [Google Scholar] [CrossRef]
- Rajamaki, K.; Taira, A.; Katainen, R.; Valimaki, N.; Kuosmanen, A.; Plaketti, R.M.; Seppala, T.T.; Ahtiainen, M.; Wirta, E.V.; Vartiainen, E.; et al. Genetic and epigenetic characteristics of inflammatory bowel disease associated colorectal cancer. Gastroenterology 2021. [Google Scholar] [CrossRef]
- Soh, J.S.; Jo, S.I.; Lee, H.; Do, E.J.; Hwang, S.W.; Park, S.H.; Ye, B.D.; Byeon, J.S.; Yang, S.K.; Kim, J.H.; et al. Immunoprofiling of Colitis-associated and Sporadic Colorectal Cancer and its Clinical Significance. Sci. Rep. 2019, 9, 6833. [Google Scholar] [CrossRef] [Green Version]
- Goltz, D.; Gevensleben, H.; Dietrich, J.; Dietrich, D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology 2017, 6, e1257454. [Google Scholar] [CrossRef] [Green Version]
- Maki-Nevala, S.; Ukwattage, S.; Olkinuora, A.; Almusa, H.; Ahtiainen, M.; Ristimaki, A.; Seppala, T.; Lepisto, A.; Mecklin, J.P.; Peltomaki, P. Somatic mutation profiles as molecular classifiers of ulcerative colitis-associated colorectal cancer. Int. J. Cancer 2021, 148, 2997–3007. [Google Scholar] [CrossRef]
- Isola, J.; DeVries, S.; Chu, L.; Ghazvini, S.; Waldman, F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am. J. Pathol. 1994, 145, 1301–1308. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Valo, S.; Kaur, S.; Ristimaki, A.; Renkonen-Sinisalo, L.; Jarvinen, H.; Mecklin, J.P.; Nystrom, M.; Peltomaki, P. DNA hypermethylation appears early and shows increased frequency with dysplasia in Lynch syndrome-associated colorectal adenomas and carcinomas. Clin. Epigenetics 2015, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Berg, M.; Hagland, H.R.; Soreide, K. Comparison of CpG island methylator phenotype (CIMP) frequency in colon cancer using different probe- and gene-specific scoring alternatives on recommended multi-gene panels. PLoS ONE 2014, 9, e86657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- UCSC Genome Browser. Available online: http://genome-euro.ucsc.edu/cgi-bin/hgGateway (accessed on 18 May 2018).
- Niskakoski, A.; Kaur, S.; Staff, S.; Renkonen-Sinisalo, L.; Lassus, H.; Jarvinen, H.J.; Mecklin, J.P.; Butzow, R.; Peltomaki, P. Epigenetic analysis of sporadic and Lynch-associated ovarian cancers reveals histology-specific patterns of DNA methylation. Epigenetics 2014, 9, 1577–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, M.W.; Ngiow, S.F.; Ribas, A.; Smyth, M.J. Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res. 2015, 75, 2139–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahlquist, T.; Lind, G.E.; Costa, V.L.; Meling, G.I.; Vatn, M.; Hoff, G.S.; Rognum, T.O.; Skotheim, R.I.; Thiis-Evensen, E.; Lothe, R.A. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol. Cancer 2008, 7, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, X.C.; Brent, T.P. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997, 57, 3672–3677. [Google Scholar] [PubMed]
- Däbritz, J.; Menheniott, T.R. Linking immunity, epigenetics, and cancer in inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 20, 1638–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasmin, R.; Siraj, S.; Hassan, A.; Khan, A.R.; Abbasi, R.; Ahmad, N. Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediat. Inflamm. 2015, 2015, 201703. [Google Scholar] [CrossRef]
- Yan, L.; Meizhi, H.; Yaoyao, Z.; Chen, Y.; Shuyi, W.; Xiaohui, B.; Odong, C.; Lang, X. The Prognostic and Clinicopathological Roles of PD-L1 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2019, 28, 139. [Google Scholar] [CrossRef] [Green Version]
- Picard, E.; Verschoor, C.P.; Ma, G.W.; Pawelec, G. Relationships between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 2020, 11, 369. [Google Scholar] [CrossRef]
- McGrail, D.J.; Pilie, P.G.; Rashid, N.U.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.B.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef]
- Gevensleben, H.; Holmes, E.E.; Goltz, D.; Dietrich, J.; Sailer, V.; Ellinger, J.; Dietrich, D.; Kristiansen, G. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget 2016, 7, 79943–79955. [Google Scholar] [CrossRef] [PubMed]
- Franzen, A.; Vogt, T.J.; Muller, T.; Dietrich, J.; Schrock, A.; Golletz, C.; Brossart, P.; Bootz, F.; Landsberg, J.; Kristiansen, G.; et al. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget 2018, 9, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Marwitz, S.; Scheufele, S.; Perner, S.; Reck, M.; Ammerpohl, O.; Goldmann, T. Epigenetic modifications of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin. Epigenetics 2017, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Kamimae, S.; Yamamoto, E.; Kai, M.; Niinuma, T.; Yamano, H.O.; Nojima, M.; Yoshikawa, K.; Kimura, T.; Takagi, R.; Harada, E.; et al. Epigenetic silencing of NTSR1 is associated with lateral and noninvasive growth of colorectal tumors. Oncotarget 2015, 6, 29975–29990. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Martinez-Fong, D.; Tredaniel, J.; Forgez, P. Neurotensin and its high affinity receptor 1 as a potential pharmacological target in cancer therapy. Front. Endocrinol. 2012, 3, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, Q.; Zhou, J.; Yang, W.; Cui, H.; Xu, M.; Yi, L. Oncogenic role of neurotensin and neurotensin receptors in various cancers. Clin. Exp. Pharmacol. Physiol. 2017, 44, 841–846. [Google Scholar] [CrossRef] [Green Version]
- Kakiuchi, N.; Yoshida, K.; Uchino, M.; Kihara, T.; Akaki, K.; Inoue, Y.; Kawada, K.; Nagayama, S.; Yokoyama, A.; Yamamoto, S.; et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 2020, 577, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.A.; Li, A.X.; Wu, X.; Yang, R.; Drew, D.A.; Rosenberg, D.W.; Pfeifer, G.P. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014, 74, 3617–3629. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.; Bae, J.H.; Han, K.; Kim, E.S.; Kim, T.O.; Yi, J.M. A Genome-Wide Methylation Approach Identifies a New Hypermethylated Gene Panel in Ulcerative Colitis. Int. J. Mol. Sci. 2016, 17, 1291. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P.; Ahuja, N.; Toyota, M.; Bronner, M.P.; Brentnall, T.A. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001, 61, 3573–3577. [Google Scholar] [PubMed]
- Svrcek, M.; Buhard, O.; Colas, C.; Coulet, F.; Dumont, S.; Massaoudi, I.; Lamri, A.; Hamelin, R.; Cosnes, J.; Oliveira, C.; et al. Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: An initiating step in the development of mismatch repair-deficient colorectal cancers. Gut 2010, 59, 1516–1526. [Google Scholar] [CrossRef]
- Scarpa, M.; Scarpa, M.; Castagliuolo, I.; Erroi, F.; Kotsafti, A.; Basato, S.; Brun, P.; D’Inca, R.; Rugge, M.; Angriman, I.; et al. Aberrant gene methylation in non-neoplastic mucosa as a predictive marker of ulcerative colitis-associated CRC. Oncotarget 2016, 7, 10322–10331. [Google Scholar] [CrossRef] [Green Version]
- Konishi, K.; Shen, L.; Wang, S.; Meltzer, S.J.; Harpaz, N.; Issa, J.P. Rare CpG island methylator phenotype in ulcerative colitis-associated neoplasias. Gastroenterology 2007, 132, 1254–1260. [Google Scholar] [CrossRef]
- Zeng, Z.; Mukherjee, A.; Zhang, H. From Genetics to Epigenetics, Roles of Epigenetics in Inflammatory Bowel Disease. Front. Genet. 2019, 10, 1017. [Google Scholar] [CrossRef] [Green Version]
| Characteristics | CA-CRC | LS Tumors Combined 1 | LS Adenomas (High-Grade Dysplasia) | LS Carcinomas | p Value CA-CRC vs. LS Tumors Combined 1 |
|---|---|---|---|---|---|
| No. of patients | 31 | 24 | 14 | 12 | NA |
| Male sex 2 | 20 (64%) | 9 (38%) | 5 (40%) | 5 (42%) | 0.060 |
| Age, years (mean ± SD) 2 | 51.4 (±10.7) | 51.2 (±14.1) | 49.2 (±15.2) | 53.6 (±12.3) | 0.948 |
| Years of colitis before CRC diagnosis (mean ± SD) 2 | 23.8 (±10.8) 3 | - | - | - | NA |
| Gene mutated in germline 2 | |||||
| MLH1 | - | 18 (75%) | 12 (86%) | 8 (67%) | NA |
| MSH2 | - | 3 (13%) | 1 (7%) | 2 (17%) | NA |
| MSH6 | - | 3 (13%) | 1 (7%) | 2 (17%) | NA |
| No. of tumors | 31 | 29 | 14 | 15 | NA |
| MSI tumors | 1 (3%) | 29 (100%) 4 | 14 (100%) 4 | 15 (100%) 4 | <0.00001 |
| CIMP(+) tumors | 12 (39%) | 7 (24%) | 2 (14%) | 5 (33%) | 0.274 |
| Tumor location | |||||
| Proximal 5 | 13 (42%) | 17 (59%) | 6 (43%) | 11 (73%) | 0.431 |
| Distal | 15 (48%) | 12 (41%) | 8 (57%) | 4 (27%) | |
| NA | 3 (10%) | 0 | 0 | 0 | |
| Stage of carcinomas | |||||
| I | 13 (42%) | 10 (67%) | - | 10 (67%) | 0.190 |
| II | 6 (19%) | 3 (20%) | - | 3 (20%) | |
| III | 9 (29%) | 1 (7%) | - | 1 (7%) | |
| IV | 3 (10%) | 0 | - | 0 | |
| NA | 0 | 1 (7%) | - | 1 (7%) |
| Ahtiainen et al. [15] | p Value | ||||
|---|---|---|---|---|---|
| Immunoprofile Characteristics | CA-CRC | pMMR-CRC | LS-CRC | CA-CRC vs. | |
| (n = 31) | (n = 100) | (n = 48) | pMMR-CRC | LS-CRC | |
| PDCD1 1 | |||||
| Low | 13 (42%) | 51 | 17 (35%) | ns | ns |
| High | 18 (58%) | 49 | 31 (65%) | ||
| CD274 on tumor cells | |||||
| Negative | 31 (100%) | 99 | 45 (94%) | ns | ns |
| Positive | 0 | 1 | 3 (6%) | ||
| CD274 on immune cells | |||||
| Negative | 15 (48%) | 70 | 18 (37%) | 0.033 | ns |
| Positive | 16 (52%) | 30 | 30 (63%) | ||
| ICS | |||||
| Low (0–2) | 14 (45%) 2 | 60 | 13 (27%) | ns | ns |
| High (3–4) | 17 (55%) 3 | 40 | 35 (73%) | ||
| ICS/CD274IC (immunological group) 4 | |||||
| ICShigh/CD274pos (type I) | 11 (35%) | 16 | 21 (44%) | 0.023 | ns |
| ICSlow/CD274neg (type II) | 9 (29%) | 52 | 9 (19%) | ||
| ICShigh/CD274neg (type III) | 6 (19%) | 25 | 14 (29%) | ||
| ICSlow/CD274pos (type IV) | 5 (16%) | 7 | 4 (8%) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mäki-Nevala, S.; Ukwattage, S.; Wirta, E.-V.; Ahtiainen, M.; Ristimäki, A.; Seppälä, T.T.; Lepistö, A.; Mecklin, J.-P.; Peltomäki, P. Immunoprofiles and DNA Methylation of Inflammatory Marker Genes in Ulcerative Colitis-Associated Colorectal Tumorigenesis. Biomolecules 2021, 11, 1440. https://doi.org/10.3390/biom11101440
Mäki-Nevala S, Ukwattage S, Wirta E-V, Ahtiainen M, Ristimäki A, Seppälä TT, Lepistö A, Mecklin J-P, Peltomäki P. Immunoprofiles and DNA Methylation of Inflammatory Marker Genes in Ulcerative Colitis-Associated Colorectal Tumorigenesis. Biomolecules. 2021; 11(10):1440. https://doi.org/10.3390/biom11101440
Chicago/Turabian StyleMäki-Nevala, Satu, Sanjeevi Ukwattage, Erkki-Ville Wirta, Maarit Ahtiainen, Ari Ristimäki, Toni T. Seppälä, Anna Lepistö, Jukka-Pekka Mecklin, and Päivi Peltomäki. 2021. "Immunoprofiles and DNA Methylation of Inflammatory Marker Genes in Ulcerative Colitis-Associated Colorectal Tumorigenesis" Biomolecules 11, no. 10: 1440. https://doi.org/10.3390/biom11101440
APA StyleMäki-Nevala, S., Ukwattage, S., Wirta, E.-V., Ahtiainen, M., Ristimäki, A., Seppälä, T. T., Lepistö, A., Mecklin, J.-P., & Peltomäki, P. (2021). Immunoprofiles and DNA Methylation of Inflammatory Marker Genes in Ulcerative Colitis-Associated Colorectal Tumorigenesis. Biomolecules, 11(10), 1440. https://doi.org/10.3390/biom11101440






