Isolation of Cysteine-Rich Peptides from Citrullus colocynthis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Extraction
2.2. Reversed-Phase High Performance Liquid Chromatography (RP-HPLC) Purification
2.3. Reduction, Alkylation and Enzymatic Digestion of Peptides
2.4. MALDI-TOF/TOF Analysis and De Novo Peptide Sequencing
2.5. Amino Acid Composition Analysis
2.6. Trypsin, α-Chymotrypsin and α-Amylase Inhibitory Assays
2.7. Sequence Homology Analysis
3. Results and Discussion
3.1. Preparation of Peptide-Enriched Citcol Extracts
3.2. Reversed-Phase HPLC Fractionation and Purification
3.3. De Novo Sequencing of Citcol Peptides by MALDI-TOF/TOF
3.4. Genome Mining and Sequence Homology Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Lei, C.; Yu, C. Mesoporous silica nanoparticles for protein protection and delivery. Front. Chem. 2019, 7, 290. [Google Scholar] [CrossRef] [PubMed]
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef] [PubMed]
- Gilad, Y.; Firer, M.; Gellerman, G. Recent innovations in peptide based targeted drug delivery to cancer cells. Biomedicines 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Hellinger, R.; Gruber, C.W. Peptide-based protease inhibitors from plants. Drug Discov. Today 2019, 24, 1877–1889. [Google Scholar] [CrossRef]
- Wang, C.K.; Craik, D.J. Designing macrocyclic disulfide-rich peptides for biotechnological applications. Nat. Chem. Biol. 2018, 14, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef]
- Lien, S.; Lowman, H.B. Therapeutic peptides. Trends Biotechnol. 2003, 21, 556–562. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- McIntosh, M.; Cruz, L.; Hunkapiller, M.; Gray, W.; Olivera, B. Isolation and structure of a peptide toxin from the marine snail Conus magus. Arch. Biochem. Biophys. 1982, 218, 329–334. [Google Scholar] [CrossRef]
- Thell, K.; Hellinger, R.; Sahin, E.; Michenthaler, P.; Gold-Binder, M.; Haider, T.; Kuttke, M.; Liutkeviciute, Z.; Goransson, U.; Grundemann, C.; et al. Oral activity of a nature-derived cyclic peptide for the treatment of multiple sclerosis. Proc. Natl. Acad. Sci. USA 2016, 113, 3960–3965. [Google Scholar] [CrossRef]
- Das, D.; Jaiswal, M.; Khan, F.N.; Ahamad, S.; Kumar, S. PlantPepDB: A manually curated plant peptide database. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Hellinger, R.; Koehbach, J.; Soltis, D.E.; Carpenter, E.J.; Wong, G.K.-S.; Gruber, C.W. Peptidomics of circular cysteine-rich plant peptides: Analysis of the diversity of cyclotides from viola tricolor by transcriptome and proteome mining. J. Proteome Res. 2015, 14, 4851–4862. [Google Scholar] [CrossRef] [PubMed]
- Colgrave, M.L.; Kotze, A.C.; Ireland, D.C.; Wang, C.K.; Craik, D.J. The anthelmintic activity of the cyclotides: Natural variants with enhanced activity. ChemBioChem 2008, 9, 1939–1945. [Google Scholar] [CrossRef]
- Baraguey, C.; Blond, A.; Cavelier, F.; Pousset, J.-L.; Bodo, B.; Auvin-Guette, C. Isolation, structure and synthesis of mahafacyclin B, a cyclic heptapeptide from the latex of Jatropha mahafalensis. J. Chem. Soc. Perkin Trans. 2001, 1, 2098–2103. [Google Scholar] [CrossRef]
- Lipkin, A.; Anisimova, V.; Nikonorova, A.; Babakov, A.; Krause, E.; Bienert, M.; Grishin, E.; Egorov, T. An antimicrobial peptide Ar-AMP from amaranth (Amaranthus retroflexus L.) seeds. Phytochemistry 2005, 66, 2426–2431. [Google Scholar] [CrossRef] [PubMed]
- Hallock, Y.F.; Sowder, R.C.; Pannell, L.K.; Hughes, C.B.; Johnson, D.G.; Gulakowski, R.; Cardellina, J.H.; Boyd, M.R. Cycloviolins A–D, Anti-HIV Macrocyclic Peptides from Leonia c ymosa. J. Org. Chem. 2000, 65, 124–128. [Google Scholar] [CrossRef]
- Blanco-Aparicio, C.; Molina, M.A.; Fernández-Salas, E.; Frazier, M.L.; Mas, J.M.; Querol, E.; Avilés, F.X.; de Llorens, R. Potato carboxypeptidase inhibitor, a T-knot protein, is an epidermal growth factor antagonist that inhibits tumor cell growth. J. Biol. Chem. 1998, 273, 12370–12377. [Google Scholar] [CrossRef]
- Hellinger, R.; Koehbach, J.; Puigpinós, A.; Clark, R.J.; Tarragó, T.; Giralt, E.; Gruber, C.W. Inhibition of human prolyl oligopeptidase activity by the cyclotide psysol 2 isolated from Psychotria solitudinum. J. Nat. Prod. 2015, 78, 1073–1082. [Google Scholar] [CrossRef]
- Lobo-Ruiz, A.; Tulla-Puche, J. Synthetic approaches of naturally and rationally designed peptides and peptidomimetics. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 23–49. [Google Scholar]
- Wang, C.K.; Gruber, C.W.; Cemazar, M.A.; Siatskas, C.; Tagore, P.; Payne, N.; Sun, G.; Wang, S.; Bernard, C.C.; Craik, D.J. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem. Biol. 2014, 9, 156–163. [Google Scholar] [CrossRef]
- Felizmenio-Quimio, M.E.; Daly, N.L.; Craik, D.J. Circular Proteins in Plants solution structure of a novel macrocyclic trypsin inhibitor frommomordica cochinchinensis. J. Biol. Chem. 2001, 276, 22875–22882. [Google Scholar] [CrossRef] [PubMed]
- He, W.-J.; Chan, L.Y.; Clark, R.J.; Tang, J.; Zeng, G.-Z.; Franco, O.L.; Cantacessi, C.; Craik, D.J.; Daly, N.L.; Tan, N.-H. Novel inhibitor cystine knot peptides from Momordica charantia. PLoS ONE 2013, 8, e75334. [Google Scholar] [CrossRef][Green Version]
- Heitz, A.; Hernandez, J.F.; Gagnon, J.; Hong, T.T.; Pham, T.T.; Nguyen, T.M.; Le-Nguyen, D.; Chiche, L. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry 2001, 40, 7973–7983. [Google Scholar] [CrossRef]
- Zhu, S.; Darbon, H.; Dyason, K.; Verdonck, F.; Tytgat, J. Evolutionary origin of inhibitor cystine knot peptides. FASEB J. 2003, 17, 1765–1767. [Google Scholar] [CrossRef]
- Marzouk, B.; Marzouk, Z.; Haloui, E.; Fenina, N.; Bouraoui, A.; Aouni, M. Screening of analgesic and anti-inflammatory activities of Citrullus colocynthis from southern Tunisia. J. Ethnopharmacol. 2010, 128, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hassan, I.A.; Abdel-Barry, J.A.; Mohammeda, S.T. The hypoglycaemic and antihyperglycaemic effect of Citrullus colocynthis fruit aqueous extract in normal and alloxan diabetic rabbits. J. Ethnopharmacol. 2000, 71, 325–330. [Google Scholar] [CrossRef]
- Ostovar, M.; Akbari, A.; Anbardar, M.H.; Iraji, A.; Salmanpour, M.; Ghoran, S.H.; Heydari, M.; Shams, M. Effects of Citrullus colocynthis L. in a rat model of diabetic neuropathy. J. Integr. Med. 2020, 18, 59–67. [Google Scholar] [CrossRef]
- Tannin-Spitz, T.; Grossman, S.; Dovrat, S.; Gottlieb, H.E.; Bergman, M. Growth inhibitory activity of cucurbitacin glucosides isolated from Citrullus colocynthis on human breast cancer cells. Biochem. Pharmacol. 2007, 73, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.E.; Boulos, J.C.; Elhaboub, G.; Rigano, D.; Saab, A.; Loizzo, M.R.; Hassan, L.E.; Sugimoto, Y.; Piacente, S.; Tundis, R. Cytotoxicity of cucurbitacin E from Citrullus colocynthis against multidrug-resistant cancer cells. Phytomedicine 2019, 62, 152945. [Google Scholar] [CrossRef]
- Gruber, C.W.; Elliott, A.G.; Ireland, D.C.; Delprete, P.G.; Dessein, S.; Göransson, U.; Trabi, M.; Wang, C.K.; Kinghorn, A.B.; Robbrecht, E. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 2008, 20, 2471–2483. [Google Scholar] [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Clark, R.J.; Daly, N.L.; Craik, D.J. Structural plasticity of the cyclic-cystine-knot framework: Implications for biological activity and drug design. Biochem. J. 2006, 394, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.M.; Kylarova, S.; Mergaert, P.; Kondorosi, E. Unexplored Arsenals of Legume Peptides With Potential for Their Applications in Medicine and Agriculture. Front. Microbiol. 2020, 11, 1307. [Google Scholar] [CrossRef]
- Parisi, K.; Shafee, T.M.A.; Quimbar, P.; van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 2019, 88, 107–118. [Google Scholar] [CrossRef]
- Mahatmanto, T.; Mylne, J.S.; Poth, A.G.; Swedberg, J.E.; Kaas, Q.; Schaefer, H.; Craik, D.J. The evolution of Momordica cyclic peptides. Molecular Biol. Evol. 2015, 32, 392–405. [Google Scholar] [CrossRef]
- Giacomelli, L.; Nanni, V.; Lenzi, L.; Zhuang, J.; Serra, M.D.; Banfield, M.J.; Town, C.D.; Silverstein, K.A.; Baraldi, E.; Moser, C. Identification and characterization of the defensin-like gene family of grapevine. Mol. Plant Microbe Interact. 2012, 25, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Jordan, T.; Schornack, S.; Lahaye, T. Alternative splicing of transcripts encoding Toll-like plant resistance proteins–what’s the functional relevance to innate immunity? Trends Plant Sci. 2002, 7, 392–398. [Google Scholar] [CrossRef]
- Lawrence, G.J.; Finnegan, E.J.; Ayliffe, M.A.; Ellis, J.G. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 1995, 7, 1195–1206. [Google Scholar]
- Moran, Y.; Weinberger, H.; Reitzel, A.M.; Sullivan, J.C.; Kahn, R.; Gordon, D.; Finnerty, J.R.; Gurevitz, M. Intron retention as a posttranscriptional regulatory mechanism of neurotoxin expression at early life stages of the starlet anemone Nematostella vectensis. J. Mol. Biol. 2008, 380, 437–443. [Google Scholar] [CrossRef]
- Schornack, S.; Ballvora, A.; Gürlebeck, D.; Peart, J.; Ganal, M.; Baker, B.; Bonas, U.; Lahaye, T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004, 37, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Favel, A.; Mattras, H.; Coletti-Previero, M.; Zwilling, R.; Robinson, E.; Castro, B. Protease inhibitors from Ecballium elaterium seeds. Int. J. Pept. Protein Res. 1989, 33, 202–208. [Google Scholar] [CrossRef]
- Hara, S.; Makino, J.; Ikenaka, T. Amino acid sequences and disulfide bridges of serine proteinase inhibitors from bitter gourd (Momordica charantia LINN.) seeds. J. Biochem. 1989, 105, 88–92. [Google Scholar] [CrossRef]
- Hayashi, K.; Takehisa, T.; Hamato, N.; Takano, R.; Hara, S.; Miyata, T.; Kato, H. Inhibition of serine proteases of the blood coagulation system by squash family protease inhibitors. J. Biochem. 1994, 116, 1013–1018. [Google Scholar] [CrossRef]
- Hernandez, J.F.; Gagnon, J.; Chiche, L.; Nguyen, T.M.; Andrieu, J.P.; Heitz, A.; Trinh Hong, T.; Pham, T.T.; Le Nguyen, D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 2000, 39, 5722–5730. [Google Scholar] [CrossRef] [PubMed]
- Hass, G.M.; Nau, H.; Biemann, K.; Grahn, D.T.; Ericsson, L.H.; Neurath, H. Amino acid sequence of a carboxypeptidase inhibitor from potatoes. Biochemistry 1975, 14, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, J.; Xu, C.; Ren, F.; Peng, C.; Wu, G.; Zhao, J. Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol. 2000, 122, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, J.; Pszczola, K.; Wilimowska-Pelc, A.; Lorenc-Kubis, I.; Zuziak, E.; Lugowski, M.; Legowska, A.; Kwiatkowska, A.; Sleszynska, M.; Lesner, A.; et al. Trypsin inhibitors from the garden four o’clock (Mirabilis jalapa) and spinach (Spinacia oleracea) seeds: Isolation, characterization and chemical synthesis. Phytochemistry 2007, 68, 1487–1496. [Google Scholar] [CrossRef]
- Chagolla-Lopez, A.; Blanco-Labra, A.; Patthy, A.; Sánchez, R.; Pongor, S. A novel alpha-amylase inhibitor from amaranth (Amaranthus hypocondriacus) seeds. J. Biol. Chem. 1994, 269, 23675–23680. [Google Scholar] [PubMed]
- Chou, M.-X.; Wei, X.-Y.; Chen, D.-S.; Zhou, J.-C. Thirteen nodule-specific or nodule-enhanced genes encoding products homologous to cysteine cluster proteins or plant lipid transfer proteins are identified in Astragalus sinicus L. by suppressive subtractive hybridization. J. Exp. Bot. 2006, 57, 2673–2685. [Google Scholar] [CrossRef]
- McBride, J.D.; Watson, E.M.; Brauer, A.B.; Jaulent, A.M.; Leatherbarrow, R.J. Peptide mimics of the Bowman–Birk inhibitor reactive site loop. Pept. Sci. Orig. Res. Biomol. 2002, 66, 79–92. [Google Scholar] [CrossRef]
- Huang, J.; Wong, K.H.; Tay, S.V.; Serra, A.; Sze, S.K.; Tam, J.P. Astratides: Insulin-Modulating, Insecticidal, and Antifungal Cysteine-Rich Peptides from Astragalus membranaceus. J. Nat. Prod. 2019, 82, 194–204. [Google Scholar] [CrossRef]
- Kam, A.; Loo, S.; Fan, J.S.; Sze, S.K.; Yang, D.; Tam, J.P. Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation. J. Biol. Chem. 2019, 294, 19604–19615. [Google Scholar] [CrossRef]
- Molesini, B.; Treggiari, D.; Dalbeni, A.; Minuz, P.; Pandolfini, T. Plant cystine-knot peptides: Pharmacological perspectives. Br. J. Clin. Pharmacol. 2017, 83, 63–70. [Google Scholar] [CrossRef]
- Shelenkov, A.; Slavokhotova, A.; Odintsova, T. Predicting Antimicrobial and Other Cysteine-Rich Peptides in 1267 Plant Transcriptomes. Antibiotics (Basel) 2020, 9, 60. [Google Scholar] [CrossRef]
- Lavergne, V.; Taft, R.J.; Alewood, P.F. Cysteine-rich mini-proteins in human biology. Curr. Top. Med. Chem. 2012, 12, 1514–1533. [Google Scholar] [CrossRef] [PubMed]
- Gruber, C.W.; Muttenthaler, M. Discovery of defense- and neuropeptides in social ants by genome-mining. PLoS ONE 2012, 7, e32559. [Google Scholar] [CrossRef] [PubMed]
- Koehbach, J.; Craik, D.J. The Vast Structural Diversity of Antimicrobial Peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef]
- de Veer, S.J.; Wang, C.K.; Harris, J.M.; Craik, D.J.; Swedberg, J.E. Improving the Selectivity of Engineered Protease Inhibitors: Optimizing the P2 Prime Residue Using a Versatile Cyclic Peptide Library. J. Med. Chem. 2015, 58, 8257–8268. [Google Scholar] [CrossRef] [PubMed]
- Legowska, A.; Debowski, D.; Lesner, A.; Wysocka, M.; Rolka, K. Introduction of non-natural amino acid residues into the substrate-specific P1 position of trypsin inhibitor SFTI-1 yields potent chymotrypsin and cathepsin G inhibitors. Bioorg. Med. Chem. 2009, 17, 3302–3307. [Google Scholar] [CrossRef]
- Gruber, C.W.; Cemazar, M.; Anderson, M.A.; Craik, D.J. Insecticidal plant cyclotides and related cystine knot toxins. Toxicon 2007, 49, 561–575. [Google Scholar] [CrossRef]


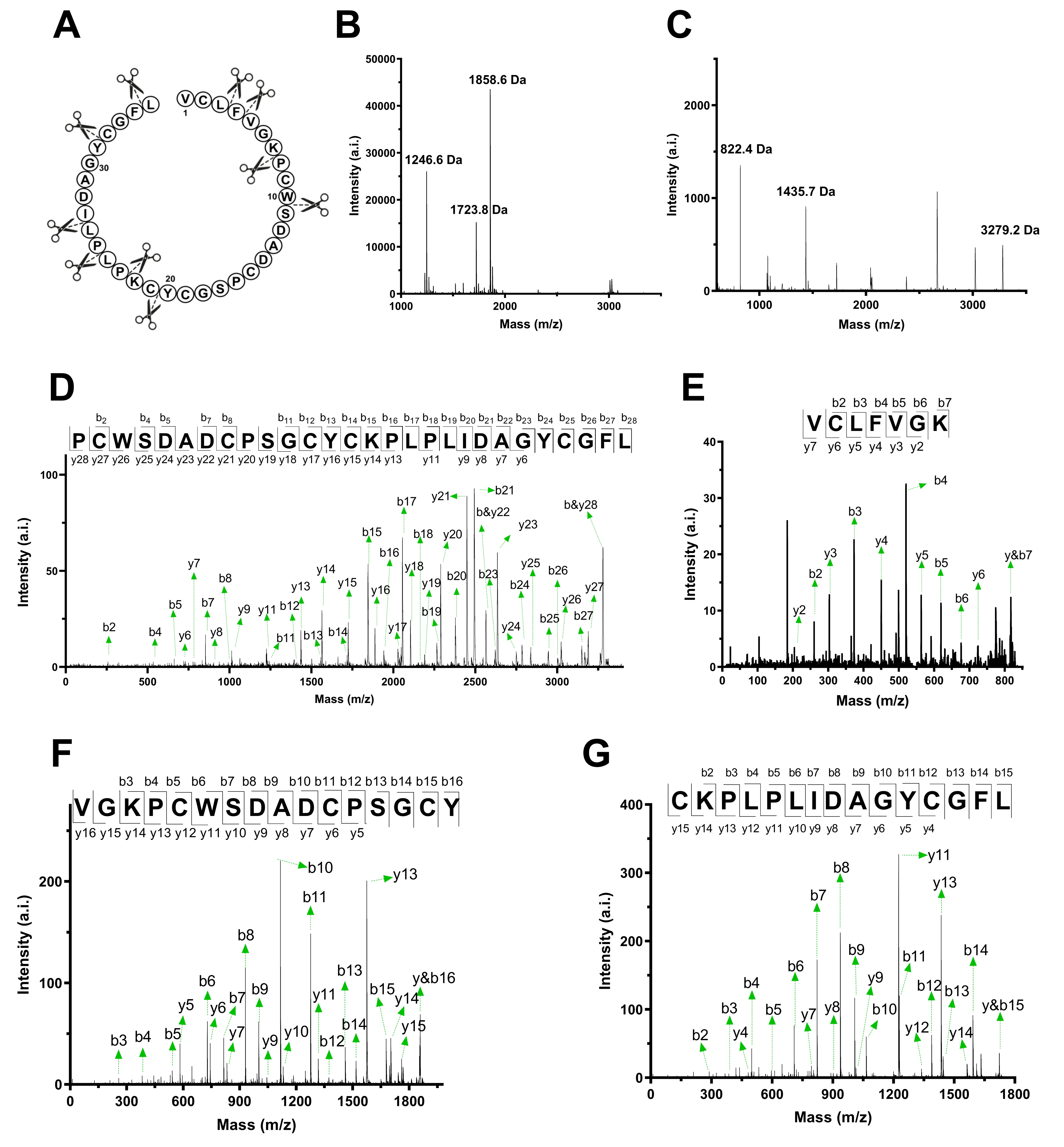

| Name | [M + H]+1 | Sequence 2 | Length |
|---|---|---|---|
| loop 1 2 3 4 5 | |||
| citcol-1 | 3658.1 | ----VCLFVGKPCWSDADCPSGCSCKPLPLIDAGYCGFL | 35 aa |
| citcol-2 | 3734.1 | ----VCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | 35 aa |
| citcol-3 | 3814.4 | --VGVCLFVGKPCWSDADCPSGCSCKPLPLIDAGYCGFL | 37 aa |
| citcol-4 | 3890.2 | --VGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | 37 aa |
| citcol-5 | 4046.9 | -RVGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | 38 aa |
| citcol-6 | 4084.1 | NRVGVCLFVGKPCWSDADCPSGCSCKPLPLIDAGYCGFL | 39 aa |
| citcol-7 | 4101.2 | NRVGVCLFVGKPCKSDANCPSGCYCKPLPLIDAGYCGFL | 39 aa |
| citcol-8 | 4160.3 | NRVGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | 39 aa |
| ********* ***:***** *************** |
| Species | Sequence 2 | E-Value 3 | Accession No |
|---|---|---|---|
| C. colocynthis (citcol-8) | NRVGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | n.a. | this work |
| Citrullus lanatus (chr10 4) | NRVGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | 3 × 10−23 | VOOL01000010.1 |
| Cucumis melo (chr7 4) | -RLGVCLLVGKPCMSDADCPSGCYCKPVPLLDIGYCGFL | 5 × 10−16 | LN713261.1 |
| *:****:***** *************:**:* ****** |
| Species | Sequence | Uniprot ID | Function | Cys Loops Pattern | Similarity (%) 1 | Reference |
|---|---|---|---|---|---|---|
| C. colocynthis (citcol-8) | NRVGVCLFVGKPCWSDADCPSGCYCKPLPLIDAGYCGFL | n.a. | n.a. | CX6CX5CX3CX1CX10C 2 | n.a. | this work |
| Momordica cochinchinensis | QRACPRILKKCRRDSDCPGECICKENGYCG | P82410 | trypsin inhibitor | CX6CX5CX3CX1CX5C | 43.6 | [51] |
| Solanum tuberosum | HADPICNKPCKTHDDCSGAWFCQACWNSARTCGPY | P01075 | metallocarboxy peptidase inhibitor | CX3CX5CX5CX2CX6C | 38.5 | [52] |
| Phytolacca americana | AGCIKNGGRCNASAGPPYCCSSYCFQIAGQSYGVCKNR | P81418 | antimicrobial peptide | CX6CX8CCX3CX10C | 31.7 | [53] |
| Spinacia oleracea | EDKCSPSGAICSGFGPPEQCCSGACVPHPILRIFVCQ | P84781 | trypsin inhibitor | CX6CX8CCX3CX10C | 29.5 | [54] |
| Amaranthus hypochondriacus | CIPKWNRCGPKMDGVPCCEPYTCTSDYYGNCS | P80403 | α-amylase inhibitor | CX6CX8CCX4CX7C | 27.3 | [55] |
| Amaranthus retroflexus | AGECVQGRCPSGMCCSQFGYCGRGPKYCGR | Q5I2B2 | antimicrobial peptide | CX4CX4CCX5CX6C | 26.7 | [17] |
| Astragalus sinicus | TYSCGGHIDCKDFCKSEGYRGFKCTPKKTCTCFH | Q07A30 | nodule specific protein | CX5CX3CX9CX5CX1C | 20.0 | [56] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahin-Kaleybar, B.; Niazi, A.; Afsharifar, A.; Nematzadeh, G.; Yousefi, R.; Retzl, B.; Hellinger, R.; Muratspahić, E.; Gruber, C.W. Isolation of Cysteine-Rich Peptides from Citrullus colocynthis. Biomolecules 2020, 10, 1326. https://doi.org/10.3390/biom10091326
Shahin-Kaleybar B, Niazi A, Afsharifar A, Nematzadeh G, Yousefi R, Retzl B, Hellinger R, Muratspahić E, Gruber CW. Isolation of Cysteine-Rich Peptides from Citrullus colocynthis. Biomolecules. 2020; 10(9):1326. https://doi.org/10.3390/biom10091326
Chicago/Turabian StyleShahin-Kaleybar, Behzad, Ali Niazi, Alireza Afsharifar, Ghorbanali Nematzadeh, Reza Yousefi, Bernhard Retzl, Roland Hellinger, Edin Muratspahić, and Christian W. Gruber. 2020. "Isolation of Cysteine-Rich Peptides from Citrullus colocynthis" Biomolecules 10, no. 9: 1326. https://doi.org/10.3390/biom10091326
APA StyleShahin-Kaleybar, B., Niazi, A., Afsharifar, A., Nematzadeh, G., Yousefi, R., Retzl, B., Hellinger, R., Muratspahić, E., & Gruber, C. W. (2020). Isolation of Cysteine-Rich Peptides from Citrullus colocynthis. Biomolecules, 10(9), 1326. https://doi.org/10.3390/biom10091326