Circulating MicroRNAs as Novel Potential Diagnostic Biomarkers for Osteosarcoma: A Systematic Review
Abstract
1. Introduction
2. Material and Methods
2.1. Literature Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction and Statistical Analysis
2.4. Quality Assessment of the Included Studies
2.5. Study Registration
3. Results
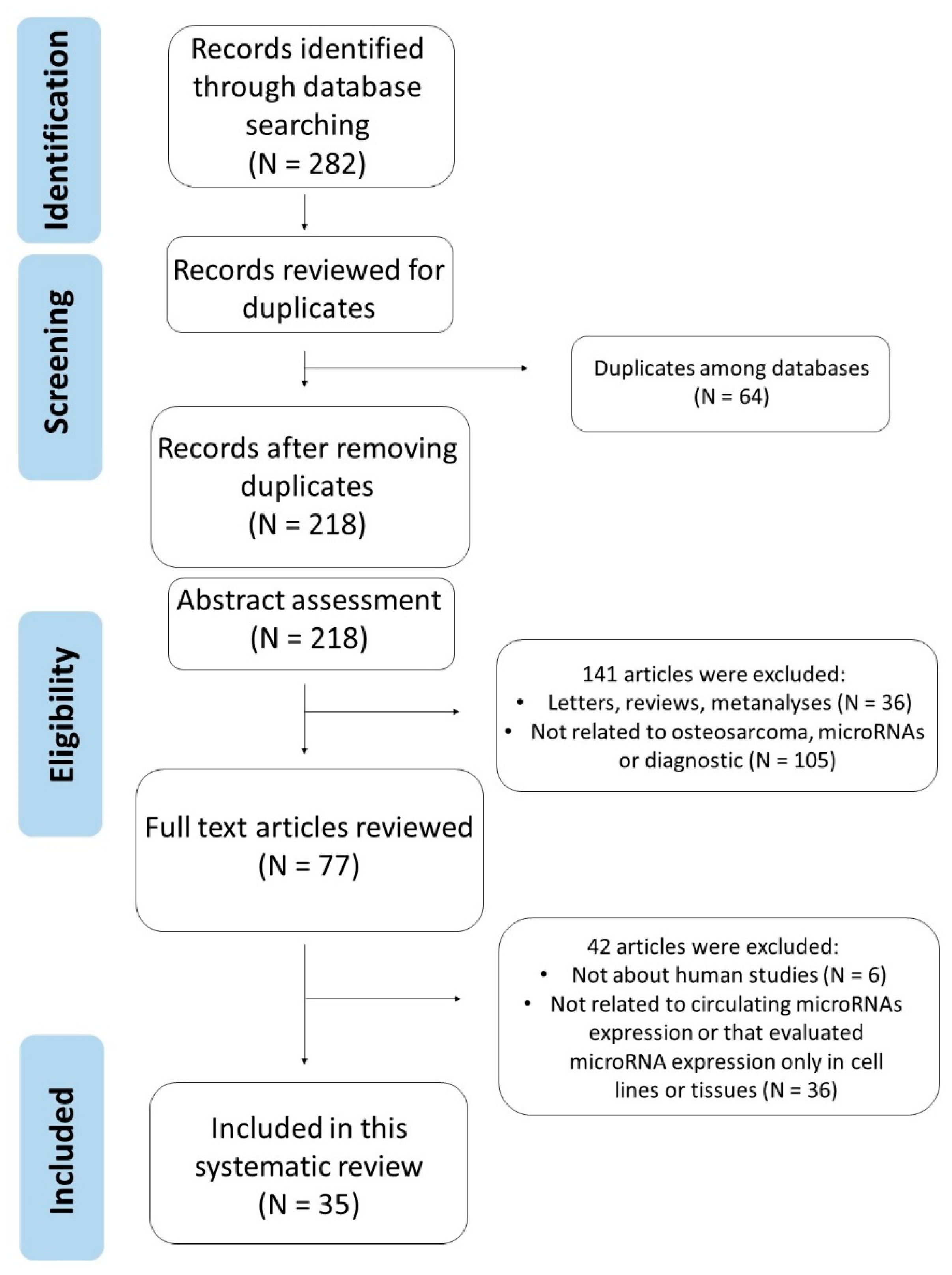
3.1. Literature Search Results
3.2. Main Results and Study Quality Assessment
| Control Group | Case Group | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Ethnicity | N | Sex | Mean Age (y) | N | Sex | Mean Age (y) | Metast | Specim | Det Met | Normaliz. | Method for Expression Level Calculation | Differentially Expressed MicroRNAs | Up- or Downregulation Description Only | AUC | SEN | SPE |
| Allen-Rhoades et al. [12] | 2015 | American | 30 | N/I | 18 | 40 | 2M 17F | 13.41 | 20 yes, 19 no | plasma | qPCR | miR-320a + miR-15a- 5p + CqUniSp2)/3 | 2−ΔCt | miR-205-5p miR-214 miR-335-5p miR-574-3p | No | MiR- 205-5p: 0.70, MiR-214: 0.8, MiR-335-5p: 0.78, MiR-574-3p: 0.88 | N/I | N/I |
| Cai et al. [13] | 2015 | Asian | 60 | N/I | N/I | 166 | 96M 70F | <55:72 ≥55:94 | 42 yes, 124 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-195 | No | 0.892 | 88.0% | 83.3% |
| Hui et al. [14] | 2015 | Asian | 20 | 12M 8F | 14.3 | 20 | 13M 7F | 13 | 2 yes, 11 no | serum | qPCR | cel-miR-39 | 2−ΔΔCt | miR-106a-5p miR-16-5p miR-20a-5p miR-25-3p miR-425-5p miR-451a miR-139-5p | No | miR-106a-5p: 0.7255 miR-16-5p: 0.7686 miR-20a-5p: 0.8471 miR-25-3p: 0.7961 miR-425-5p: 0.7765 miR-451a: 0.7961 miR-139-5p: 0.7098 | N/I | N/I |
| Lian et al. [15] | 2015 | Asian | 90 | 44M 46 F | 16.2 | 90 | 43M 47F | 15.8 | 18 yes, 72 no | plasma | qPCR |
comparison of the miRNA concentrat. to the serum volume |
comparison of the miRNA concentrat. to the serum volume | miR-195-5p miR-199a-3p miR-320a miR-374a-5p | No | miR-195–5p: 0.9029 miR-199a-3p: 0.9025 miR-320a: 0.9188 miR-374a-5p: 0.9173 4-miRNAs: 0.608 | 4-miRNAs: 91.1% | 4-miRNAs: 94.4% |
| Tang et al. [16] | 2015 | Asian | 60 | N/I | N/I | 166 | 96M 70F | <55:72 ≥55:94 | 42 yes, 124 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-27a | No | 0.867 | 70.01% | 98.30% |
| Wang et al. [17] | 2015 | Asian | 20 | N/I | N/I | 80 | 40M 40F | ≤19:40 >19:40 | 12 yes, 68 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-152 | No | 0.956 | 96.2% | 92.5% |
| Wang et al. [18] | 2015 | Asian | 20 | N/I | N/I | 100 | 66M 34F | <20:69 ≥20:31 | 42 yes, 58 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-191 | No | 0.858 | 74.00% | 100.0% |
| Yang et al. [19] | 2015 | Asian | 50 | N/I | N/I | 108 | 78M 30F | <20:40 ≥20:68 | 40 yes, 68 no | serum | qPCR | RNU6 | 2−ΔΔCt | MiR-221 | No | 0.844 | 65.7% | 100.0% |
| Zhou et al. [20] | 2015 | Asian | 60 | 38M 22F | ≥20:23 <20:37 | 60 | 38M 22F | ≥20:23 <20:37 | 8 yes, 52 no | serum | qPCR |
comparison of the miRNA concentrat. to the serum volume |
comparison of the miRNA concentrat. to the serum volume | MiR-199a-5p | No | 0.8606 | 88.33% | 76.67% |
| Cao et al. [21] | 2016 | Asian | 20 | N/I | N/I | 60 | 32M 28F | ≤18:37 >18:23 | 9 yes, 51 no | serum | qPCR | RNU48 | 2−ΔΔCt | MiR-326 | No | 0.897 | 83.7% | 94.5% |
| Li et al. [3] | 2016 | Asian | 46 | 27M 19F | 19.6 | 46 | 27M 19F | 19.6 | N/I | serum | qPCR | U6 | 2−ΔΔCt | MiR-17 | Yes | N/I | N/I | N/I |
| Niu et al. [22] | 2016 | Asian | 133 | 71M 62F | ≤15:59 >15:74 | 133 | 71M 62F | ≤15:59 >15:74 | 68 yes, 65 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-95-3p | No | 0.863 | N/I | N/I |
| Pang et al. [23] | 2016 | Asian | 130 | N/I | N/I | 185 | 110M 75F | <55:73 ≥55:112 | 57 yes, 128 no | serum | qPCR | U6 | 2−ΔCt | MiR-497 | No | 0.848 | N/I | N/I |
| Sun et al. [24] | 2016 | Asian | 62 | N/I | N/I | 62 | N/I | N/I | N/I | serum | qPCR | U6 | 2−ΔΔCt | MiR-24 | Yes | N/C | N/C | N/C |
| Zhou et al. [25] | 2016 | Asian | 40 | N/I | N/I | 40 | 25M 15F | ≥15:27 <15:13 | N/I | serum | qPCR | U6 | 2−ΔΔCt | MiR-421 | Yes | N/C | N/C | N/C |
| Fujiwara et al. [26] | 2017 | Asian | 8 | 4M 4F | N/I | 14 | 7M 7F | 0–10:2 11–20:8 ≥21:4 | 1 yes, 13 no | serum | qPCR | N/I | 2−ΔΔCt | miR-25-3p miR-17-3p | No | MiR-25-3p: 0.868 MiR-17-3p: 0.720 | MiR-25-3p: 71.4% MiR-17-3p: 64.3% | MiR-25-3p: 92.3%; MiR-17-3p: 84.6% |
| Liu et al. [27] | 2017 | Asian | 10 | N/I | N/I | 20 | N/I | N/I | N/I | serum | qPCR | N/I | N/I | MiR-598 | Yes | N/C | N/C | N/C |
| Wang et al. [28] | 2017 | Asian | 20 | 8M 12F | 24.5 | 102 | 54M 48F | Low: 17.3 High: 16.4 | 36 yes, 66 no | serum | qPCR | RNU6B | N/I | MiR-491 | Yes | N/I | N/I | N/I |
| Xie et al. [29] | 2017 | Asian | 3 | N/I | N/I | 3 | N/I | N/I | N/I | PBMC | qPCR | U6 | 2−ΔCt | hsa-miR-221-5p hsa-miR-26b-5p hsa-miR-21-5p hsamiR-5706 hsa-miR-656-3p | Yes | N/C | N/C | N/C |
| Cong et al. [30] | 2018 | Asian | 50 | N/I | N/I | 114 | 62M 52F | ≥18: 71 <18: 43 | 60 yes, 54 no | serum | qPCR | RNU6 | 2−ΔΔCt | MiR-124 | No | 0.846 | 79.8% | 86% |
| Li, Song et al. [31] | 2018 | Asian | 76 | N/I | N/I | 76 | N/I | N/I | N/I | plasma | qPCR | U6 | 2−ΔΔCt | MiR-542-3p | No | 0.841 | 77.8% | 93.6% |
| Liu, Zhao et al. [32] | 2018 | Asian | 95 | N/I | N/I | 95 | 63M 32F | <20: 69 ≥20: 26 | 37 yes, 58 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-375 | No | 0.89 | 82.1% | 74.7% |
| Monterde-Cruz et al. [2] | 2018 | Mexican | 15 | 9M 6F | 20 | 15 | 9M6F | 20 | 13 yes, 2 no | serum | qPCR | RNU6 | 2−ΔΔCt | miR-215-5p miR-642a-5p | No | miR-215-5p: 0.8667, miR-642a-5p: 0.8413, 2-miRNAs: 0.8520 | N/I | N/I |
| Tian et al. [33] | 2018 | Asian | 30 | N/I | N/I | 65 | 35M 30F | ≤12:35 >12:30 | No | serum | qPCR | U6 | N/I | MiR-337-5p | No | 0.7761 | N/I | N/I |
| Xu et al. [34] | 2018 | Asian | 30 | N/I | N/I | 30 | N/I | N/I | N/I | serum | qPCR | U6 | N/I | MiR-411 | Yes | N/C | N/C | N/C |
| Yao et al. [35] | 2018 | Asian | 70 | N/I | N/I | 152 | 8M 65F | <55: 84 ≥55: 68 | 21 yes, 131 no | serum | qPCR | U6 | 2−ΔΔCt | MiR-101 | No | 0.850 | 78.95% | 82.86% |
| Zhao et al. [36] | 2018 | Asian | N/I | N/I | N/I | N/I | N/I | N/I | N/I | serum | qPCR | N/I | N/I | MiR-95-3p | Yes | N/C | N/C | N/C |
| Zhou et al. [37] | 2018 | Asian | 50 | N/I | N/I | 98 | 62M 36F | <19: 47 ≥19: 51 | 30 yes, 68 no | serum | qPCR | cel-MiR-39 | 2−ΔΔCt | MiR-139-5p | No | 0.846 | 76.5% | 80% |
| Zhou et al. [38] | 2018 | Asian | 7 | 4M 3F | N/I | 7 | 4M 3F | N/I | N/I | serum | qPCR | U6 | 2−ΔΔCt | MiR-22 | Yes | N/C | N/C | N/C |
| Cuscino et al. [9] | 2019 | Italian | 3 | N/I | N/I | 5 | M | 16.8 | 2 yes, 3 no | plasma | Digital PCR | U6 | 2−ΔCt | 5 new microRNA candidates | Yes | N/I | N/I | N/I |
| Huang et al. [4] | 2019 | Asian | 30 | 22M 28F | ≤16: 26 >16: 24 | 50 | 22M 28F | ≤16: 26 >16: 24 | 18 yes, 32 no | serum | qPCR | U6 and cel-MiR-39 | ΔCt = CtmiRNA− CtmiR—39/U6 | MiR-487-a MiR-493-5p MiR-501-3p MiR-502-5p | No | miR-487a: 0.83, miR-493-5p: 0.79, miR-501-3p: 0.82, miR-502-5p: 0.83, 4-miRNAs: 0.89 | N/I | N/I |
| Huang, Sun et al. [39] | 2019 | Asian | 50 | 32M 18F | ≤14: 30 >14: 20 | 50 | 3M 14F | ≤14:31 >14: 19 | 11 yes, 39 no | plasma | qPCR | U6, cel-MiR-39 | ΔCt = CtmiRNA− CtmiR—39/U6; ΔCtCt = ΔCtpatient− Ctcontrol | MiR-663a | No | 0.86 | 67.35% | 89.8% |
| Zhu et al. [1] | 2019 | Asian | 25 | N/I | N/I | 55 | N/I | N/I | N/I | serum | qPCR | GAPDH | 2−ΔΔCt | hsa_circ_0000885 | No | 0.783 | N/I | N/I |
| Shi et al. [40] | 2020 | Asian | 60 | N/I | N/I | 124 | 79H e 45 M | <50:72 ≥50:52 | 33 yes, 91 no | serum | qPCR | cel-miR-39 | 2−ΔΔCt | MiR-194 | No | 0.855 | 84.2% | 79.1% |
| Zhang et al. [41] | 2020 | Asian | 20 | 12M 8F | 18.5 | 41 | 27M 14F | 16 | 14 yes, 27 no | Extrac. Vesicul. | qPCR | U6, Cel-mir-39 e let-7i-5p | 2−ΔΔCt | MiR-101 | No | 0.7957 | N/I | N/I |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, K.; Niu, L.; Wang, J.; Wang, Y.; Zhou, J.; Wang, F.; Cheng, Y.; Zhang, Q.; Li, H. Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med. Sci. Monit. 2019, 25, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Monterde-Cruz, L.; Ramírez-Salazar, E.G.; Rico-Martínez, G.; Linares-González, L.M.; Guzmán-González, R.; Delgado-Cedillo, E.; Estrada-Villaseñor, E.; Valdés-Flores, M.; Velázquez-Cruz, R.; Hidalgo-Bravo, A. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum. Cell 2018, 31, 292–299. [Google Scholar] [CrossRef]
- Li, S.; Gao, Y.; Wang, Y.; Wang, K.; Dai, Z.P.; Xu, D.; Liu, W.; Li, Z.L.; Zhang, Z.D.; Yang, S.H.; et al. Serum microRNA-17 functions as a prognostic biomarker in osteosarcoma. Oncol. Lett. 2016, 12, 4905–4910. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Q.; Ma, S.; Sun, Y.; Vadamootoo, A.S.; Jin, C. A four serum-miRNA panel serves as a potential diagnostic biomarker of osteosarcoma. Int. J. Clin. Oncol. 2019, 24, 976–982. [Google Scholar] [CrossRef]
- Kong, Y.W.; Ferland-McCollough, D.; Jackson, T.J.; Bushell, M. microRNAs in cancer management. Lancet Oncol. 2012, 13, e249–e258. [Google Scholar] [CrossRef]
- Rossi, M.; Amodio, N.; Martino, M.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. From Target Therapy to miRNA Therapeutics of Human Multiple Myeloma: Theoretical and Technological Issues in the Evolving Scenario. Curr. Drug Targets 2013, 14, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A.M.; Kerin, M.J.; Miller, N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wu, Y.; Huang, J.; Li, Q.; Kang, K.; Qu, J.; Li, F.; Gou, D. Identification of reference genes for circulating microRNA analysis in colorectal cancer. Sci. Rep. 2016, 6, 1–9. [Google Scholar]
- Cuscino, N.; Raimondi, L.; De Luca, A.; Carcione, C.; Russelli, G.; Conti, L.; Baldi, J.; Giulio Conaldi, P.; Giavaresi, G.; Gallo, A. Gathering novel circulating exosomal microRNA in osteosarcoma cell lines and possible implications for the disease. Cancers 2019, 11, 1–11. [Google Scholar] [CrossRef]
- Whiting, P.F. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Allen-Rhoades, W.; Kurenbekova, L.; Satterfield, L.; Parikh, N.; Fuja, D.; Shuck, R.L.; Rainusso, N.; Trucco, M.; Barkauskas, D.A.; Jo, E.; et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015, 4, 977–988. [Google Scholar] [CrossRef]
- Cai, H.; Zhao, H.; Tang, J.; Wu, H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J. Surg. Res. 2015, 194, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, K.; Liu, L.H.; Ouyang, Y.; Guo, H.B.; Zhang, H.; Bu, J.; Xiao, T. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol. Lett. 2015, 10, 1662–1668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lian, F.; Cui, Y.; Zhou, C.; Gao, K.; Wu, L. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PLoS ONE 2015, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhao, H.; Cai, H.; Wu, H. Diagnostic and prognostic potentials of microRNA-27a in osteosarcoma. Biomed. Pharmacother. 2015, 71, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.G.; Wang, D.C.; Tan, B.Y.; Wang, F.; Yuan, Z.N. Down-regulation of microRNA152 is associated with the diagnosis and prognosis of patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 9314–9319. [Google Scholar]
- Wang, T.; Ji, F.; Dai, Z.; Xie, Y.; Yuan, D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015, 15, 543–550. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zhang, X.; Zhang, M.; Liu, H.; Zhang, S.; Qi, B.; Sun, X. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed. Pharmacother. 2015, 75, 153–158. [Google Scholar] [CrossRef]
- Zhou, G.; Lu, M.; Chen, J.; Li, C.; Zhang, J.; Chen, J.; Shi, X.; Wu, S. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumor Biol. 2015, 36, 8845–8852. [Google Scholar] [CrossRef]
- Cao, L.; Wang, J.; Wang, P.Q. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed. Pharmacother. 2016, 84, 828–835. [Google Scholar] [CrossRef]
- Niu, J.; Sun, Y.; Guo, Q.; Niu, D.; Liu, B. Serum miR-95-3p is a diagnostic and prognostic marker for osteosarcoma. Springerplus 2016, 5, 1947. [Google Scholar] [CrossRef]
- Pang, P.-C.; Shi, X.-Y.; Huang, W.-L.; Sun, K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3765–3769. [Google Scholar]
- Sun, Y.; He, N.; Dong, Y.; Jiang, C. MiR-24-BIM-Smac/DIABLO axis controls the sensitivity to doxorubicin treatment in osteosarcoma. Sci. Rep. 2016, 6, 34238. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, B.; Hu, J.; Zhou, Y.; Jiang, M.; Wu, M.; Qin, L.; Yang, X. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumor Biol. 2016, 37, 9001–9007. [Google Scholar] [CrossRef]
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Yoshida, A.; Uehara, T.; Omori, T.; Sugiu, K.; et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 2017, 8, 33375–33392. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Sun, X.; Zhang, Y.; Liu, L.; Yuan, Q. MiR-598: A tumor suppressor with biomarker significance in osteosarcoma. Life Sci. 2017, 188, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.N.; Luo, S.; Liu, C.; Piao, Z.; Gou, W.; Wang, Y.; Guan, W.; Li, Q.; Zou, H.; Yang, Z.Z.; et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting αB-crystallin. Mol. Ther. 2017, 25, 2140–2149. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Liao, Y.; Shen, L.; Hu, F.; Yu, S.; Zhou, Y.; Zhang, Y.; Yang, Y.; Li, D.; Ren, M.; et al. Identification of the miRNA-mRNA regulatory network of small cell osteosarcoma based on RNA-seq. Oncotarget 2017, 8, 42525–42536. [Google Scholar] [CrossRef]
- Cong, C.; Wang, W.; Tian, J.; Gao, T.; Zheng, W.; Zhou, C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark. 2018, 21, 449–454. [Google Scholar] [CrossRef]
- Li, Q.; Song, S.; Ni, G.; Li, Y.; Wang, X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer Biomark. 2018, 21, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, X.T.; Zhang, Y.J.; Fang, G.W.; Xue, Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J. Int. Med. Res. 2018, 46, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.G.; Zhuang, Y.; Jin, Z.; Zhou, F.; Zhu, L.F.; Shen, P.C. MicroRNA-337-5p participates in the development and progression of osteosarcoma via ERBB, MAPK and VEGF pathways. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5460–5470. [Google Scholar] [PubMed]
- Xu, N.; Yang, W.; Liu, Y.; Yan, F.; Yu, Z. MicroRNA-411 promoted the osteosarcoma progression by suppressing MTSS1 expression. Environ. Sci. Pollut. Res. 2018, 25, 12064–12071. [Google Scholar] [CrossRef]
- Yao, Z.S.; Li, C.; Liang, D.; Jiang, X.B.; Tang, J.J.; Ye, L.Q.; Yuan, K.; Ren, H.; Yang, Z.D.; Jin, D.X.; et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark. 2018, 22, 127–133. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Xu, J.; Luo, Y.; Xin, Y.; Wang, Y. Downregulation of microRNA-95-3p suppresses cell growth of osteosarcoma via CDKN1A/p21 expression. Oncol. Rep. 2018, 39, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Natino, D.; Zhai, X.; Gao, Z.; He, X. MicroRNA-22 inhibits the proliferation and migration, and increases the cisplatin sensitivity, of osteosarcoma cells. Mol. Med. Rep. 2018, 17, 7209–7217. [Google Scholar] [CrossRef]
- Zhou, L.; Ma, X.; Yue, J.; Chen, T.; Wang, X.Y.; Wang, Z.W.; Pan, J.; Lin, Y. The diagnostic effect of serum miR-139-5p as an indicator in osteosarcoma. Cancer Biomark. 2018, 23, 561–567. [Google Scholar] [CrossRef]
- Huang, C.; Sun, Y.; Ma, S.; Vadamootoo, A.S.; Wang, L.; Jin, C. Identification of circulating miR-663a as a potential biomarker for diagnosing osteosarcoma. Pathol. Res. Pract. 2019, 215, 1–6. [Google Scholar] [CrossRef]
- Shi, L.; Xie, C.; Zhu, J.; Chen, X. Downregulation of serum miR-194 predicts poor prognosis in osteosarcoma patients. Ann. Diagn. Pathol. 2020, 46, 151488. [Google Scholar] [CrossRef]
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular vesicle-mediated delivery of miR-101 inhibits lung metastasis in osteosarcoma. Theranostics 2020, 10, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Kopp, L.M.; Womer, R.B.; Schwartz, C.L.; Ebb, D.H.; Franco, V.I.; Hall, D.; Barkauskas, D.A.; Krailo, M.D.; Grier, H.E.; Meyers, P.A.; et al. Effects of dexrazoxane on doxorubicin-related cardiotoxicity and second malignant neoplasms in children with osteosarcoma: A report from the Children’s Oncology Group. Cardio-Oncology 2019, 5, 1–12. [Google Scholar] [CrossRef]
- Klein, M.J.; Siegal, G.P. Osteosarcoma. Am. J. Clin. Pathol. 2006, 125, 555–581. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, M.; Malhotra, K.; Patel, S. Primary Osteosarcoma in the Elderly Revisited: Current Concepts in Diagnosis and Treatment. Curr. Oncol. Rep. 2018, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Zamborsky, R.; Kokavec, M.; Harsanyi, S.; Danisovic, L. Identification of Prognostic and Predictive Osteosarcoma Biomarkers. Med. Sci. 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, L.; De Luca, A.; Costa, V.; Amodio, N.; Carina, V.; Bellavia, D.; Tassone, P.; Pagani, S.; Fini, M.; Alessandro, R.; et al. Circulating biomarkers in osteosarcoma: New translational tools for diagnosis and treatment. Oncotarget 2017, 8, 100831–100851. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of Circulating MicroRNA: Preanalytical and Analytical Challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Guttery, D.S.; Zahra, N.; Primrose, L.; Elshaw, S.R.; Pringle, J.H.; Blighe, K.; Marchese, S.D.; Hills, A.; Woodley, L.; et al. Influence of Plasma Processing on Recovery and Analysis of Circulating Nucleic Acids. PLoS ONE 2013, 8, e77963. [Google Scholar] [CrossRef]
- Li, Y.; Kowdley, K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012, 431, 69–75. [Google Scholar] [CrossRef]
- Kroh, E.M.; Parkin, R.K.; Mitchell, P.S.; Tewari, M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010, 50, 298–301. [Google Scholar] [CrossRef]
- Prado, M.S.J.G.; de Goes, T.C.; de Jesus, M.L.; Mendonça, L.S.O.; Nascimento, J.S.; Kaneto, C.M. Identification of miR-328-3p as an endogenous reference gene for the normalization of miRNA expression data from patients with Diabetic Retinopathy. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]

| Author | Year | Type of Global MicroRNA Expression Profiling | Were Samples Pooled? | If Yes, What Was the Number of Samples Per Pool? | If Samples Were Not Pooled, How Many Samples Per Group Were Analyzed in Large-Scale Analysis? | Were Candidate MicroRNAs Selected by Analysis of Public MicroRNAs Datasets? | Were Candidate MicroRNAs Selected by Literature Review? |
|---|---|---|---|---|---|---|---|
| Allen-Rhoades et al. [12] | 2015 | MicroRNA PCR panel | Conducted using non-human samples | N/A | N/A | No | No |
| Cai et al. [13] | 2015 | N/A | N/A | N/A | N/A | No | Yes |
| Hui et al. [14] | 2015 | MicroRNA PCR panel | No | N/A | 3 per group | No | No |
| Lian et al. [15] | 2015 | MicroRNA PCR panel | Yes | 2 pools with 10 samples each | N/A | No | Yes |
| Tang et al. [16] | 2015 | N/A | N/A | N/A | N/A | No | Yes |
| Wang et al. [17] | 2015 | N/A | N/A | N/A | N/A | No | Yes |
| Wang et al. [18] | 2015 | N/A | N/A | N/A | N/A | No | Yes |
| Yang et al. [19] | 2015 | N/A | N/A | N/A | N/A | No | Yes |
| Zhou et al. [20] | 2015 | MicroRNA PCR panel | Yes | 3 pools with 10 samples each | N/A | No | No |
| Cao et al. [21] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Li et al. [3] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Niu et al. [22] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Pang et al. [23] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Sun et al. [24] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Zhou et al. [25] | 2016 | N/A | N/A | N/A | N/A | No | Yes |
| Fujiwara et al. [26] | 2017 | Microarray | No | N/A | 10 per group | No | No |
| Liu et al. [27] | 2017 | N/A | N/A | N/A | N/A | No | Yes |
| Wang et al. [28] | 2017 | N/A | N/A | N/A | N/A | No | Yes |
| Xie et al. [29] | 2017 | Sequencing | No | N/A | 3 OS and 10 control subjects | No | No |
| Cong et al. [30] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Li, Song et al. [31] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Liu, Zhao et al. [32] | 2018 | N/A | N/A | N/A | N/A | No | No |
| Monterde-Cruz et al. [2] | 2018 | MicroRNA PCR panel | Yes | 4 pools with 5 samples each | N/A | No | No |
| Tian et al. [33] | 2018 | N/A | N/A | N/A | N/A | Yes | No |
| Xu et al. [34] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Yao et al. [35] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Zhao et al. [36] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Zhou et al. [37] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Zhou et al. [38] | 2018 | N/A | N/A | N/A | N/A | No | Yes |
| Cuscino et al. [9] | 2019 | Sequencing | Conducted using cell lineages samples | N/A | N/A | No | No |
| Huang et al. [4] | 2019 | N/A | N/A | N/A | N/A | Yes | No |
| Huang, Sun et al. [39] | 2019 | N/A | N/A | N/A | N/A | No | No |
| Zhu et al. [1] | 2019 | N/A | N/A | N/A | N/A | No | Yes |
| Shi et al. [40] | 2020 | N/A | N/A | N/A | N/A | No | Yes |
| Zhang et al. [41] | 2020 | Sequencing | No | N/A | 1 per group | No | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gally, T.B.; Aleluia, M.M.; Borges, G.F.; Kaneto, C.M. Circulating MicroRNAs as Novel Potential Diagnostic Biomarkers for Osteosarcoma: A Systematic Review. Biomolecules 2021, 11, 1432. https://doi.org/10.3390/biom11101432
Gally TB, Aleluia MM, Borges GF, Kaneto CM. Circulating MicroRNAs as Novel Potential Diagnostic Biomarkers for Osteosarcoma: A Systematic Review. Biomolecules. 2021; 11(10):1432. https://doi.org/10.3390/biom11101432
Chicago/Turabian StyleGally, Thaís Borges, Milena Magalhães Aleluia, Grasiely Faccin Borges, and Carla Martins Kaneto. 2021. "Circulating MicroRNAs as Novel Potential Diagnostic Biomarkers for Osteosarcoma: A Systematic Review" Biomolecules 11, no. 10: 1432. https://doi.org/10.3390/biom11101432
APA StyleGally, T. B., Aleluia, M. M., Borges, G. F., & Kaneto, C. M. (2021). Circulating MicroRNAs as Novel Potential Diagnostic Biomarkers for Osteosarcoma: A Systematic Review. Biomolecules, 11(10), 1432. https://doi.org/10.3390/biom11101432






