Epigenome-Wide Association Study of Prostate Cancer in African Americans Identifies DNA Methylation Biomarkers for Aggressive Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Whole Genome Methylation Profiling Using Illumina Human MethylationEPIC Beadchip
2.3. Bioinformatics and Data Analyses
3. Results
3.1. Patient Characteristics
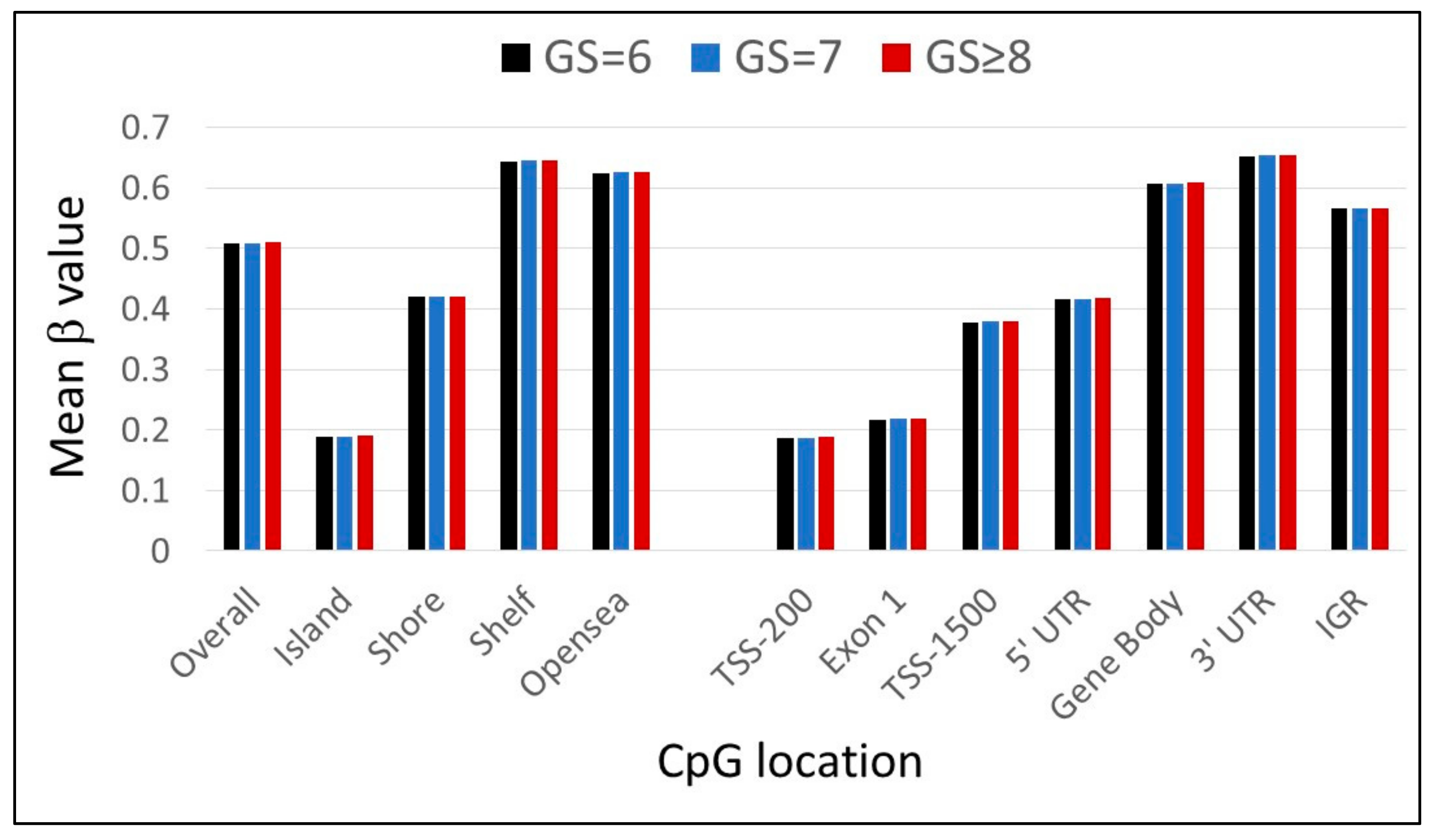
3.2. Leukocyte CpG Methylation Pattern in African American PCa Patients
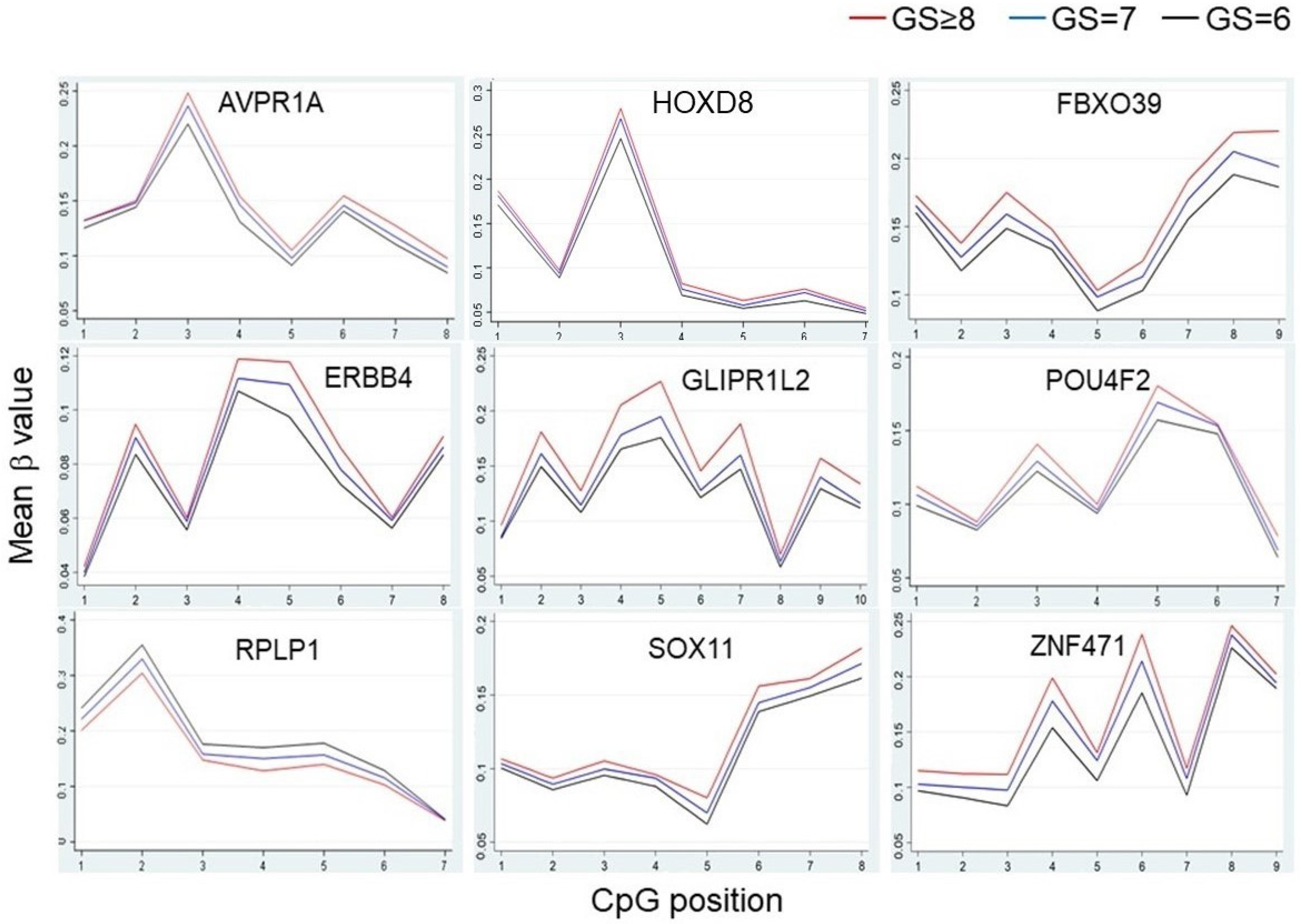
3.3. Differentially Methylated Regions/Genes (DMRs) in High-Grade African American PCa Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- McGinley, K.F.; Tay, K.J.; Moul, J.W. Prostate cancer in men of African origin. Nat. Rev. Urol. 2016, 13, 99–107. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Prostate Cancer Disparities by Race and Ethnicity: From Nucleotide to Neighborhood. Cold Spring Harb. Perspect. Med. 2018, 8, a030387. [Google Scholar] [CrossRef] [PubMed]
- Kan, Z.; Jaiswal, B.S.; Stinson, J.; Janakiraman, V.; Bhatt, D.; Stern, H.M.; Yue, P.; Haverty, P.M.; Bourgon, R.; Zheng, J.; et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nat. Cell Biol. 2010, 466, 869–873. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Fraser, M.; Sabelnykova, V.Y.; Yamaguchi, T.; Heisler, L.; Livingstone, J.; Huang, V.; Shiah, Y.-J.; Yousif, F.; Lin, X.; Masella, A.P.; et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nat. Cell Biol. 2017, 541, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Blattner, M.; Lee, D.J.; O’Reilly, C.; Park, K.; MacDonald, T.Y.; Khani, F.; Turner, K.R.; Chiu, Y.-L.; Wild, P.J.; Dolgalev, I.; et al. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia 2014, 16, 14–20. [Google Scholar] [CrossRef]
- Goering, W.; Kloth, M.; Schulz, W.A. DNA Methylation Changes in Prostate Cancer. Springer Protocols Handbooks 2012, 863, 47–66. [Google Scholar] [CrossRef]
- Jerónimo, C.; Bastian, P.J.; Bjartell, A.; Carbone, G.M.; Catto, J.; Clark, S.; Henrique, R.; Nelson, W.G.; Shariat, S.F. Epigenetics in Prostate Cancer: Biologic and Clinical Relevance. Eur. Urol. 2011, 60, 753–766. [Google Scholar] [CrossRef]
- Boström, P.J.; Bjartell, A.S.; Catto, J.; Eggener, S.E.; Lilja, H.; Loeb, S.; Schalken, J.; Schlomm, T.; Cooperberg, M.R. Genomic Predictors of Outcome in Prostate Cancer. Eur. Urol. 2015, 68, 1033–1044. [Google Scholar] [CrossRef]
- Van Neste, L.; Partin, A.W.; Stewart, G.; Epstein, J.I.; Harrison, D.; Van Criekinge, W. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 2016, 76, 1078–1087. [Google Scholar] [CrossRef]
- Waterhouse, R.L.; Van Neste, L.; Moses, K.A.; Barnswell, C.; Silberstein, J.L.; Jalkut, M.; Tutrone, R.; Sylora, J.; Anglade, R.; Murdock, M.; et al. Evaluation of an Epigenetic Assay for Predicting Repeat Prostate Biopsy Outcome in African American Men. Urology 2019, 128, 62–65. [Google Scholar] [CrossRef]
- Flanagan, J.M.; Wilson, A.; Koo, C.; Masrour, N.; Gallon, J.; Loomis, E.; Flower, K.; Wilhelm-Benartzi, C.; Hergovich, A.; Cunnea, P.; et al. Platinum-Based Chemotherapy Induces Methylation Changes in Blood DNA Associated with Overall Survival in Patients with Ovarian Cancer. Clin. Cancer Res. 2016, 23, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.E.; Fab, K.C.; Dowty, J.G.; Milne, R.L.; Wong, E.M.; Dugue, P.-A.; English, D.; Hopper, J.L.; Goldgar, D.E.; Giles, G.G.; et al. Heritable DNA methylation marks associated with susceptibility to breast cancer. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Dugué, P.-A.; Dowty, J.G.; Joo, J.E.; Wong, E.M.; Makalic, E.; Schmidt, D.F.; English, D.R.; Hopper, J.L.; Pedersen, J.; Severi, G.; et al. Heritable methylation marks associated with breast and prostate cancer risk. Prostate 2018, 78, 962–969. [Google Scholar] [CrossRef]
- Barry, K.H.; Moore, L.E.; Sampson, J.N.; Koutros, S.; Yan, L.; Meyer, A.; Reddy, M.; Oler, A.J.; Cook, M.B.; Jr, J.F.F.; et al. Prospective study of DNA methylation at chromosome 8q24 in peripheral blood and prostate cancer risk. Br. J. Cancer 2017, 116, 1470–1479. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xu, J.; Kim, J.; Wu, X.; Gu, J. Methylation of subtelomeric repeat D4Z4 in peripheral blood leukocytes is associated with biochemical recurrence in localized prostate cancer patients. Carcinoenesis 2017, 38, 821–826. [Google Scholar] [CrossRef]
- Han, Y.; Xu, J.; Kim, J.; Wu, X.; Gu, J. LINE-1 methylation in peripheral blood leukocytes and clinical characteristics and prognosis of prostate cancer patients. Oncotarget 2017, 8, 94020–94027. [Google Scholar] [CrossRef]
- Xu, J.; Tsai, C.-W.; Chang, W.-S.; Han, Y.; Bau, D.-T.; Pettaway, C.A.; Gu, J. Methylation of global DNA repeat LINE-1 and subtelomeric DNA repeats D4Z4 in leukocytes is associated with biochemical recurrence in African American prostate cancer patients. Carcinog. 2019, 40, 1055–1060. [Google Scholar] [CrossRef]
- Moses-Fynn, E.; Tang, W.; Beyene, D.; Apprey, V.; Copeland, R.; Kanaan, Y.; Kwabi-Addo, B. Correlating blood-based DNA methylation markers and prostate cancer risk in African-American men. PLoS ONE 2018, 13, e0203322. [Google Scholar] [CrossRef]
- FitzGerald, L.M.; Naeem, H.; Makalic, E.; Schmidt, D.F.; Dowty, J.G.; Joo, J.E.; Jung, C.-H.; Bassett, J.K.; Dugue, P.-A.; Chung, J.; et al. Genome-Wide Measures of Peripheral Blood Dna Methylation and Prostate Cancer Risk in a Prospective Nested Case-Control Study. Prostate 2017, 77, 471–478. [Google Scholar] [CrossRef]
- Mehdi, A.; Cheishvili, D.; Arakelian, A.; Bismar, T.A.; Szyf, M.; Rabbani, S.A. DNA methylation signatures of Prostate Cancer in peripheral T-cells. BMC Cancer 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, M.; Xu, J.; Li, J.; Xu, Y.; Thompson, T.C.; Logothetis, C.J.; Sun, D.; Gu, J. Genome-wide DNA methylation profiling of leukocytes identifies CpG methylation signatures of aggressive prostate cancer. Am. J. Cancer Res. 2021, 11, 968–978. [Google Scholar]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.; Parker, H.S.; Jaffe, A.; Storey, J. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Jaffe, A.E.; Irizarry, R.A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014, 15, R31. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, G.; Qian, G.W.J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.W.H.; Booth, H.A.F.; Bruford, E. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 1–28. [Google Scholar] [CrossRef]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Haria, D.; Naora, H. Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer. Cancer Hallm. 2013, 1, 67–76. [Google Scholar] [CrossRef][Green Version]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef]
- Pai, P.; Sukumar, S. HOX genes and the NF-κB pathway: A convergence of developmental biology, inflammation and cancer biology. Biochim. Biophys. Acta (BBA) Bioenerg. 2020, 1874, 188450. [Google Scholar] [CrossRef]
- Northcott, J.M.; Northey, J.J.; Barnes, J.M.; Weaver, V.M. Fighting the force: Potential of homeobox genes for tumor microenvironment regulation. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1855, 248–253. [Google Scholar] [CrossRef]
- Scott, C.L.; Omilusik, K.D. ZEBs: Novel Players in Immune Cell Development and Function. Trends Immunol. 2019, 40, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.F.S.D.; Esteves, C.M.; Xavier, F.C.A.; Nunes, F.D. Methylation status of homeobox genes in common human cancers. Genomics 2016, 108, 185–193. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Li, Y.; Ma, S.; Cao, W.; Guan, F. HOXD1 functions as a novel tumor suppressor in kidney renal clear cell carcinoma. Cell Biol. Int. 2021, 45, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.A.; Senga, T. HOXD8 exerts a tumor-suppressing role in colorectal cancer as an apoptotic inducer. Int. J. Biochem. Cell Biol. 2017, 88, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, Y.; Su, X.; Lu, Y. HOXD8 inhibits the proliferation and migration of triple-negative breast cancer cells and induces apoptosis in them through regulation of AKT/mTOR pathway. Reprod. Biol. 2021, 21, 100544. [Google Scholar] [CrossRef] [PubMed]
- Leshchenko, V.V.; Kuo, P.-Y.; Shaknovich, R.; Yang, D.T.; Gellen, T.; Petrich, A.M.; Yu, Y.; Remache, Y.; Weniger, M.A.; Rafiq, S.; et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood 2010, 116, 1025–1034. [Google Scholar] [CrossRef]
- Zhao, F.; Olkhov-Mitsel, E.; Van der Kwast, T.; Sykes, J.; Zdravic, D.; Venkateswaran, V.; Zlotta, A.R.; Loblaw, A.; Fleshner, N.E.; Klotz, L.; et al. Urinary DNA Methylation Biomarkers for Noninvasive Prediction of Aggressive Disease in Patients with Prostate Cancer on Active Surveillance. J. Urol. 2017, 197, 335–341. [Google Scholar] [CrossRef]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: SURVEY AND SUMMARY. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Rakhra, G. Zinc finger proteins: Insights into the transcriptional and post transcriptional regulation of immune response. Mol. Biol. Rep. 2021, 48, 5735–5743. [Google Scholar] [CrossRef]
- Jen, J.; Wang, Y.-C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, S.; Zhang, Y.; Wong, K.-C.; Nakatsu, G.; Wang, X.; Wong, S.; Ji, J.; Yu, J. Zinc-finger protein 471 suppresses gastric cancer through transcriptionally repressing downstream oncogenic PLS3 and TFAP2A. Oncogene 2018, 37, 3601–3616. [Google Scholar] [CrossRef]
- Sun, R.; Xiang, T.; Tang, J.; Peng, W.; Luo, J.; Li, L.; Qiu, Z.; Tan, Y.; Ye, L.; Zhang, M.; et al. 19q13 KRAB zinc-finger protein ZNF471 activates MAPK10/JNK3 signaling but is frequently silenced by promoter CpG methylation in esophageal cancer. Theranostics 2020, 10, 2243–2259. [Google Scholar] [CrossRef]
- Tao, C.; Luo, J.; Tang, J.; Zhou, D.; Feng, S.; Qiu, Z.; Putti, T.C.; Xiang, T.; Tao, Q.; Li, L.; et al. The tumor suppressor Zinc finger protein 471 suppresses breast cancer growth and metastasis through inhibiting AKT and Wnt/β-catenin signaling. Clin. Epigenet. 2020, 12, 173. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Kabekkodu, S.P.; Adiga, D.; Fernandes, R.; Shukla, V.; Bhandari, P.; Pandey, D.; Sharan, K.; Satyamoorthy, K. ZNF471 modulates EMT and functions as methylation regulated tumor suppressor with diagnostic and prognostic significance in cervical cancer. Cell Biol. Toxicol. 2021, 37, 731–749. [Google Scholar] [CrossRef]
- Bhat, S.; Kabekkodu, S.; Jayaprakash, C.; Radhakrishnan, R.; Ray, S.; Satyamoorthy, K. Gene promoter-associated CpG island hypermethylation in squamous cell carcinoma of the tongue. Virchows Archiv. 2017, 470, 445–454. [Google Scholar] [CrossRef]
- Lorenzo, P.M.; Izquierdo, A.G.; Diaz-Lagares, A.; Carreira, M.C.; Macias-Gonzalez, M.; Sandoval, J.; Cueva, J.; Lopez-Lopez, R.; Casanueva, F.F.; Crujeiras, A.B. ZNF577 Methylation Levels in Leukocytes From Women With Breast Cancer Is Modulated by Adiposity, Menopausal State, and the Mediterranean Diet. Front. Endocrinol. 2020, 11, 245. [Google Scholar] [CrossRef]
- Allott, E.H.; Masko, E.M.; Freedland, S.J. Obesity and Prostate Cancer: Weighing the Evidence. Eur. Urol. 2013, 63, 800–809. [Google Scholar] [CrossRef]
- Peters, F.S.; Peeters, A.M.A.; Mandaviya, P.R.; Van Meurs, J.B.J.; Hofland, L.J.; Van De Wetering, J.; Betjes, M.G.H.; Baan, C.C.; Boer, K. Differentially methylated regions in T cells identify kidney transplant patients at risk for de novo skin cancer. Clin. Epigenet. 2018, 10, 81. [Google Scholar] [CrossRef]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, S.; Lu, C.; Ji, T.; Yang, W.; Li, T.; Lv, J.; Hu, W.; Yang, Y.; Jin, Z. SOX11: Friend or foe in tumor prevention and carcinogenesis? Ther. Adv. Med Oncol. 2019, 11, 1758835919853449. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Sun, B.; Hong, Q.; Yan, J.; Mu, D.; Li, J.; Sheng, H.; Guo, H. The role of tumor suppressor gene SOX11 in prostate cancer. Tumor Biol. 2015, 36, 6133–6138. [Google Scholar] [CrossRef] [PubMed]
- Pugongchai, A.; Bychkov, A.; Sampatanukul, P. Promoter hypermethylation ofSOX11correlates with adverse clinicopathological features of human prostate cancer. Int. J. Exp. Pathol. 2017, 98, 341–346. [Google Scholar] [CrossRef]
- Sun, H.; Zhou, H.; Zhang, Y.; Chen, J.; Han, X.; Huang, D.; Ren, X.; Jia, Y.; Fan, Q.; Tian, W.; et al. Aberrant methylation of FAT4 and SOX11 in peripheral blood leukocytes and their association with gastric cancer risk. J. Cancer 2018, 9, 2275–2283. [Google Scholar] [CrossRef]
- Kelsey, K.T.; Wiencke, J.K. Immunomethylomics: A Novel Cancer Risk Prediction Tool. Ann. Am. Thorac. Soc. 2018, 15, S76–S80. [Google Scholar] [CrossRef]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Just, A.; Marioni, R.; Pilling, L.C.; Reynolds, L.; Mandaviya, P.R.; Guan, W.; Xu, T.; Elks, C.E.; Aslibekyan, S.; et al. Epigenetic Signatures of Cigarette Smoking. Circ. Cardiovasc. Genet. 2016, 9, 436–447. [Google Scholar] [CrossRef]
- Park, S.L.; Patel, Y.M.; Loo, L.W.M.; Mullen, D.J.; Offringa, I.A.; Maunakea, A.; Stram, D.O.; Siegmund, K.; Murphy, S.E.; Tiirikainen, M.; et al. Association of internal smoking dose with blood DNA methylation in three racial/ethnic populations. Clin. Epigenet. 2018, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Koestler, D.C.; Usset, J.L.; Christensen, B.C.; Marsit, C.J.; Karagas, M.R.; Kelsey, K.T.; Wiencke, J.K. DNA Methylation-Derived Neutrophil-to-Lymphocyte Ratio: An Epigenetic Tool to Explore Cancer Inflammation and Outcomes. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Ambatipudi, S.; Langdon, R.; Richmond, R.C.; Suderman, M.; Koestler, D.C.; Kelsey, K.T.; Kazmi, N.; Penfold, C.; Ho, K.M.; McArdle, W.; et al. DNA methylation derived systemic inflammation indices are associated with head and neck cancer development and survival. Oral Oncol. 2018, 85, 87–94. [Google Scholar] [CrossRef]
- Grieshober, L.; Graw, S.; Barnett, M.J.; Thornquist, M.D.; Goodman, G.E.; Chen, C.; Koestler, D.C.; Marsit, C.; Doherty, J.A. Methylation-derived Neutrophil-to-Lymphocyte Ratio and Lung Cancer Risk in Heavy Smokers. Cancer Prev. Res. 2018, 11, 727–734. [Google Scholar] [CrossRef]
- Ligthart, S.; WHI-EMPC Investigators; Marzi, C.; Aslibekyan, S.; Mendelson, M.; Conneely, K.N.; Tanaka, T.; Colicino, E.; Waite, L.L.; Joehanes, R.; et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016, 17, 1–15. [Google Scholar] [CrossRef]
| Variables | N (%) |
|---|---|
| Age, median (range) | 58 (39–83) |
| Smoking status | |
| Never smoker | 162 (57.9) |
| Former smoker | 84 (30.0) |
| Current smoker | 34 (12.1) |
| Body mass index (BMI) | |
| <25 | 33 (14.1) |
| 25 ≤ BMI < 30 | 86 (36.8) |
| ≥30 | 115 (49.1) |
| Gleason Score | |
| 6 | 82 (29.3) |
| 7 | 162 (58.8) |
| ≥8 | 36 (12.9) |
| Stage | |
| T1 | 162 (57.9) |
| T2 | 101 (36.1) |
| T3 | 11 (3.9) |
| T4 | 6 (2.1) |
| PSA at diagnosis | |
| <10 ng/mL | 216 (77.2) |
| ≤10 and <20 ng/mL | 39 (13.9) |
| ≥20 ng/mL | 25 (8.9) |
| CpG ID | β Value | P for Trend | Chr. | Gene | CpG Location | Cox Analysis for BCR | |||
|---|---|---|---|---|---|---|---|---|---|
| GS = 6 | GS = 7 | GS ≥ 8 | HR (95% CI) | p Value | |||||
| cg11039604 | 0.926 | 0.94 | 0.952 | 1.17 × 10−6 | 3 | N/A | IGR-Open sea | 1.16 (0.43–3.09) | 0.768 |
| cg15610955 | 0.843 | 0.85 | 0.865 | 3.32 × 10−6 | 2 | GPR39 | Body-Open sea | 1.1 (0.43–2.8) | 0.834 |
| cg01097384 | 0.313 | 0.339 | 0.395 | 3.42 × 10−6 | 11 | N/A | IGR-Island | 1.14 (0.45–2.91) | 0.782 |
| cg02919721 | 0.674 | 0.692 | 0.712 | 3.84 × 10−6 | 1 | N/A | Open sea | 1.51 (0.59–3.84) | 0.387 |
| cg10283070 | 0.803 | 0.801 | 0.773 | 4.46 × 10−6 | 9 | OR1B1 | TSS1500-Open sea | 0.74 (0.31–1.75) | 0.488 |
| cg22367191 | 0.259 | 0.284 | 0.3 | 5.73 × 10−6 | 6 | N/A | IGR-Shore | 0.74 (0.28–1.91) | 0.529 |
| cg18092924 | 0.809 | 0.82 | 0.833 | 7.27 × 10−6 | 20 | DIDO1 | 5′UTR-Shelf | 1.02 (0.42–2.46) | 0.968 |
| cg02073492 | 0.845 | 0.859 | 0.866 | 7.48 × 10−6 | 6 | N/A | IGR-Open sea | 0.94 (0.38–2.34) | 0.901 |
| cg03890037 | 0.157 | 0.168 | 0.197 | 7.50 × 10−6 | 5 | LVRN | TSS200-Island | 1.09 (0.45–2.66) | 0.853 |
| cg24086405 | 0.679 | 0.707 | 0.728 | 1.38 × 10−5 | 8 | DERL1 | TSS1500-Shore | 1.22 (0.46–3.29) | 0.687 |
| cg14299177 | 0.035 | 0.034 | 0.031 | 1.55 × 10−5 | 8 | MTFR1 | 5′UTR-Shore | 1.1 (0.44–2.7) | 0.844 |
| cg02512123 | 0.825 | 0.843 | 0.849 | 1.61 × 10−5 | 1 | ZC3H11A | 5′UTR-Shelf | 2.05 (0.84–5.01) | 0.115 |
| cg03828224 | 0.036 | 0.041 | 0.045 | 1.75 × 10−5 | 5 | PPIC | TSS1500-Island | 0.95 (0.37–2.44) | 0.910 |
| cg16570133 | 0.08 | 0.09 | 0.098 | 1.85 × 10−5 | 8 | N/A | IGR-Shore | 0.97 (0.4–2.32) | 0.942 |
| cg19614456 | 0.855 | 0.862 | 0.872 | 1.99 × 10−5 | 1 | GABPB2 | 5′UTR-Open sea | 1.51 (0.63–3.64) | 0.360 |
| cg27362302 | 0.181 | 0.197 | 0.209 | 2.20 × 10−5 | 18 | GNAL | Exon 1-Island | 1.14 (0.46–2.82) | 0.779 |
| cg21811204 | 0.396 | 0.429 | 0.44 | 2.31 × 10−5 | 5 | SHROOM1 | Body-Island | 1.15 (0.49–2.7) | 0.748 |
| cg16432885 | 0.691 | 0.71 | 0.721 | 2.53 × 10−5 | 3 | GSK3B | Body-Open sea | 3.66 (1.33–10.11) | 0.012 |
| cg00915676 | 0.651 | 0.663 | 0.678 | 2.95 × 10−5 | 7 | N/A | IGR-Open sea | 3.24 (1.12–9.4) | 0.030 |
| cg10481023 | 0.753 | 0.774 | 0.783 | 3.01 × 10−5 | 6 | GABRR2 | Body-Open sea | 1.31 (0.54–3.19) | 0.546 |
| cg15419054 | 0.087 | 0.081 | 0.073 | 3.15 × 10−5 | 17 | NPEPPS | Body-Shore | 0.7 (0.29–1.67) | 0.418 |
| cg19078430 | 0.672 | 0.689 | 0.708 | 3.22 × 10−5 | 17 | SSH2 | Body-Open sea | 0.89 (0.38–2.11) | 0.797 |
| cg10183781 | 0.719 | 0.733 | 0.756 | 3.65 × 10−5 | 18 | ATP9B | Body-Open sea | 1.27 (0.52–3.11) | 0.603 |
| cg20171236 | 0.464 | 0.484 | 0.498 | 3.68 × 10−5 | 4 | N/A | IGR-Open sea | 2.37 (0.9–6.26) | 0.082 |
| cg07160783 | 0.666 | 0.662 | 0.645 | 3.83 × 10−5 | 16 | N/A | IGR-Open sea | 0.99 (0.4–2.46) | 0.977 |
| cg02747319 | 0.306 | 0.313 | 0.335 | 3.84 × 10−5 | 2 | N/A | IGR-Shore | 2.01 (0.81–5.02) | 0.134 |
| cg07665241 | 0.7 | 0.723 | 0.736 | 3.89 × 10−5 | X | CXorf36 | TSS1500-Open sea | 1.06 (0.45–2.47) | 0.898 |
| cg12058586 | 0.123 | 0.113 | 0.102 | 4.23 × 10−5 | 12 | N/A | IGR-Open sea | 0.39 (0.14–1.1) | 0.074 |
| cg05135861 | 0.426 | 0.418 | 0.403 | 4.29 × 10−5 | 18 | DLGAP1 | 5′UTR-Open sea | 0.72 (0.28–1.88) | 0.503 |
| cg00157515 | 0.079 | 0.087 | 0.1 | 4.32 × 10−5 | 2 | LOC100132215 | TSS1500-Island | 2.06 (0.77–5.52) | 0.151 |
| cg26470340 | 0.02 | 0.023 | 0.027 | 4.33 × 10−5 | 10 | ARHGAP21 | 5′UTR-Island | 1.02 (0.4–2.61) | 0.965 |
| cg11945022 | 0.576 | 0.594 | 0.642 | 4.51 × 10−5 | 7 | DYNC1I1 | 5′UTR-Open sea | 0.62 (0.25–1.55) | 0.308 |
| cg18303466 | 0.753 | 0.76 | 0.776 | 4.57 × 10−5 | 1 | SLC9A1 | TSS1500-Shore | 1.19 (0.5–2.84) | 0.694 |
| cg11650926 | 0.781 | 0.78 | 0.764 | 4.59 × 10−5 | 5 | N/A | IGR-Open sea | 0.67 (0.28–1.61) | 0.370 |
| cg15954675 | 0.723 | 0.744 | 0.771 | 4.65 × 10−5 | 3 | SYNPR | Body-Open sea | 3.11 (1.08–8.95) | 0.036 |
| cg25595028 | 0.842 | 0.852 | 0.86 | 4.72 × 10−5 | 11 | RNF214 | Body-Open sea | 1.21 (0.49–2.97) | 0.676 |
| cg20981146 | 0.846 | 0.856 | 0.871 | 5.02 × 10−5 | 1 | RSBN1 | Body-Open sea | 1.02 (0.42–2.5) | 0.959 |
| cg19734896 | 0.825 | 0.812 | 0.802 | 5.12 × 10−5 | 1 | ILDR2 | Body-Shore | 0.55 (0.22–1.36) | 0.196 |
| cg12911208 | 0.744 | 0.731 | 0.719 | 5.13 × 10−5 | 6 | RP11-73O6.4 | 5′UTR-Open sea | 0.89 (0.36–2.19) | 0.793 |
| cg13928649 | 0.13 | 0.138 | 0.156 | 5.21 × 10−5 | 9 | PRDM12 | Body-Island | 1.35 (0.51–3.55) | 0.546 |
| cg05492453 | 0.101 | 0.087 | 0.082 | 5.32 × 10−5 | 22 | FAM19A5 | Body-Open sea | 0.83 (0.33–2.1) | 0.699 |
| cg04492396 | 0.073 | 0.057 | 0.05 | 5.32 × 10−5 | 8 | N/A | IGR-Open sea | 0.59 (0.23–1.51) | 0.270 |
| cg00290605 | 0.024 | 0.028 | 0.029 | 5.33 × 10−5 | 22 | CTA-342B11.2 | TSS200-Island | 0.85 (0.35–2.06) | 0.721 |
| cg01382153 | 0.058 | 0.063 | 0.066 | 5.34 × 10−5 | 2 | CCDC108 | Exon 1-Island | 1.11 (0.44–2.79) | 0.822 |
| cg00903099 | 0.131 | 0.144 | 0.15 | 5.37 × 10−5 | 7 | HTR5A | TSS200-Shore | 1.14 (0.47–2.78) | 0.774 |
| cg18285105 | 0.631 | 0.652 | 0.677 | 5.37 × 10−5 | 9 | RP11-87N24.3 | TSS1500-Open sea | 1.89 (0.77–4.64) | 0.167 |
| cg01049417 | 0.06 | 0.064 | 0.07 | 5.50 × 10−5 | 3 | SCHIP1 | Body-Island | 1.3 (0.5–3.39) | 0.595 |
| cg07744841 | 0.191 | 0.208 | 0.221 | 5.62 × 10−5 | 3 | SLC6A11 | TSS1500-Island | 3.5 (1.21–10.17) | 0.021 |
| cg14530623 | 0.308 | 0.326 | 0.343 | 5.66 × 10−5 | 2 | ITSN2 | 5′UTR-Open sea | 1.7 (0.69–4.22) | 0.250 |
| cg25252658 | 0.623 | 0.644 | 0.67 | 5.67 × 10−5 | 7 | FKBP6 | Body-Open sea | 2.08 (0.83–5.24) | 0.120 |
| cg03052956 | 0.252 | 0.268 | 0.306 | 5.70 × 10−5 | X | ARHGAP36 | TSS1500-Shore | 1.45 (0.57–3.68) | 0.430 |
| cg01909140 | 0.836 | 0.842 | 0.853 | 6.07 × 10−5 | 9 | C9orf3 | Body-Open sea | 1.57 (0.64–3.81) | 0.324 |
| cg11179997 | 0.137 | 0.144 | 0.152 | 6.42 × 10−5 | X | N/A | IGR-Island | 0.85 (0.34–2.17) | 0.737 |
| cg13453374 | 0.222 | 0.236 | 0.261 | 6.44 × 10−5 | 3 | RP11-649A16.1 | 3′UTR-Open sea | 1.15 (0.48–2.78) | 0.753 |
| cg21081034 | 0.177 | 0.192 | 0.203 | 6.57 × 10−5 | 2 | LOC100132215 | TSS1500-Island | 0.73 (0.27–1.98) | 0.532 |
| cg16372976 | 0.494 | 0.531 | 0.57 | 6.74 × 10−5 | 1 | RFWD2 | Body-Shelf | 1.11 (0.45–2.76) | 0.824 |
| cg00088299 | 0.013 | 0.016 | 0.018 | 6.80 × 10−5 | 4 | N/A | IGR-Island | 2.37 (0.89–6.32) | 0.084 |
| cg25339368 | 0.809 | 0.817 | 0.828 | 6.92 × 10−5 | 17 | TBCD | Body-Shore | 1.22 (0.49–3.06) | 0.673 |
| cg25215047 | 0.804 | 0.807 | 0.837 | 6.97 × 10−5 | 13 | N/A | IGR-Open sea | 1.29 (0.54–3.07) | 0.565 |
| cg07629204 | 0.484 | 0.507 | 0.534 | 7.14 × 10−5 | 10 | N/A | IGR-Open sea | 2.41 (0.8–7.23) | 0.118 |
| cg21377071 | 0.495 | 0.521 | 0.541 | 7.31 × 10−5 | 15 | ADAMTSL3 | 5′UTR-Shore | 1.94 (0.77–4.89) | 0.160 |
| cg23099959 | 0.622 | 0.643 | 0.665 | 7.54 × 10−5 | 15 | RP11-66B24.2 | 5′UTR-Shore | 0.6 (0.22–1.61) | 0.309 |
| cg14824107 | 0.73 | 0.721 | 0.683 | 7.58 × 10−5 | 17 | TBCD | Body-Shore | 0.63 (0.25–1.59) | 0.326 |
| cg01993946 | 0.699 | 0.688 | 0.671 | 8.00 × 10−5 | 10 | N/A | IGR-Open sea | 0.52 (0.21–1.28) | 0.156 |
| cg11875624 | 0.06 | 0.084 | 0.083 | 8.08 × 10−5 | X | FGF13 | 5′UTR-Island | 0.93 (0.36–2.41) | 0.882 |
| cg19004134 | 0.02 | 0.021 | 0.024 | 8.12 × 10−5 | 6 | VTA1 | TSS200-Island | 1.58 (0.65–3.82) | 0.310 |
| cg03854198 | 0.087 | 0.093 | 0.104 | 8.14 × 10−5 | 12 | NTF3 | Exon 1-Island | 1.49 (0.53–4.15) | 0.448 |
| cg13786089 | 0.092 | 0.103 | 0.117 | 8.28 × 10−5 | 19 | ZFR2 | TSS200-Island | 1.68 (0.67–4.26) | 0.271 |
| cg05917797 | 0.775 | 0.782 | 0.792 | 8.40 × 10−5 | 14 | N/A | IGR-Open sea | 1.58 (0.64–3.87) | 0.318 |
| cg12574406 | 0.813 | 0.822 | 0.834 | 8.60 × 10−5 | 8 | N/A | IGR-Open sea | 1.2 (0.48–3.04) | 0.693 |
| cg04855249 | 0.788 | 0.782 | 0.77 | 8.97 × 10−5 | 15 | OCA2 | Body-Open sea | 0.92 (0.37–2.29) | 0.859 |
| cg23409289 | 0.018 | 0.02 | 0.023 | 9.42 × 10−5 | 5 | C5orf56 | TSS1500-Island | 1.46 (0.59–3.62) | 0.410 |
| cg12949141 | 0.593 | 0.612 | 0.635 | 9.48 × 10−5 | 5 | PCBD2 | Body-Open sea | 1.56 (0.61–3.98) | 0.354 |
| cg00170540 | 0.768 | 0.785 | 0.791 | 9.55 × 10−5 | 16 | HSDL1 | 5′UTR-Open sea | 1.18 (0.49–2.79) | 0.715 |
| cg22851420 | 0.22 | 0.238 | 0.25 | 9.58 × 10−5 | 1 | HPCAL4 | Body-Island | 0.67 (0.26–1.68) | 0.391 |
| cg04921989 | 0.07 | 0.077 | 0.084 | 9.84 × 10−5 | 2 | N/A | IGR-Island | 1.23 (0.51–2.95) | 0.651 |
| cg13551368 | 0.508 | 0.497 | 0.484 | 9.87 × 10−5 | 5 | N/A | IGR-Open sea | 0.5 (0.19–1.3) | 0.154 |
| cg06465285 | 0.776 | 0.796 | 0.815 | 9.95 × 10−5 | 8 | N/A | IGR-Open sea | 0.79 (0.32–1.94) | 0.608 |
| ALX1, ANKHD1-EIF4EBP3, ARHGAP15, ATXN7, AVPR1A, BACH2, C6orf174, CD24, CD247, |
| CLDN11, CLVS2, CSDAP1, DLX6AS, DNASE1L2, ERBB4, ESRP2, FAM171A2, FAM179A, |
| FBN2, FBXO39, FOXG1, GABRB2, GALNT13, GJB6, GLIPR1L2, GNAS, GNMT, GRIK2, HERC2, |
| HOXC11, HOXD1, HOXD11, HOXD8, HPSE2, KHDRBS2, KSR2, LHX8, LRP5, MAPK8IP3, |
| MIR2277, MSX2, NKAIN3, NKX6-2, PAX7, PCDH10, PEG10, PF4, PHYHIPL, PITX2, POU4F2 |
| PPP2R5E, PRH1, PTPN13, RBM33, RPLP1, RSPO2, SFRP2, SGCE, SHROOM1, SLC6A11, |
| SOX11, SPATA18, SPTBN1, TRIM2, TWIST1, UPB1, WDR60, WDR8, WNT6, WT1, WWP2, |
| ZBTB16, ZNF471, ZNF577, ZNF714, ZNF83, ZSCAN1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Tsai, C.-W.; Chang, W.-S.; Han, Y.; Huang, M.; Pettaway, C.A.; Bau, D.-T.; Gu, J. Epigenome-Wide Association Study of Prostate Cancer in African Americans Identifies DNA Methylation Biomarkers for Aggressive Disease. Biomolecules 2021, 11, 1826. https://doi.org/10.3390/biom11121826
Xu Y, Tsai C-W, Chang W-S, Han Y, Huang M, Pettaway CA, Bau D-T, Gu J. Epigenome-Wide Association Study of Prostate Cancer in African Americans Identifies DNA Methylation Biomarkers for Aggressive Disease. Biomolecules. 2021; 11(12):1826. https://doi.org/10.3390/biom11121826
Chicago/Turabian StyleXu, Yifan, Chia-Wen Tsai, Wen-Shin Chang, Yuyan Han, Maosheng Huang, Curtis A. Pettaway, Da-Tian Bau, and Jian Gu. 2021. "Epigenome-Wide Association Study of Prostate Cancer in African Americans Identifies DNA Methylation Biomarkers for Aggressive Disease" Biomolecules 11, no. 12: 1826. https://doi.org/10.3390/biom11121826
APA StyleXu, Y., Tsai, C.-W., Chang, W.-S., Han, Y., Huang, M., Pettaway, C. A., Bau, D.-T., & Gu, J. (2021). Epigenome-Wide Association Study of Prostate Cancer in African Americans Identifies DNA Methylation Biomarkers for Aggressive Disease. Biomolecules, 11(12), 1826. https://doi.org/10.3390/biom11121826







