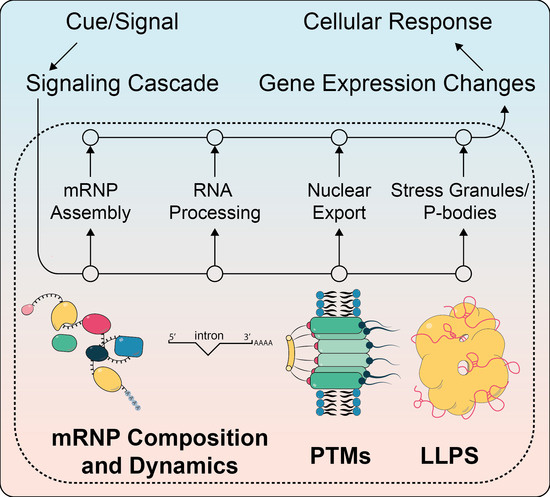
Dynamic mRNP Remodeling in Response to Internal and External Stimuli
Abstract
1. Introduction
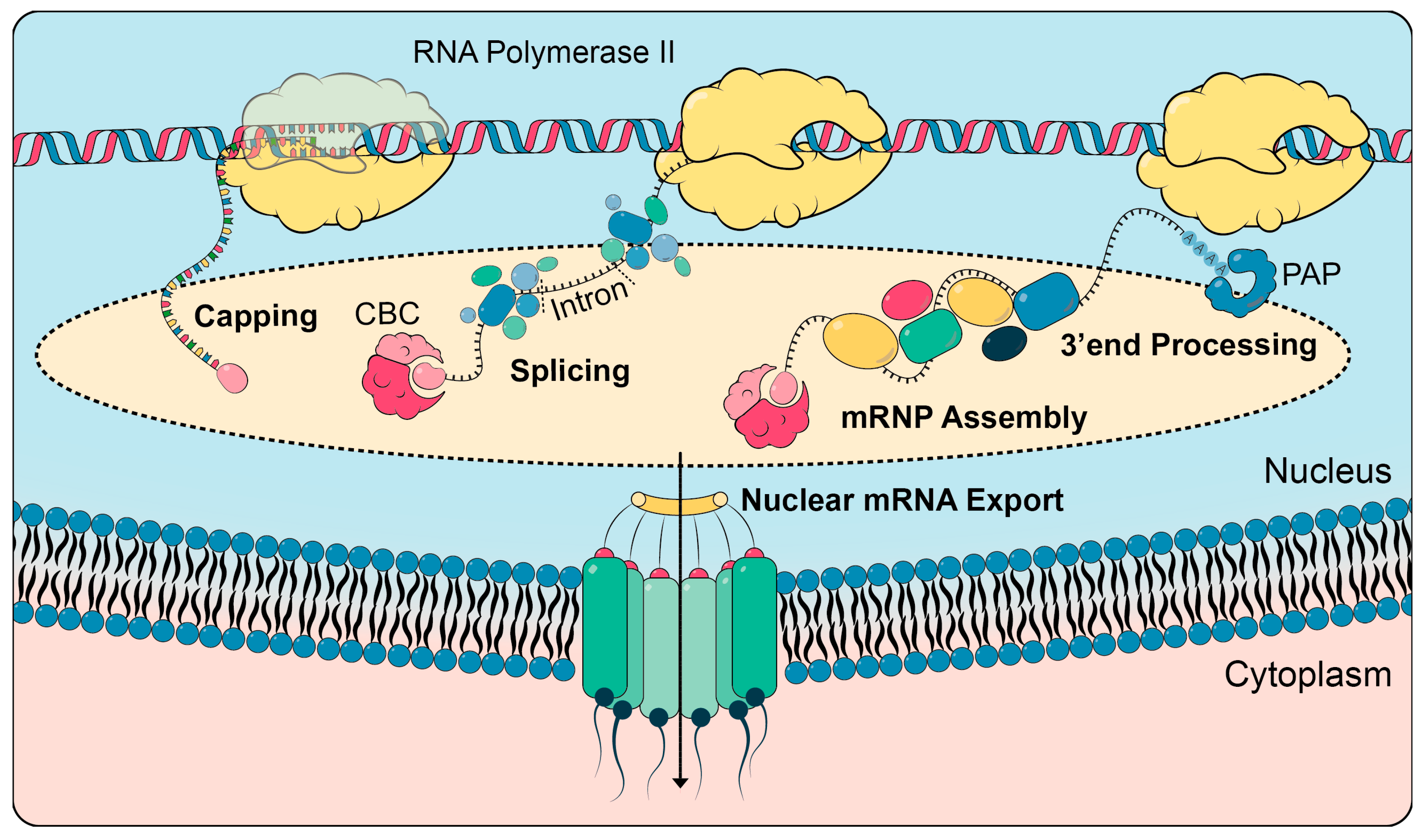
1.1. Nuclear mRNA Processing
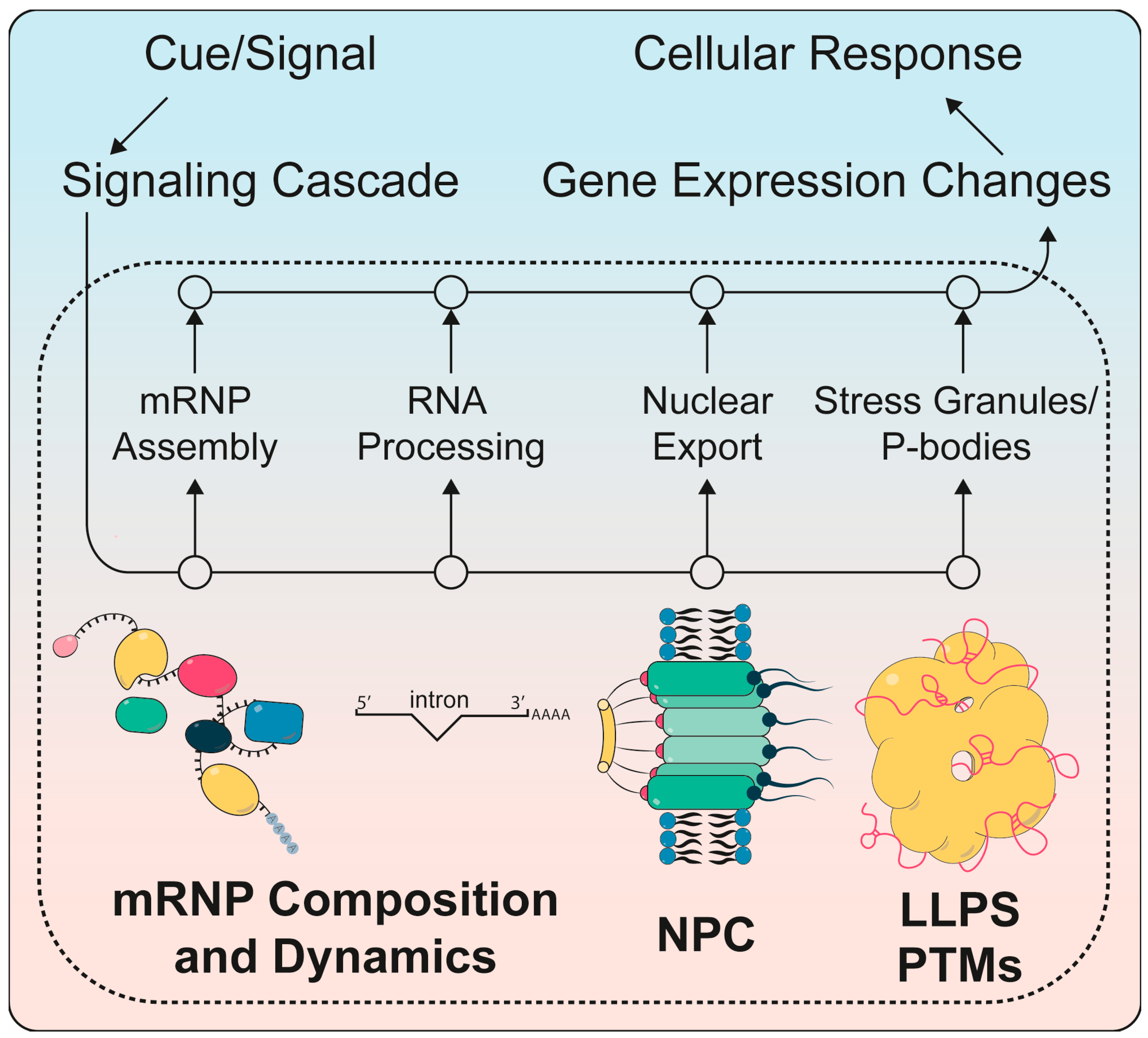
1.2. mRNP Assembly and Dynamics
1.3. Nuclear mRNA Export and Its Regulation
1.4. mRNA Translation and Decay in the Cytoplasm
1.5. Principles of Signal Transduction
2. Selected Examples
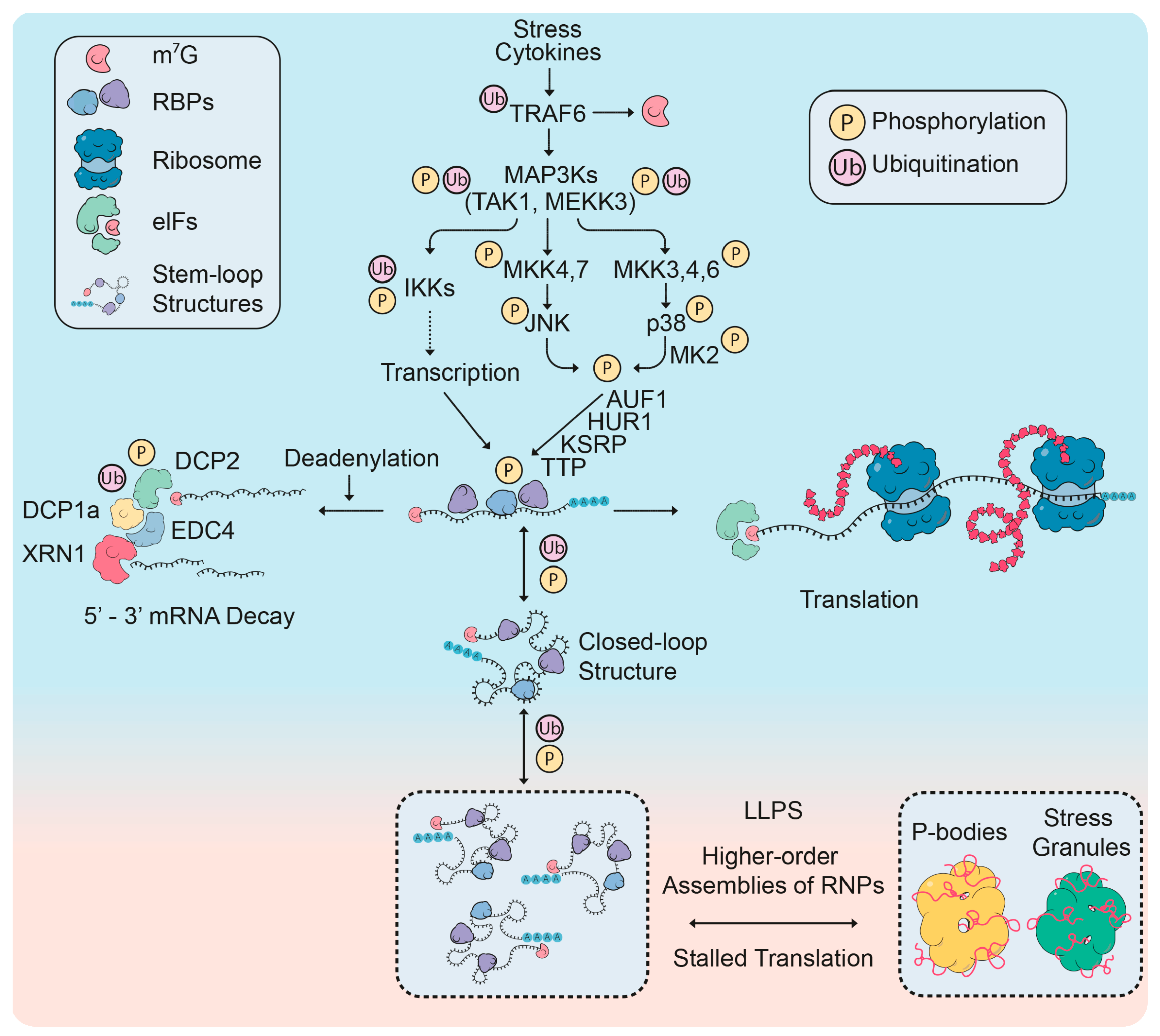
2.1. mRNP Remodeling Through Protein Modifications
2.1.1. Roles of Post-Translational Modifications in mRNP Remodeling
2.1.2. Regulation of P-Body Factors by Post-Translational Modifications
2.2. RNA Processing Changes in Response to External Signals
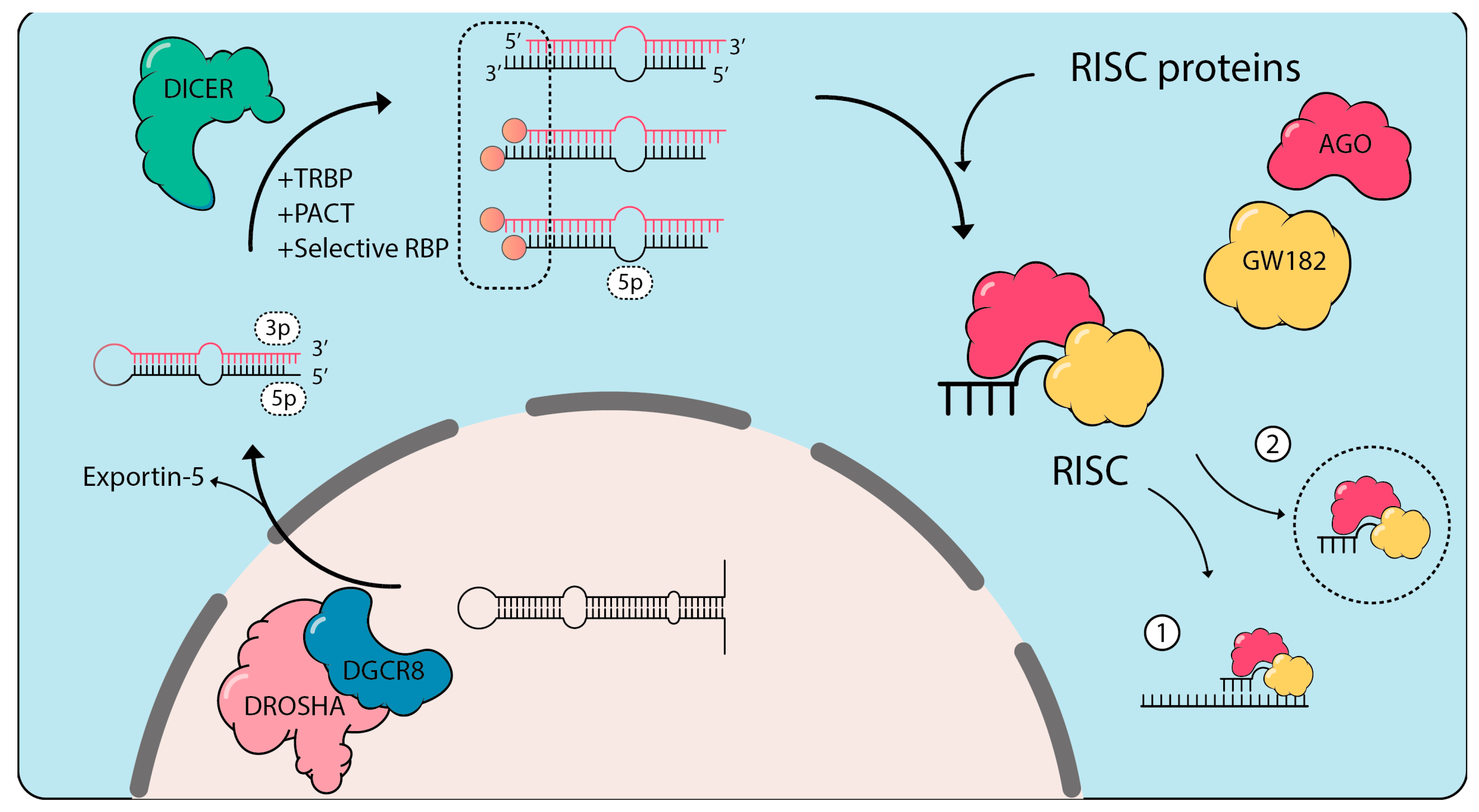
2.2.1. Differential Regulation of microRNAs Upon Stress
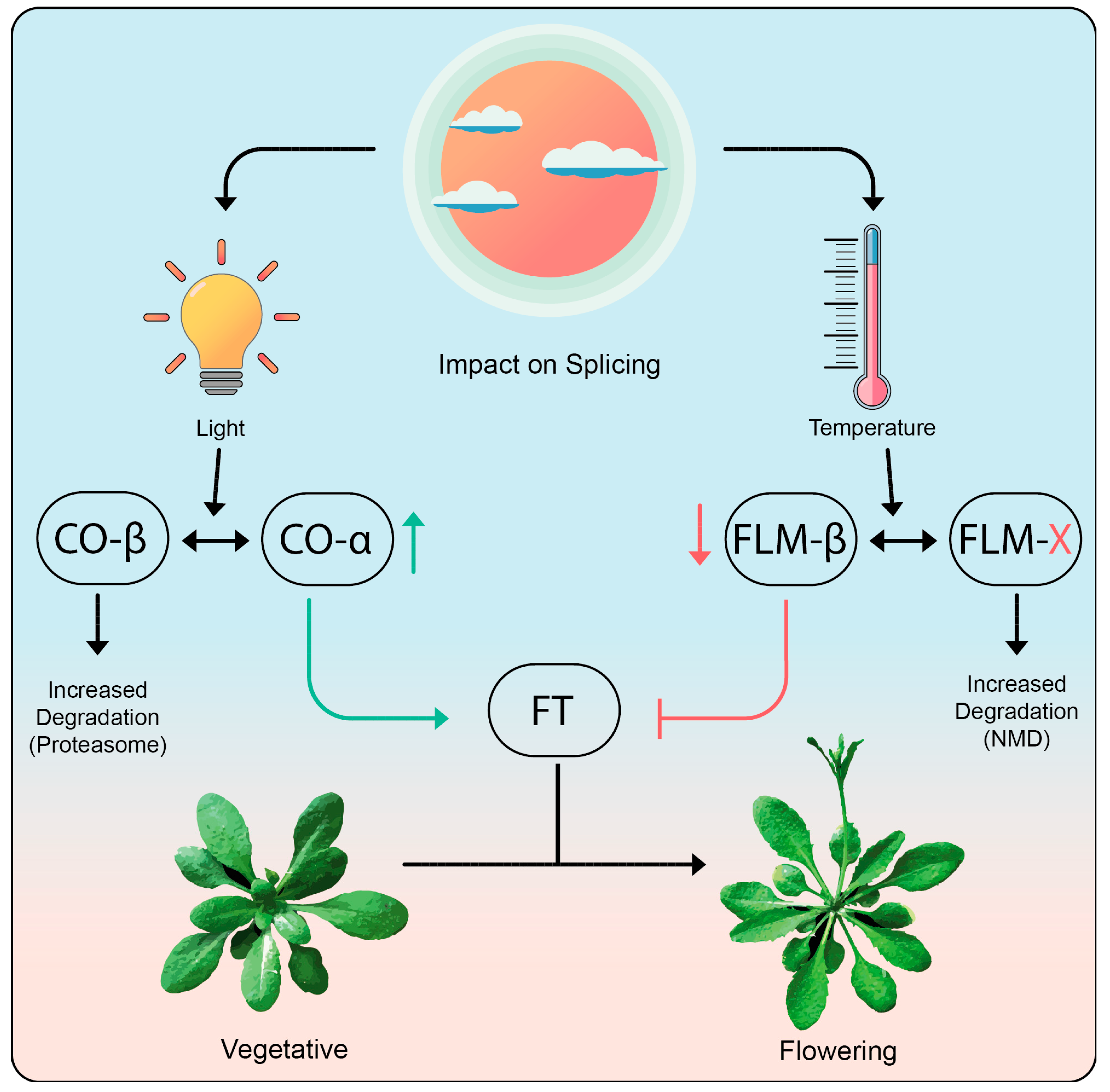
2.2.2. Responding to Signaling via Alternative Splicing
3. Conclusions
Funding
Conflicts of Interest
References
- Mitchell, S.F.; Parker, R. Principles and properties of eukaryotic mRNPs. Mol. Cell 2014, 54, 547–558. [Google Scholar] [CrossRef]
- Labbadia, J.; Morimoto, R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015, 84, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Battich, N.; Stoeger, T.; Pelkmans, L. Control of Transcript Variability in Single Mammalian Cells. Cell 2015, 163, 1596–1610. [Google Scholar] [CrossRef] [PubMed]
- Haimovich, G.; Choder, M.; Singer, R.H.; Trcek, T. The fate of the messenger is pre-determined: A new model for regulation of gene expression. Biochim. Biophys. Acta 2013, 1829, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Weake, V.M.; Workman, J.L. Inducible gene expression: Diverse regulatory mechanisms. Nat. Rev. Genet. 2010, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef]
- Sanchez de Groot, N.; Armaos, A.; Grana-Montes, R.; Alriquet, M.; Calloni, G.; Vabulas, R.M.; Tartaglia, G.G. RNA structure drives interaction with proteins. Nat. Commun. 2019, 10, 3246. [Google Scholar] [CrossRef]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Galloway, A.; Cowling, V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, H.; Wu, X.; He, Z.; Wang, L.; Yin, S.; Tian, B.; Li, G.; Cheng, H. ALYREF mainly binds to the 5’ and the 3’ regions of the mRNA in vivo. Nucleic Acids Res. 2017, 45, 9640–9653. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Müller-McNicoll, M. Nuclear retention of mRNAs - quality control, gene regulation and human disease. Semin. Cell Dev. Biol. 2018, 79, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Xiong, J.; Wang, D.; Fu, X.D. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol. 2011, 21, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Papasaikas, P.; Valcárcel, J. The Spliceosome: The Ultimate RNA Chaperone and Sculptor. Trends Biochem. Sci. 2016, 41, 33–45. [Google Scholar] [CrossRef]
- Enculescu, M.; Braun, S.; Thonta Setty, S.; Busch, A.; Zarnack, K.; König, J.; Legewie, S. Exon Definition Facilitates Reliable Control of Alternative Splicing in the RON Proto-Oncogene. Biophys. J. 2020, 118, 2027–2041. [Google Scholar] [CrossRef]
- Parenteau, J.; Maignon, L.; Berthoumieux, M.; Catala, M.; Gagnon, V.; Abou Elela, S. Introns are mediators of cell response to starvation. Nature 2019, 565, 612–617. [Google Scholar] [CrossRef]
- Bergkessel, M.; Whitworth, G.B.; Guthrie, C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA 2011, 17, 1461–1478. [Google Scholar]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Änkö, M.L. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin. Cell Dev. Biol. 2014, 32, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Sutandy, F.X.R.; Ebersberger, S.; Huang, L.; Busch, A.; Bach, M.; Kang, H.S.; Fallmann, J.; Maticzka, D.; Backofen, R.; Stadler, P.F.; et al. In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 2018, 28, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Dassi, E. Handshakes and Fights: The Regulatory Interplay of RNA-Binding Proteins. Front Mol. Biosci. 2017, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Müller-McNicoll, M. View from an mRNP: The Roles of SR Proteins in Assembly, Maturation and Turnover. Adv. Exp. Med. Biol. 2019, 1203, 83–112. [Google Scholar]
- Naro, C.; Sette, C. Phosphorylation-mediated regulation of alternative splicing in cancer. Int. J. Cell Biol. 2013, 2013, 151839. [Google Scholar] [CrossRef]
- Herzel, L.; Ottoz, D.S.M.; Alpert, T.; Neugebauer, K.M. Splicing and transcription touch base: Co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol. 2017, 18, 637–650. [Google Scholar] [CrossRef]
- Shi, Y.; Manley, J.L. The end of the message: Multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015, 29, 889–897. [Google Scholar] [CrossRef]
- Turner, R.E.; Pattison, A.D.; Beilharz, T.H. Alternative polyadenylation in the regulation and dysregulation of gene expression. Semin. Cell Dev. Biol. 2018, 75, 61–69. [Google Scholar] [CrossRef]
- Shen, T.; Li, H.; Song, Y.; Li, L.; Lin, J.; Wei, G.; Ni, T. Alternative polyadenylation dependent function of splicing factor SRSF3 contributes to cellular senescence. Aging (Albany NY) 2019, 11, 1356–1388. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; de Jesus Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Meinel, D.M.; Sträßer, K. Co-transcriptional mRNP formation is coordinated within a molecular mRNP packaging station in S. cerevisiae. Bioessays 2015, 37, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Björk, P.; Wieslander, L. Integration of mRNP formation and export. Cell Mol. Life Sci. 2017, 74, 2875–2897. [Google Scholar]
- Lei, E.P.; Krebber, H.; Silver, P.A. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001, 15, 1771–1782. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Pratt, G.; Yeo, G.W.; Moore, M.J. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 2015, 84, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Wende, W.; Friedhoff, P.; Strässer, K. Mechanism and Regulation of Co-transcriptional mRNP Assembly and Nuclear mRNA Export. Adv. Exp. Med. Biol. 2019, 1203, 1–31. [Google Scholar] [PubMed]
- Kilchert, C.; Sträßer, K.; Kunetsky, V.; Änkö, M.L. From parts lists to functional significance-RNA-protein interactions in gene regulation. Wiley Interdiscip. Rev. RNA 2020, 11, e1582. [Google Scholar] [CrossRef]
- Gehring, N.H.; Wahle, E.; Fischer, U. Deciphering the mRNP Code: RNA-Bound Determinants of Post-Transcriptional Gene Regulation. Trends Biochem. Sci. 2017, 42, 369–382. [Google Scholar] [CrossRef]
- Haimovich, G.; Medina, D.A.; Causse, S.Z.; Garber, M.; Millan-Zambrano, G.; Barkai, O.; Chavez, S.; Perez-Ortin, J.E.; Darzacq, X.; Choder, M. Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 2013, 153, 1000–1011. [Google Scholar] [CrossRef]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef]
- Boehm, V.; Gehring, N.H. Exon Junction Complexes: Supervising the Gene Expression Assembly Line. Trends Genet. 2016, 32, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Schlautmann, L.P.; Gehring, N.H. A Day in the Life of the Exon Junction Complex. Biomolecules 2020, 10, 866. [Google Scholar] [CrossRef]
- Woodward, L.A.; Mabin, J.W.; Gangras, P.; Singh, G. The exon junction complex: A lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Tange, T.O.; Shibuya, T.; Jurica, M.S.; Moore, M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 2005, 11, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, H.; Saulière, J.; Wang, Z. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016, 17, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Kishor, A.; Fritz, S.E.; Hogg, J.R. Nonsense-mediated mRNA decay: The challenge of telling right from wrong in a complex transcriptome. Wiley Interdiscip. Rev. RNA 2019, 10, e1548. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Karousis, E.D.; Mühlemann, O. Nonsense-Mediated mRNA Decay Begins Where Translation Ends. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- McMahon, J.J.; Miller, E.E.; Silver, D.L. The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci. 2016, 55, 117–123. [Google Scholar] [CrossRef]
- Chuang, T.W.; Lee, K.M.; Tarn, W.Y. Function and pathological implications of exon junction complex factor Y14. Biomolecules 2015, 5, 343–355. [Google Scholar] [CrossRef]
- Strässer, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodriguez-Navarro, S.; Rondón, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar]
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The role of TREX in gene expression and disease. Biochem. J. 2016, 473, 2911–2935. [Google Scholar] [CrossRef] [PubMed]
- Hurt, E.; Luo, M.J.; Röther, S.; Reed, R.; Strässer, K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 2004, 101, 1858–1862. [Google Scholar] [CrossRef]
- Abruzzi, K.C.; Lacadie, S.; Rosbash, M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004, 23, 2620–2631. [Google Scholar] [CrossRef]
- Meinel, D.M.; Burkert-Kautzsch, C.; Kieser, A.; O’Duibhir, E.; Siebert, M.; Mayer, A.; Cramer, P.; Söding, J.; Holstege, F.C.; Strässer, K. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 2013, 9, e1003914. [Google Scholar] [CrossRef]
- Chanarat, S.; Seizl, M.; Strässer, K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011, 25, 1147–1158. [Google Scholar] [CrossRef]
- Minocha, R.; Popova, V.; Kopytova, D.; Misiak, D.; Hüttelmaier, S.; Georgieva, S.; Strässer, K. Mud2 functions in transcription by recruiting the Prp19 and TREX complexes to transcribed genes. Nucleic Acids Res. 2018, 46, 9749–9763. [Google Scholar] [CrossRef] [PubMed]
- Gromadzka, A.M.; Steckelberg, A.L.; Singh, K.K.; Hofmann, K.; Gehring, N.H. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 2016, 44, 2348–2361. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S.; Das, R.; Cheng, H.; Hurt, E.; Dorman, N.; Reed, R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005, 19, 1512–1517. [Google Scholar] [CrossRef]
- Valencia, P.; Dias, A.P.; Reed, R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 3386–3391. [Google Scholar] [CrossRef]
- Cheng, H.; Dufu, K.; Lee, C.S.; Hsu, J.L.; Dias, A.; Reed, R. Human mRNA export machinery recruited to the 5’ end of mRNA. Cell 2006, 127, 1389–1400. [Google Scholar] [CrossRef]
- Nojima, T.; Hirose, T.; Kimura, H.; Hagiwara, M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007, 282, 15645–15651. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.A.; Cubberley, G.; Bentley, D.L. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3’ end processing factor Pcf11. Mol. Cell 2009, 33, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.L.; Pasquinelli, A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Soucek, S.; Corbett, A.H.; Fasken, M.B. The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim. Biophys. Acta 2012, 1819, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Leung, S.W.; Pak, C.; Banerjee, A.; Moberg, K.H.; Corbett, A.H. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA 2014, 20, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, M.; Martani, F.; Bertacchi, S.; Vitangeli, I.; Branduardi, P. The Saccharomyces cerevisiae poly (A) binding protein (Pab1): Master regulator of mRNA metabolism and cell physiology. Yeast 2019, 36, 23–34. [Google Scholar] [CrossRef]
- Jimeno, S.; Luna, R.; Garcia-Rubio, M.; Aguilera, A. Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol. Cell Biol. 2006, 26, 4387–4398. [Google Scholar] [CrossRef]
- Dufu, K.; Livingstone, M.J.; Seebacher, J.; Gygi, S.P.; Wilson, S.A.; Reed, R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010, 24, 2043–2053. [Google Scholar] [CrossRef]
- Bucheli, M.E.; Buratowski, S. Npl3 is an antagonist of mRNA 3’ end formation by RNA polymerase II. EMBO J. 2005, 24, 2150–2160. [Google Scholar] [CrossRef]
- Dermody, J.L.; Dreyfuss, J.M.; Villen, J.; Ogundipe, B.; Gygi, S.P.; Park, P.J.; Ponticelli, A.S.; Moore, C.L.; Buratowski, S.; Bucheli, M.E. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 2008, 3, e3273. [Google Scholar] [CrossRef] [PubMed]
- Kress, T.L.; Krogan, N.J.; Guthrie, C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 2008, 32, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Henry, M.; Silver, P.A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996, 10, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Batisse, J.; Batisse, C.; Budd, A.; Böttcher, B.; Hurt, E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 2009, 284, 34911–34917. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M.; Marfatia, K.A.; Crafton, E.B.; Zhang, X.; Cheng, X.; Corbett, A.H. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2002, 277, 7752–7760. [Google Scholar] [CrossRef]
- Hector, R.E.; Nykamp, K.R.; Dheur, S.; Anderson, J.T.; Non, P.J.; Urbinati, C.R.; Wilson, S.M.; Minvielle-Sebastia, L.; Swanson, M.S. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002, 21, 1800–1810. [Google Scholar] [CrossRef]
- Jeong, S. SR Proteins: Binders, Regulators, and Connectors of RNA. Mol. Cells 2017, 40, 1–9. [Google Scholar] [CrossRef]
- Gilbert, W.; Guthrie, C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 2004, 13, 201–212. [Google Scholar] [CrossRef]
- Gwizdek, C.; Iglesias, N.; Rodriguez, M.S.; Ossareh-Nazari, B.; Hobeika, M.; Divita, G.; Stutz, F.; Dargemont, C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc. Natl. Acad. Sci. USA 2006, 103, 16376–16381. [Google Scholar] [CrossRef]
- Iglesias, N.; Tutucci, E.; Gwizdek, C.; Vinciguerra, P.; Von Dach, E.; Corbett, A.H.; Dargemont, C.; Stutz, F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010, 24, 1927–1938. [Google Scholar] [CrossRef]
- Luo, M.L.; Zhou, Z.; Magni, K.; Christoforides, C.; Rappsilber, J.; Mann, M.; Reed, R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 2001, 413, 644–647. [Google Scholar] [CrossRef]
- Hackmann, A.; Wu, H.; Schneider, U.M.; Meyer, K.; Jung, K.; Krebber, H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat. Commun. 2014, 5, 3123. [Google Scholar] [CrossRef]
- Huang, Y.; Gattoni, R.; Stevenin, J.; Steitz, J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 2003, 11, 837–843. [Google Scholar] [CrossRef]
- Strässer, K.; Bassler, J.; Hurt, E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 2000, 150, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Hochberg-Laufer, H.; Schwed-Gross, A.; Neugebauer, K.M.; Shav-Tal, Y. Uncoupling of nucleo-cytoplasmic RNA export and localization during stress. Nucleic Acids Res. 2019, 47, 4778–4797. [Google Scholar] [CrossRef]
- Zander, G.; Krebber, H. Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol. 2017, 14, 1642–1648. [Google Scholar] [CrossRef]
- Carmody, S.R.; Tran, E.J.; Apponi, L.H.; Corbett, A.H.; Wente, S.R. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol. Cell Biol. 2010, 30, 5168–5179. [Google Scholar] [CrossRef] [PubMed]
- Béthune, J.; Jansen, R.P.; Feldbrügge, M.; Zarnack, K. Membrane-Associated RNA-Binding Proteins Orchestrate Organelle-Coupled Translation. Trends Cell Biol. 2019, 29, 178–188. [Google Scholar] [CrossRef]
- Glock, C.; Heumüller, M.; Schuman, E.M. mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 2017, 45, 169–177. [Google Scholar]
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef]
- Kahvejian, A.; Roy, G.; Sonenberg, N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Khong, A.; Parker, R. The landscape of eukaryotic mRNPs. RNA 2020, 26, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Pierron, G.; Weil, D. Re-viewing the 3D Organization of mRNPs. Mol. Cell 2018, 72, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Adivarahan, S.; Livingston, N.; Nicholson, B.; Rahman, S.; Wu, B.; Rissland, O.S.; Zenklusen, D. Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol. Cell 2018, 72, 727–738.e5. [Google Scholar] [CrossRef] [PubMed]
- Khong, A.; Parker, R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J. Cell Biol. 2018, 217, 4124–4140. [Google Scholar] [CrossRef]
- Archer, S.K.; Shirokikh, N.E.; Hallwirth, C.V.; Beilharz, T.H.; Preiss, T. Probing the closed-loop model of mRNA translation in living cells. RNA Biol. 2015, 12, 248–254. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef]
- Harvey, R.F.; Smith, T.S.; Mulroney, T.; Queiroz, R.M.L.; Pizzinga, M.; Dezi, V.; Villenueva, E.; Ramakrishna, M.; Lilley, K.S.; Willis, A.E. Trans-acting translational regulatory RNA binding proteins. Wiley Interdiscip. Rev. RNA 2018, 9, e1465. [Google Scholar] [CrossRef]
- Webster, M.W.; Chen, Y.H.; Stowell, J.A.W.; Alhusaini, N.; Sweet, T.; Graveley, B.R.; Coller, J.; Passmore, L.A. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol. Cell 2018, 70, 1089–1100.e8. [Google Scholar] [CrossRef]
- Siwaszek, A.; Ukleja, M.; Dziembowski, A. Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 2014, 11, 1122–1136. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, H.; Fukao, A.; Funakami, Y.; Duncan, K.E.; Fujiwara, T. Emerging Evidence of Translational Control by AU-Rich Element-Binding Proteins. Front. Genet. 2019, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Neininger, A.; Kontoyiannis, D.; Kotlyarov, A.; Winzen, R.; Eckert, R.; Volk, H.D.; Holtmann, H.; Kollias, G.; Gaestel, M. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 2002, 277, 3065–3068. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.; Pasparakis, M.; Pizarro, T.T.; Cominelli, F.; Kollias, G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity 1999, 10, 387–398. [Google Scholar] [CrossRef]
- Banner, D.W.; D’Arcy, A.; Janes, W.; Gentz, R.; Schoenfeld, H.J.; Broger, C.; Loetscher, H.; Lesslauer, W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: Implications for TNF receptor activation. Cell 1993, 73, 431–445. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Hsu, H.; Xiong, J.; Goeddel, D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 1995, 81, 495–504. [Google Scholar] [CrossRef]
- Hsu, H.; Huang, J.; Shu, H.B.; Baichwal, V.; Goeddel, D.V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 1996, 4, 387–396. [Google Scholar] [CrossRef]
- Shu, H.B.; Takeuchi, M.; Goeddel, D.V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc. Natl. Acad. Sci. USA 1996, 93, 13973–13978. [Google Scholar] [CrossRef]
- Dondelinger, Y.; Darding, M.; Bertrand, M.J.; Walczak, H. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell Mol. Life Sci. 2016, 73, 2165–2176. [Google Scholar] [CrossRef]
- Gaestel, M.; Kotlyarov, A.; Kracht, M. Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug Discov. 2009, 8, 480–499. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Wasiliew, P.; Kracht, M. Interleukin-1 (IL-1) pathway. Sci. Signal 2010, 3, cm1. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya-Tsuji, J.; Kishimoto, K.; Hiyama, A.; Inoue, J.; Cao, Z.; Matsumoto, K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 1999, 398, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Ajibade, A.A.; Wang, H.Y.; Wang, R.F. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013, 34, 307–316. [Google Scholar] [CrossRef]
- Dai, L.; Aye Thu, C.; Liu, X.Y.; Xi, J.; Cheung, P.C. TAK1, more than just innate immunity. IUBMB Life 2012, 64, 825–834. [Google Scholar] [CrossRef]
- Takaesu, G.; Kishida, S.; Hiyama, A.; Yamaguchi, K.; Shibuya, H.; Irie, K.; Ninomiya-Tsuji, J.; Matsumoto, K. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 2000, 5, 649–658. [Google Scholar] [CrossRef]
- Holtmann, H.; Winzen, R.; Holland, P.; Eickemeier, S.; Hoffmann, E.; Wallach, D.; Malinin, N.L.; Cooper, J.A.; Resch, K.; Kracht, M. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell Biol. 1999, 19, 6742–6753. [Google Scholar] [CrossRef]
- Winzen, R.; Kracht, M.; Ritter, B.; Wilhelm, A.; Chen, C.Y.; Shyu, A.B.; Müller, M.; Gaestel, M.; Resch, K.; Holtmann, H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999, 18, 4969–4980. [Google Scholar] [CrossRef]
- Huang, Q.; Yang, J.; Lin, Y.; Walker, C.; Cheng, J.; Liu, Z.G.; Su, B. Differential regulation of interleukin 1 receptor and Toll-like receptor signaling by MEKK3. Nat. Immunol. 2004, 5, 98–103. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Jurida, L.; Soelch, J.; Bartkuhn, M.; Handschick, K.; Muller, H.; Newel, D.; Weber, A.; Dittrich-Breiholz, O.; Schneider, H.; Bhuju, S.; et al. The Activation of IL-1-Induced Enhancers Depends on TAK1 Kinase Activity and NF-kappaB p65. Cell Rep. 2015, 10, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Holtmann, H.; Enninga, J.; Kalble, S.; Thiefes, A.; Dorrie, A.; Broemer, M.; Winzen, R.; Wilhelm, A.; Ninomiya-Tsuji, J.; Matsumoto, K.; et al. The MAPK kinase kinase TAK1 plays a central role in coupling the interleukin-1 receptor to both transcriptional and RNA-targeted mechanisms of gene regulation. J. Biol. Chem. 2001, 276, 3508–3516. [Google Scholar] [CrossRef] [PubMed]
- Thiefes, A.; Wolter, S.; Mushinski, J.F.; Hoffmann, E.; Dittrich-Breiholz, O.; Graue, N.; Dorrie, A.; Schneider, H.; Wirth, D.; Luckow, B.; et al. Simultaneous blockade of NFkappaB, JNK, and p38 MAPK by a kinase-inactive mutant of the protein kinase TAK1 sensitizes cells to apoptosis and affects a distinct spectrum of tumor necrosis factor [corrected] target genes. J. Biol. Chem. 2005, 280, 27728–27741. [Google Scholar] [CrossRef]
- Briata, P.; Forcales, S.V.; Ponassi, M.; Corte, G.; Chen, C.Y.; Karin, M.; Puri, P.L.; Gherzi, R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 2005, 20, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.L.; Sully, G.; Wait, R.; Rawlinson, L.; Clark, A.R.; Saklatvala, J. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem. J. 2002, 366, 709–719. [Google Scholar] [CrossRef]
- Dhamija, S.; Kuehne, N.; Winzen, R.; Doerrie, A.; Dittrich-Breiholz, O.; Thakur, B.K.; Kracht, M.; Holtmann, H. Interleukin-1 activates synthesis of interleukin-6 by interfering with a KH-type splicing regulatory protein (KSRP)-dependent translational silencing mechanism. J. Biol. Chem. 2011, 286, 33279–33288. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Akool el, S.; Huwiler, A.; Muller, R.; Radeke, H.H.; Pfeilschifter, J.; Eberhardt, W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell Biol. 2008, 28, 2608–2625. [Google Scholar] [CrossRef]
- Ronkina, N.; Shushakova, N.; Tiedje, C.; Yakovleva, T.; Tollenaere, M.A.X.; Scott, A.; Batth, T.S.; Olsen, J.V.; Helmke, A.; Bekker-Jensen, S.H.; et al. The Role of TTP Phosphorylation in the Regulation of Inflammatory Cytokine Production by MK2/3. J. Immunol. 2019, 203, 2291–2300. [Google Scholar] [CrossRef]
- Tenekeci, U.; Poppe, M.; Beuerlein, K.; Buro, C.; Muller, H.; Weiser, H.; Kettner-Buhrow, D.; Porada, K.; Newel, D.; Xu, M.; et al. K63-Ubiquitylation and TRAF6 Pathways Regulate Mammalian P-Body Formation and mRNA Decapping. Mol. Cell 2016, 62, 943–957. [Google Scholar] [CrossRef]
- Winzen, R.; Thakur, B.K.; Dittrich-Breiholz, O.; Shah, M.; Redich, N.; Dhamija, S.; Kracht, M.; Holtmann, H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell Biol. 2007, 27, 8388–8400. [Google Scholar] [CrossRef]
- Eick, D.; Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef] [PubMed]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef]
- Lee, S.R.; Lykke-Andersen, J. Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol. 2013, 23, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Hardman, G.; Perkins, S.; Brownridge, P.J.; Clarke, C.J.; Byrne, D.P.; Campbell, A.E.; Kalyuzhnyy, A.; Myall, A.; Eyers, P.A.; Jones, A.R.; et al. Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation. EMBO J. 2019, 38, e100847. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.R.; Trelle, M.B.; Thingholm, T.E.; Jensen, O.N. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques 2006, 40, 790–798. [Google Scholar] [CrossRef]
- Richard, P.; Vethantham, V.; Manley, J.L. Roles of Sumoylation in mRNA Processing and Metabolism. Adv. Exp. Med. Biol. 2017, 963, 15–33. [Google Scholar] [PubMed]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef]
- Dimitrova-Paternoga, L.; Jagtap, P.K.A.; Chen, P.C.; Hennig, J. Integrative Structural Biology of Protein-RNA Complexes. Structure 2020, 28, 6–28. [Google Scholar] [CrossRef]
- Garcia-Jove Navarro, M.; Kashida, S.; Chouaib, R.; Souquere, S.; Pierron, G.; Weil, D.; Gueroui, Z. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 2019, 10, 3230. [Google Scholar] [CrossRef]
- Duncan, K.; Umen, J.G.; Guthrie, C. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 2000, 10, 687–696. [Google Scholar] [CrossRef]
- Bretes, H.; Rouviere, J.O.; Leger, T.; Oeffinger, M.; Devaux, F.; Doye, V.; Palancade, B. Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Res. 2014, 42, 5043–5058. [Google Scholar] [CrossRef] [PubMed]
- Durairaj, G.; Lahudkar, S.; Bhaumik, S.R. A new regulatory pathway of mRNA export by an F-box protein, Mdm30. RNA 2014, 20, 133–142. [Google Scholar] [CrossRef]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef]
- Lin, Y.; Currie, S.L.; Rosen, M.K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017, 292, 19110–19120. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef]
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 2018, 69, 465–479.e7. [Google Scholar] [CrossRef]
- Hofweber, M.; Hutten, S.; Bourgeois, B.; Spreitzer, E.; Niedner-Boblenz, A.; Schifferer, M.; Ruepp, M.D.; Simons, M.; Niessing, D.; Madl, T.; et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 2018, 173, 706–719.e13. [Google Scholar] [CrossRef]
- Scaramuzzino, C.; Monaghan, J.; Milioto, C.; Lanson, N.A., Jr.; Maltare, A.; Aggarwal, T.; Casci, I.; Fackelmayer, F.O.; Pennuto, M.; Pandey, U.B. Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS ONE 2013, 8, e61576. [Google Scholar] [CrossRef]
- Monahan, Z.; Ryan, V.H.; Janke, A.M.; Burke, K.A.; Rhoads, S.N.; Zerze, G.H.; O’Meally, R.; Dignon, G.L.; Conicella, A.E.; Zheng, W.; et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017, 36, 2951–2967. [Google Scholar] [CrossRef]
- Arribas-Layton, M.; Dennis, J.; Bennett, E.J.; Damgaard, C.K.; Lykke-Andersen, J. The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation. Mol. Cell Biol. 2016, 36, 2226–2235. [Google Scholar] [CrossRef]
- Calvanese, V.; Lara, E.; Suarez-Alvarez, B.; Abu Dawud, R.; Vazquez-Chantada, M.; Martinez-Chantar, M.L.; Embade, N.; Lopez-Nieva, P.; Horrillo, A.; Hmadcha, A.; et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 13736–13741. [Google Scholar] [CrossRef]
- Chen, Y.C.; Milliman, E.J.; Goulet, I.; Cote, J.; Jackson, C.A.; Vollbracht, J.A.; Yu, M.C. Protein arginine methylation facilitates cotranscriptional recruitment of pre-mRNA splicing factors. Mol. Cell Biol. 2010, 30, 5245–5256. [Google Scholar] [CrossRef]
- Desterro, J.M.; Keegan, L.P.; Jaffray, E.; Hay, R.T.; O’Connell, M.A.; Carmo-Fonseca, M. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 2005, 16, 5115–5126. [Google Scholar] [CrossRef]
- Kim, T.H.; Tsang, B.; Vernon, R.M.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 2019, 365, 825–829. [Google Scholar] [CrossRef]
- Tsang, B.; Arsenault, J.; Vernon, R.M.; Lin, H.; Sonenberg, N.; Wang, L.Y.; Bah, A.; Forman-Kay, J.D. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. USA 2019, 116, 4218–4227. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Conicella, A.E.; Schmidt, H.B.; Martin, E.W.; Rhoads, S.N.; Reeb, A.N.; Nourse, A.; Ramirez Montero, D.; Ryan, V.H.; Rohatgi, R.; et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Bulek, K.; Li, X.; Herjan, T.; Yu, M.; Qian, W.; Wang, H.; Zhou, G.; Chen, X.; Yang, H.; et al. IRAK2 directs stimulus-dependent nuclear export of inflammatory mRNAs. Elife 2017, 6. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007, 8, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Panse, V.G.; Hardeland, U.; Werner, T.; Kuster, B.; Hurt, E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 2004, 279, 41346–41351. [Google Scholar] [CrossRef] [PubMed]
- Alkalay, I.; Yaron, A.; Hatzubai, A.; Orian, A.; Ciechanover, A.; Ben-Neriah, Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 1995, 92, 10599–10603. [Google Scholar] [CrossRef]
- Schmitz, M.L.; Grishina, I. Regulation of the tumor suppressor PML by sequential post-translational modifications. Front. Oncol. 2012, 2, 204. [Google Scholar] [CrossRef][Green Version]
- Sharma, K.; D’Souza, R.C.; Tyanova, S.; Schaab, C.; Wisniewski, J.R.; Cox, J.; Mann, M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, S.; Gu, J.; Tong, Y.; Li, Y.; Gui, X.; Long, H.; Wang, C.; Zhao, C.; Lu, J.; et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 2020, 27, 363–372. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef]
- Jain, S.; Parker, R. The discovery and analysis of p bodies. Adv. Exp. Med. Biol. 2013, 768, 23–43. [Google Scholar]
- Lui, J.; Castelli, L.M.; Pizzinga, M.; Simpson, C.E.; Hoyle, N.P.; Bailey, K.L.; Campbell, S.G.; Ashe, M.P. Granules Harboring Translationally Active mRNAs Provide a Platform for P-Body Formation following Stress. Cell Rep. 2014, 9, 944–954. [Google Scholar] [CrossRef]
- Sheth, U.; Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003, 300, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Rzeczkowski, K.; Beuerlein, K.; Muller, H.; Dittrich-Breiholz, O.; Schneider, H.; Kettner-Buhrow, D.; Holtmann, H.; Kracht, M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 2011, 194, 581–596. [Google Scholar] [CrossRef]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005, 11, 371–382. [Google Scholar] [CrossRef]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef]
- Simpson, C.E.; Lui, J.; Kershaw, C.J.; Sims, P.F.; Ashe, M.P. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 2014, 127, 1254–1262. [Google Scholar] [CrossRef]
- Standart, N.; Weil, D. P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet. 2018, 34, 612–626. [Google Scholar] [CrossRef]
- Courel, M.; Clement, Y.; Bossevain, C.; Foretek, D.; Vidal Cruchez, O.; Yi, Z.; Benard, M.; Benassy, M.N.; Kress, M.; Vindry, C.; et al. GC content shapes mRNA storage and decay in human cells. eLife 2019, 8. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Blumenthal, J.; Behar, L.; Elliott, E.; Ginzburg, I. Dcp1a phosphorylation along neuronal development and stress. FEBS Lett. 2009, 583, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Aizer, A.; Kafri, P.; Kalo, A.; Shav-Tal, Y. The P Body Protein Dcp1a Is Hyper-phosphorylated during Mitosis. PLoS ONE 2013, 8, e49783. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.Y.; Shen, Y.F.; Su, Y.L.; Kao, C.H.; Lin, N.Y.; Hsu, P.H.; Tsai, M.D.; Wang, S.C.; Chang, G.D.; Lee, S.C.; et al. Phosphorylation of mRNA decapping protein Dcp1a by the ERK signaling pathway during early differentiation of 3T3-L1 preadipocytes. PLoS ONE 2013, 8, e61697. [Google Scholar] [CrossRef] [PubMed]
- Dickey, L.L.; Duncan, J.K.; Hanley, T.M.; Fearns, R. Decapping protein 1 phosphorylation modulates IL-8 expression during respiratory syncytial virus infection. Virology 2015, 481, 199–209. [Google Scholar] [CrossRef]
- Bai, R.Y.; Koester, C.; Ouyang, T.; Hahn, S.A.; Hammerschmidt, M.; Peschel, C.; Duyster, J. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFbeta signalling. Nat. Cell Biol. 2002, 4, 181–190. [Google Scholar] [CrossRef]
- Mayr-Buro, C.; Schlereth, E.; Beuerlein, K.; Tenekeci, U.; Meier-Soelch, J.; Schmitz, M.L.; Kracht, M. Single-Cell Analysis of Multiple Steps of Dynamic NF-kappaB Regulation in Interleukin-1alpha-Triggered Tumor Cells Using Proximity Ligation Assays. Cancers (Basel) 2019, 11, 1199. [Google Scholar] [CrossRef]
- Skaug, B.; Jiang, X.; Chen, Z.J. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009, 78, 769–796. [Google Scholar] [CrossRef]
- Yoon, J.H.; Choi, E.J.; Parker, R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell. Biol. 2010, 189, 813–827. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol. Rev. 2012, 92, 689–737. [Google Scholar] [CrossRef]
- Chadee, D.N.; Yuasa, T.; Kyriakis, J.M. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol. Cell Biol. 2002, 22, 737–749. [Google Scholar] [CrossRef]
- Chang, C.T.; Bercovich, N.; Loh, B.; Jonas, S.; Izaurralde, E. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 2014, 42, 5217–5233. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Erickson, S.L.; Corpuz, E.O.; Maloy, J.P.; Fillman, C.; Webb, K.; Bennett, E.J.; Lykke-Andersen, J. Competition between Decapping Complex Formation and Ubiquitin-Mediated Proteasomal Degradation Controls Human Dcp2 Decapping Activity. Mol. Cell Biol. 2015, 35, 2144–2153. [Google Scholar] [CrossRef]
- Singh, S.; Singh, P.K.; Bhadauriya, P.; Ganesh, S. Lafora disease E3 ubiquitin ligase malin is recruited to the processing bodies and regulates the microRNA-mediated gene silencing process via the decapping enzyme Dcp1a. RNA Biol. 2012, 9, 1440–1449. [Google Scholar] [CrossRef]
- Zheng, D.; Chen, C.Y.; Shyu, A.B. Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA 2011, 17, 1619–1634. [Google Scholar] [CrossRef]
- Cargnello, M.; Tcherkezian, J.; Dorn, J.F.; Huttlin, E.L.; Maddox, P.S.; Gygi, S.P.; Roux, P.P. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol. Cell Biol. 2012, 32, 4572–4584. [Google Scholar] [CrossRef]
- Rodriguez-Gil, A.; Ritter, O.; Hornung, J.; Stekman, H.; Krüger, M.; Braun, T.; Kremmer, E.; Kracht, M.; Schmitz, M.L. HIPK family kinases bind and regulate the function of the CCR4-NOT complex. Mol. Biol. Cell 2016, 27, 1969–1980. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Latham, V.; Murray, B.; Nandhikonda, V.; Nord, A.; Skrzypek, E.; Wheeler, T.; Zhang, B.; Gnad, F. 15 years of PhosphoSitePlus(R): Integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 2019, 47, D433–D441. [Google Scholar] [CrossRef]
- Gantier, M.P. New perspectives in MicroRNA regulation of innate immunity. J. Interferon. Cytokine Res. 2010, 30, 283–289. [Google Scholar] [CrossRef]
- Pepin, G.; Gantier, M.P. microRNA Decay: Refining microRNA Regulatory Activity. MicroRNA 2016, 5, 167–174. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Wu, D.; Hu, Y.; Tong, S.; Williams, B.R.; Smyth, G.K.; Gantier, M.P. The use of miRNA microarrays for the analysis of cancer samples with global miRNA decrease. RNA 2013, 19, 876–888. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiss, J.L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef]
- Karlsen, T.A.; Aae, T.F.; Brinchmann, J.E. Robust profiling of microRNAs and isomiRs in human plasma exosomes across 46 individuals. Sci. Rep. 2019, 9, 19999. [Google Scholar] [CrossRef]
- Yu, F.; Pillman, K.A.; Neilsen, C.T.; Toubia, J.; Lawrence, D.M.; Tsykin, A.; Gantier, M.P.; Callen, D.F.; Goodall, G.J.; Bracken, C.P. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Res. 2017, 45, 11371–11385. [Google Scholar] [CrossRef]
- Siddle, K.J.; Tailleux, L.; Deschamps, M.; Loh, Y.H.; Deluen, C.; Gicquel, B.; Antoniewski, C.; Barreiro, L.B.; Farinelli, L.; Quintana-Murci, L. bacterial infection drives the expression dynamics of microRNAs and their isomiRs. PLoS Genet. 2015, 11, e1005064. [Google Scholar] [CrossRef]
- Kim, B.; Jeong, K.; Kim, V.N. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol. Cell 2017, 66, 258–269.e5. [Google Scholar] [CrossRef]
- Nejad, C.; Stunden, H.J.; Gantier, M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018, 285, 3695–3716. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Yang, K.; Wen, X.; Mudunuri, S.B.; Sablok, G. Plant IsomiR Atlas: Large Scale Detection, Profiling, and Target Repertoire of IsomiRs in Plants. Front. Plant Sci. 2018, 9, 1881. [Google Scholar] [CrossRef]
- Trontti, K.; Vaananen, J.; Sipila, T.; Greco, D.; Hovatta, I. Strong conservation of inbred mouse strain microRNA loci but broad variation in brain microRNAs due to RNA editing and isomiR expression. RNA 2018, 24, 643–655. [Google Scholar]
- Wright, C.; Rajpurohit, A.; Burke, E.E.; Williams, C.; Collado-Torres, L.; Kimos, M.; Brandon, N.J.; Cross, A.J.; Jaffe, A.E.; Weinberger, D.R.; et al. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genomics 2019, 20, 513. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.; Kim, K.; Chang, H.; You, K.; Kim, V.N. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3’ end modification. Nucleic Acids Res. 2019, 47, 2630–2640. [Google Scholar] [CrossRef]
- Juzenas, S.; Venkatesh, G.; Hubenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A.; et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Kasprzak, W.K.; Bhandari, Y.; Fan, L.; Cavanaugh, Q.; Jiang, M.; Dai, L.; Yang, A.; Shao, T.J.; Shapiro, B.A.; et al. Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires. Cell Rep. 2019, 26, 447–459.e4. [Google Scholar] [CrossRef]
- Pullagura, S.R.N.; Buaas, B.; Gray, N.; Krening, L.C.; Srivastava, A.; Braun, R.E. Functional Redundancy of DICER Cofactors TARBP2 and PRKRA During Murine Embryogenesis Does Not Involve miRNA Biogenesis. Genetics 2018, 208, 1513–1522. [Google Scholar] [CrossRef]
- Pillman, K.A.; Goodall, G.J.; Bracken, C.P.; Gantier, M.P. miRNA length variation during macrophage stimulation confounds the interpretation of results: Implications for miRNA quantification by RT-qPCR. RNA 2019, 25, 232–238. [Google Scholar] [CrossRef]
- Nejad, C.; Pillman, K.A.; Siddle, K.J.; Pepin, G.; Änkö, M.L.; McCoy, C.E.; Beilharz, T.H.; Quintana-Murci, L.; Goodall, G.J.; Bracken, C.P.; et al. miR-222 isoforms are differentially regulated by type-I interferon. RNA 2018, 24, 332–341. [Google Scholar] [CrossRef]
- Nejad, C.; Pepin, G.; Behlke, M.A.; Gantier, M.P. Modified Polyadenylation-Based RT-qPCR Increases Selectivity of Amplification of 3’-MicroRNA Isoforms. Front Genet. 2018, 9, 11. [Google Scholar] [CrossRef]
- Das, S.K.; Sokhi, U.K.; Bhutia, S.K.; Azab, B.; Su, Z.Z.; Sarkar, D.; Fisher, P.B. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11948–11953. [Google Scholar] [CrossRef]
- Muiwo, P.; Pandey, P.; Ahmad, H.M.; Ramachandran, S.S.; Bhattacharya, A. IsomiR processing during differentiation of myelogenous leukemic cell line K562 by phorbol ester PMA. Gene 2018, 641, 172–179. [Google Scholar] [CrossRef]
- Fernandez-Perez, D.; Brieno-Enriquez, M.A.; Isoler-Alcaraz, J.; Larriba, E.; Del Mazo, J. MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA 2018, 24, 287–303. [Google Scholar] [CrossRef]
- Ndika, J.; Seemab, U.; Poon, W.L.; Fortino, V.; El-Nezami, H.; Karisola, P.; Alenius, H. Silver, titanium dioxide, and zinc oxide nanoparticles trigger miRNA/isomiR expression changes in THP-1 cells that are proportional to their health hazard potential. Nanotoxicology 2019, 13, 1380–1395. [Google Scholar] [CrossRef]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Kashiwabara, S.; Baba, T.; Suzuki, T. Selective stabilization of mammalian microRNAs by 3’ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009, 23, 433–438. [Google Scholar] [CrossRef]
- Jones, M.R.; Quinton, L.J.; Blahna, M.T.; Neilson, J.R.; Fu, S.; Ivanov, A.R.; Wolf, D.A.; Mizgerd, J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009, 11, 1157–1163. [Google Scholar] [CrossRef]
- Tan, G.C.; Chan, E.; Molnar, A.; Sarkar, R.; Alexieva, D.; Isa, I.M.; Robinson, S.; Zhang, S.; Ellis, P.; Langford, C.F.; et al. 5’ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014, 42, 9424–9435. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, X.; Cai, Z.; Zhou, J.; Cao, R.; Zhao, Y.; Chen, Z.; Wang, D.; Ruan, W.; Zhao, Q.; et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018, 25, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Sheu-Gruttadauria, J.; Xiao, Y.; Gebert, L.F.; MacRae, I.J. Beyond the seed: Structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 2019, 38, e101153. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Bofill-De Ros, X.; Shao, T.J.; Jiang, M.; Li, K.; Villanueva, P.; Dai, L.; Gu, S. 3’ Uridylation Confers miRNAs with Non-canonical Target Repertoires. Mol. Cell 2019, 75, 511–522.e4. [Google Scholar] [CrossRef] [PubMed]
- Pai, A.A.; Baharian, G.; Page Sabourin, A.; Brinkworth, J.F.; Nedelec, Y.; Foley, J.W.; Grenier, J.C.; Siddle, K.J.; Dumaine, A.; Yotova, V.; et al. Widespread Shortening of 3’ Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS Genet. 2016, 12, e1006338. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Nakano, Y.; Onomoto, K.; Murakami, F.; Komori, C.; Suzuki, Y.; Yoneyama, M.; Ui-Tei, K. LGP2 virus sensor regulates gene expression network mediated by TRBP-bound microRNAs. Nucleic Acids Res. 2018, 46, 9134–9147. [Google Scholar] [CrossRef] [PubMed]
- Zangari, J.; Ilie, M.; Rouaud, F.; Signetti, L.; Ohanna, M.; Didier, R.; Romeo, B.; Goldoni, D.; Nottet, N.; Staedel, C.; et al. Rapid decay of engulfed extracellular miRNA by XRN1 exonuclease promotes transient epithelial-mesenchymal transition. Nucleic Acids Res. 2017, 45, 4131–4141. [Google Scholar]
- Lattin, J.E.; Schroder, K.; Su, A.I.; Walker, J.R.; Zhang, J.; Wiltshire, T.; Saijo, K.; Glass, C.K.; Hume, D.A.; Kellie, S.; et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008, 4, 5. [Google Scholar] [CrossRef]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant. J. 2017, 90, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Andres, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Sureshkumar, S.; Lempe, J.; Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006, 2, e106. [Google Scholar] [CrossRef] [PubMed]
- Barta, A.; Kalyna, M.; Lorkovic, Z.J. Plant SR proteins and their functions. Curr. Top Microbiol. Immunol. 2008, 326, 83–102. [Google Scholar] [PubMed]
- Graveley, B.R.; Hertel, K.J.; Maniatis, T. SR proteins are ‘locators’ of the RNA splicing machinery. Curr. Biol. 1999, 9, R6–R7. [Google Scholar] [CrossRef]
- Scortecci, K.C.; Michaels, S.D.; Amasino, R.M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001, 26, 229–236. [Google Scholar] [CrossRef]
- Sureshkumar, S.; Dent, C.; Seleznev, A.; Tasset, C.; Balasubramanian, S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2016, 2, 16055. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Weigel, D. Temperature Induced Flowering in Arabidopsis thaliana. Plant Signal Behav. 2006, 1, 227–228. [Google Scholar] [CrossRef]
- Arciga-Reyes, L.; Wootton, L.; Kieffer, M.; Davies, B. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006, 47, 480–489. [Google Scholar] [CrossRef]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Park, M.J.; Lee, H.J.; Park, Y.J.; Han, S.H.; Kwon, Y.J.; Seo, P.J.; Jung, J.H.; Park, C.M. Alternative splicing provides a proactive mechanism for the diurnal CONSTANS dynamics in Arabidopsis photoperiodic flowering. Plant J. 2017, 89, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, J.H.; Kim, J.Y.; Park, C.M. Alternative RNA Splicing Expands the Developmental Plasticity of Flowering Transition. Front. Plant Sci. 2019, 10, 606. [Google Scholar] [CrossRef]
- Bush, S.J.; Chen, L.; Tovar-Corona, J.M.; Urrutia, A.O. Alternative splicing and the evolution of phenotypic novelty. Philos. Trans. R Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Werner, J.D.; Borevitz, J.O.; Uhlenhaut, N.H.; Ecker, J.R.; Chory, J.; Weigel, D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 2005, 170, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Shindo, C.; Aranzana, M.J.; Lister, C.; Baxter, C.; Nicholls, C.; Nordborg, M.; Dean, C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005, 138, 1163–1173. [Google Scholar] [CrossRef]
- Michaels, S.D.; Bezerra, I.C.; Amasino, R.M. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3281–3285. [Google Scholar] [CrossRef]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef]
- Lempe, J.; Balasubramanian, S.; Sureshkumar, S.; Singh, A.; Schmid, M.; Weigel, D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005, 1, 109–118. [Google Scholar] [CrossRef]
- Gazzani, S.; Gendall, A.R.; Lister, C.; Dean, C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003, 132, 1107–1114. [Google Scholar] [CrossRef]
- Zhu, W.; Ausin, I.; Seleznev, A.; Mendez-Vigo, B.; Pico, F.X.; Sureshkumar, S.; Sundaramoorthi, V.; Bulach, D.; Powell, D.; Seemann, T.; et al. Natural Variation Identifies ICARUS1, a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in Arabidopsis thaliana. PLoS Genet. 2015, 11, e1005085. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, C.; Tung, K.S.; Amberg, D.C.; Hopper, A.K.; Cole, C.N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996, 10, 1608–1620. [Google Scholar] [CrossRef] [PubMed]
| RNP | Modification | Functional Effect | Reference |
|---|---|---|---|
| DDX4 | asymmetric dimethylation | suppression of phase separation | [147] |
| hnRNPA2 | asymmetric dimethylation | reduction of phase separation | [148] |
| FUS | arginine methylation | reduction of phase separation | [149] [150] |
| phosphorylation | reduction of phase separation | [151] | |
| LSM4 | symmetric dimethylation of | promotion of P-body formation | [152] |
| ELAVL1 | arginine methylation | downregulation of SIRT1-encoding mRNAs | [153] |
| U1-70K (SNRNP70) | arginine methylation | recruitment to spliceosome | [154] |
| Hpr1 | SUMOylation | promotes association with mRNPs | [141] |
| ADAR1 | SUMOylation | impaired RNA editing activity | [155] |
| FMRP | phosphorylation | increased interaction with CAPRIN1 increased phase separation | [156] [157] |
| CAPRIN1 | phosphorylation | increased interaction with FMRP increased phase separation | [156] |
| TDP-43 (TARDBP) | phosphorylation | decreased phase separation | [158] |
| TTP | phosphorylation | inhibition of TTP-mediated mRNA destabilization | [128] |
| SRSF1 | phosphorylation | reduced binding to target mRNAs | [159] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarnack, K.; Balasubramanian, S.; Gantier, M.P.; Kunetsky, V.; Kracht, M.; Schmitz, M.L.; Sträßer, K. Dynamic mRNP Remodeling in Response to Internal and External Stimuli. Biomolecules 2020, 10, 1310. https://doi.org/10.3390/biom10091310
Zarnack K, Balasubramanian S, Gantier MP, Kunetsky V, Kracht M, Schmitz ML, Sträßer K. Dynamic mRNP Remodeling in Response to Internal and External Stimuli. Biomolecules. 2020; 10(9):1310. https://doi.org/10.3390/biom10091310
Chicago/Turabian StyleZarnack, Kathi, Sureshkumar Balasubramanian, Michael P. Gantier, Vladislav Kunetsky, Michael Kracht, M. Lienhard Schmitz, and Katja Sträßer. 2020. "Dynamic mRNP Remodeling in Response to Internal and External Stimuli" Biomolecules 10, no. 9: 1310. https://doi.org/10.3390/biom10091310
APA StyleZarnack, K., Balasubramanian, S., Gantier, M. P., Kunetsky, V., Kracht, M., Schmitz, M. L., & Sträßer, K. (2020). Dynamic mRNP Remodeling in Response to Internal and External Stimuli. Biomolecules, 10(9), 1310. https://doi.org/10.3390/biom10091310