Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2
Abstract
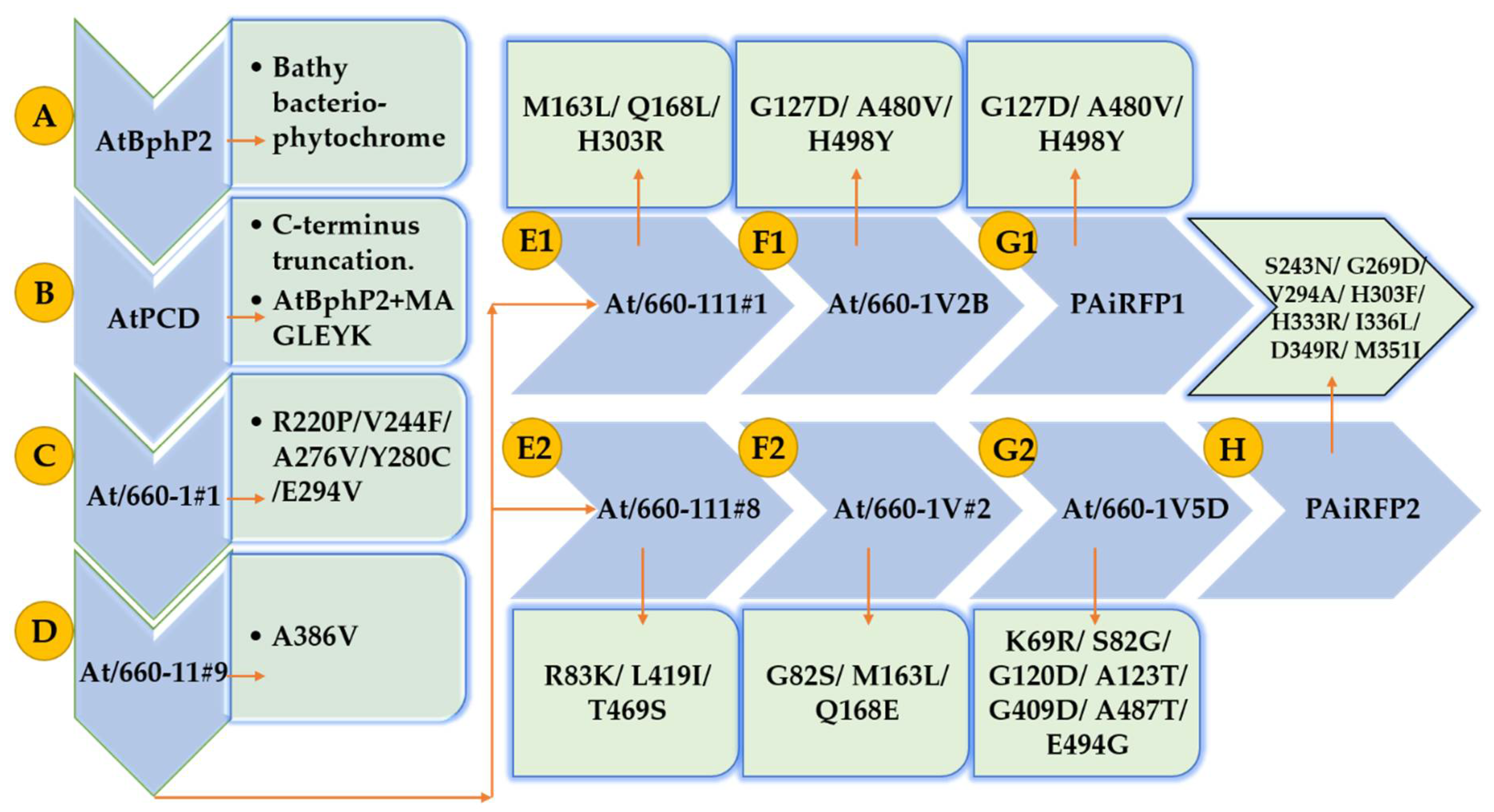
1. Introduction
2. Materials and Methods
2.1. Protein Expression, Purification, and In Vitro Assembly with BV
2.2. Spectroscopic Measurements
2.3. 3D Structure Modelling
2.4. Mutation Analysis
2.5. Docking Studies
2.6. MD Simulations
2.7. Essential Dynamics
2.8. Gibbs Free Energy Landscape
3. Results
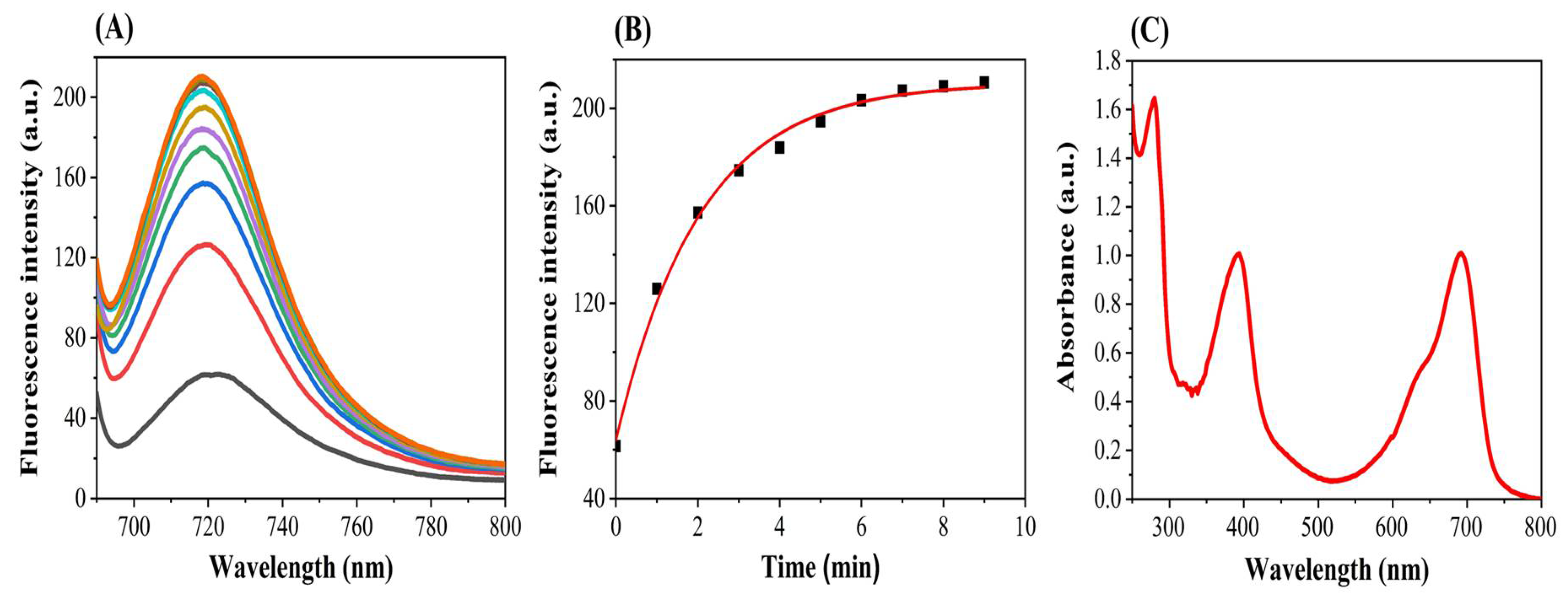
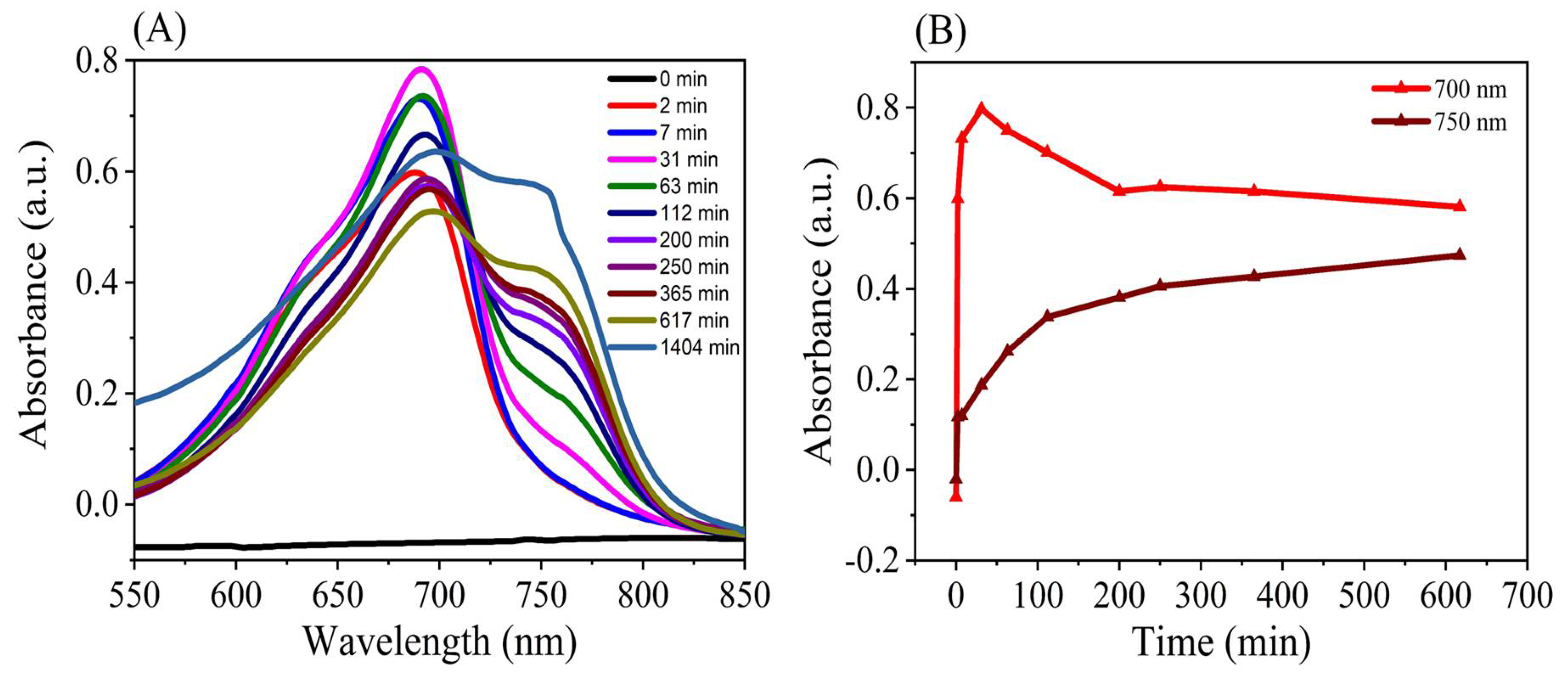
3.1. Stability of NIR Fluorescence Emission
3.2. Structure Analysis
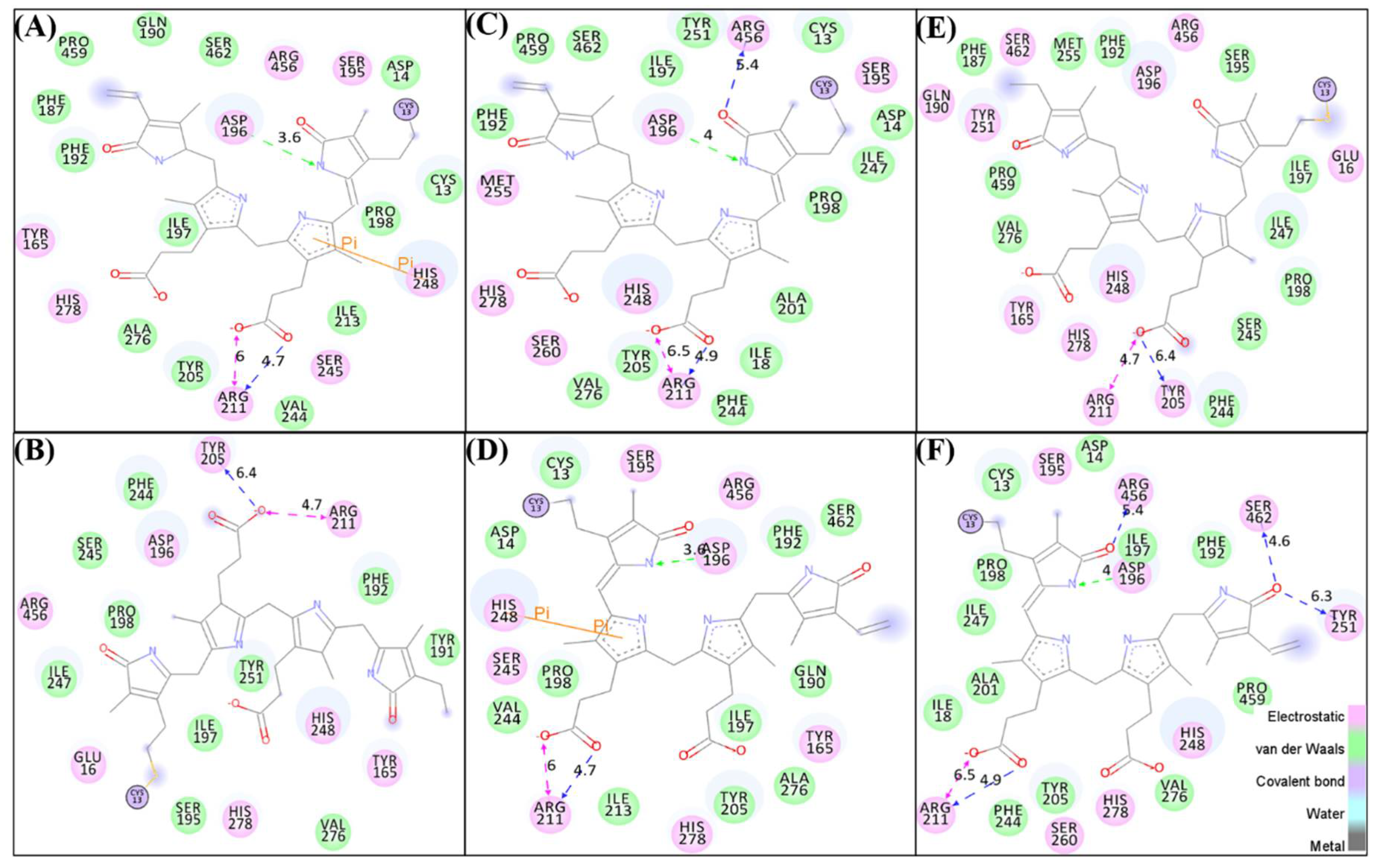
3.3. Hydrogen Bonds and van der Waals Interactions
3.4. Stability and Flexibility Analysis
3.5. Interaction of BV in Cis and Trans Forms
3.6. Structural Dynamics
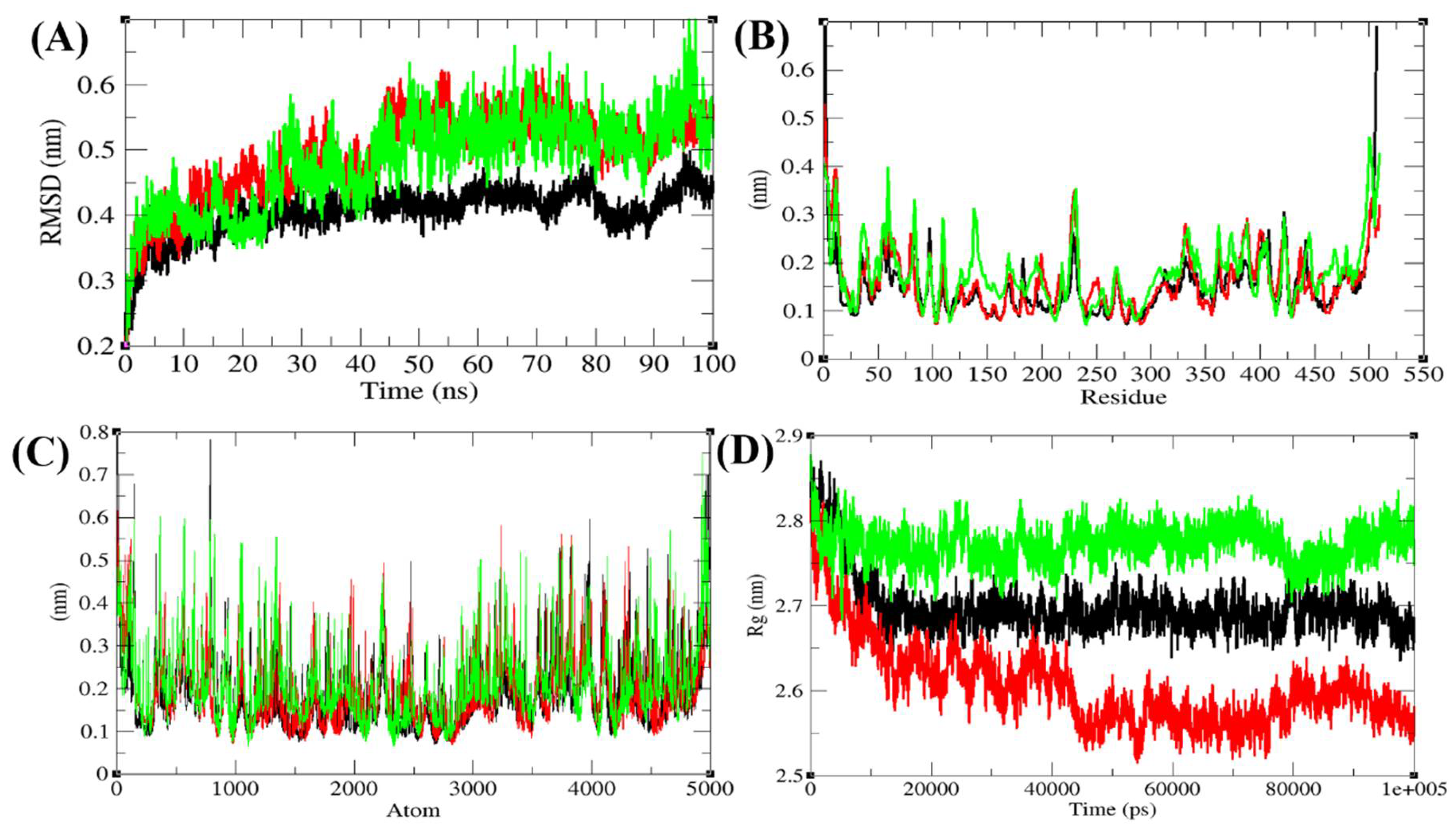
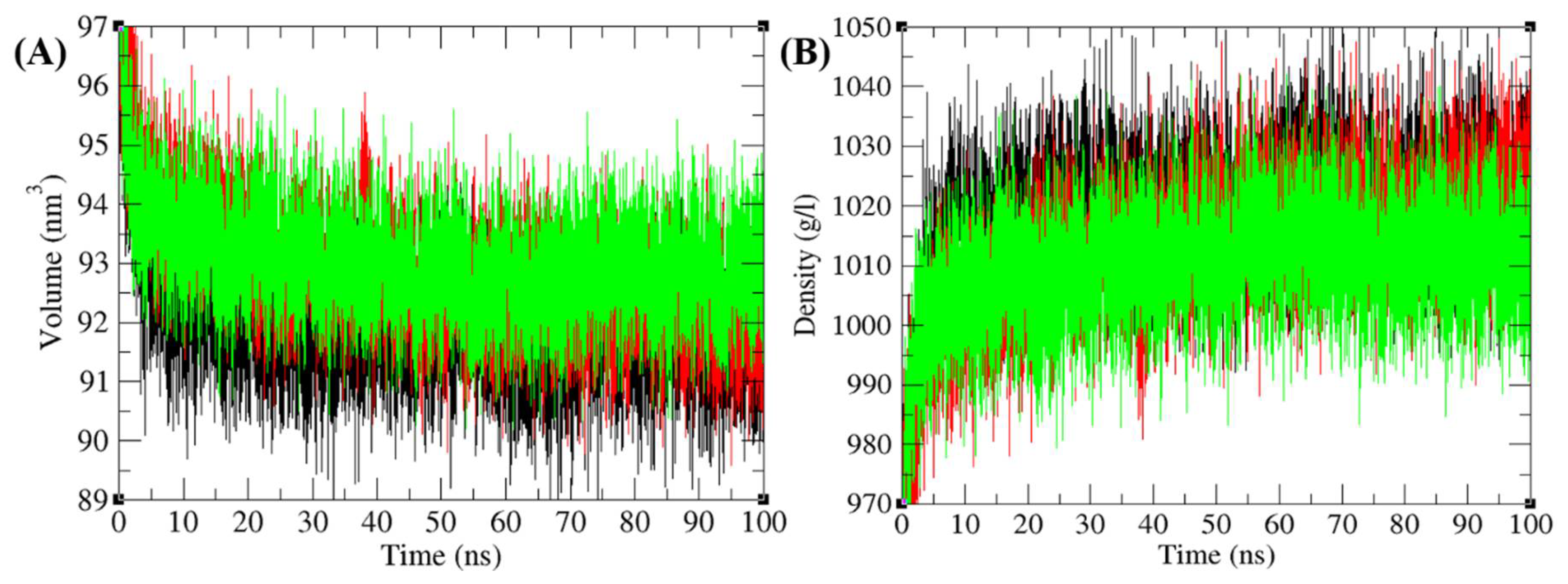
3.6.1. Structural Deviations and Compactness
3.6.2. Solvent Accessible Surface Area
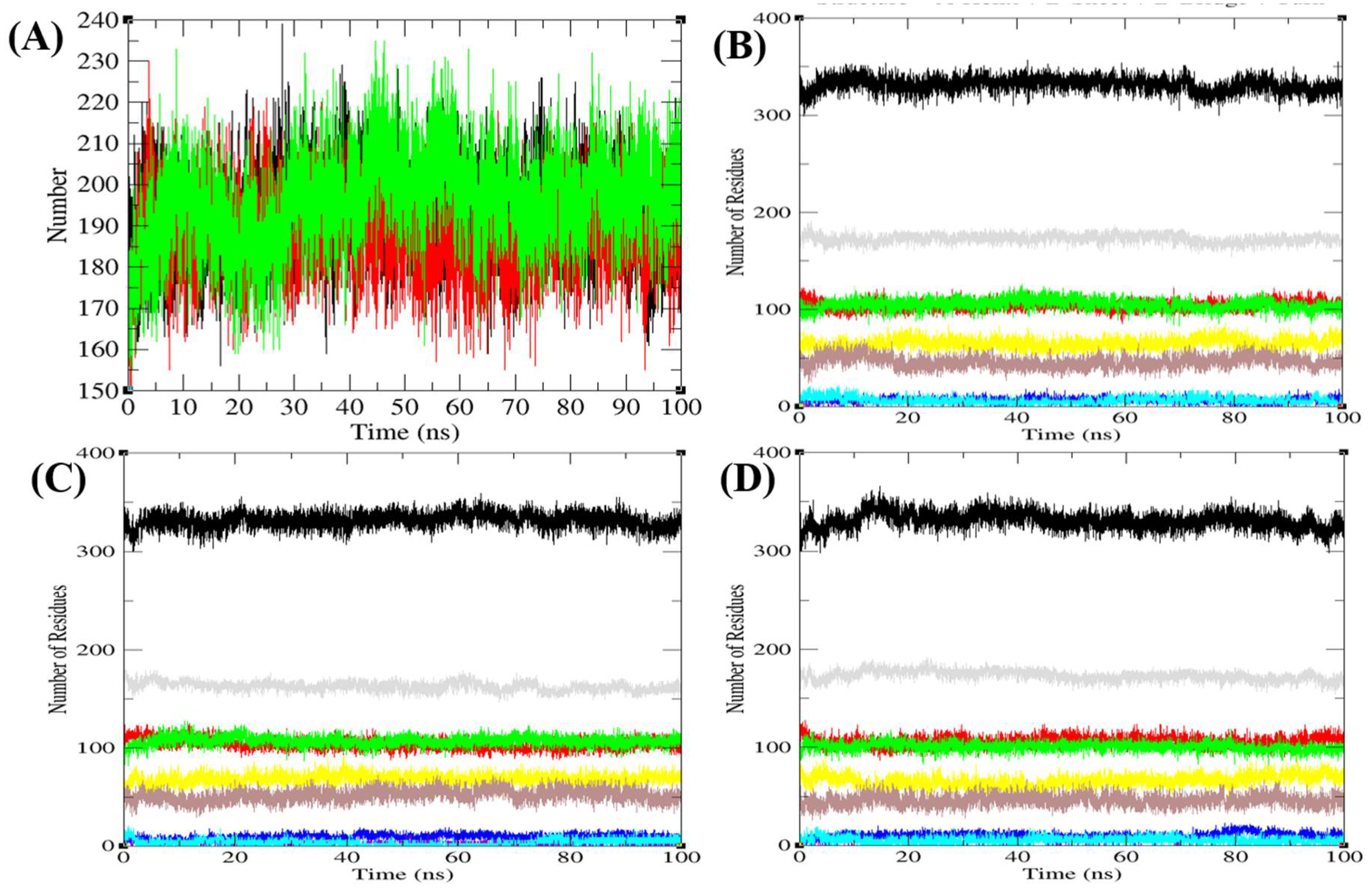
3.6.3. Hydrogen Bonds and Secondary Structure Analysis
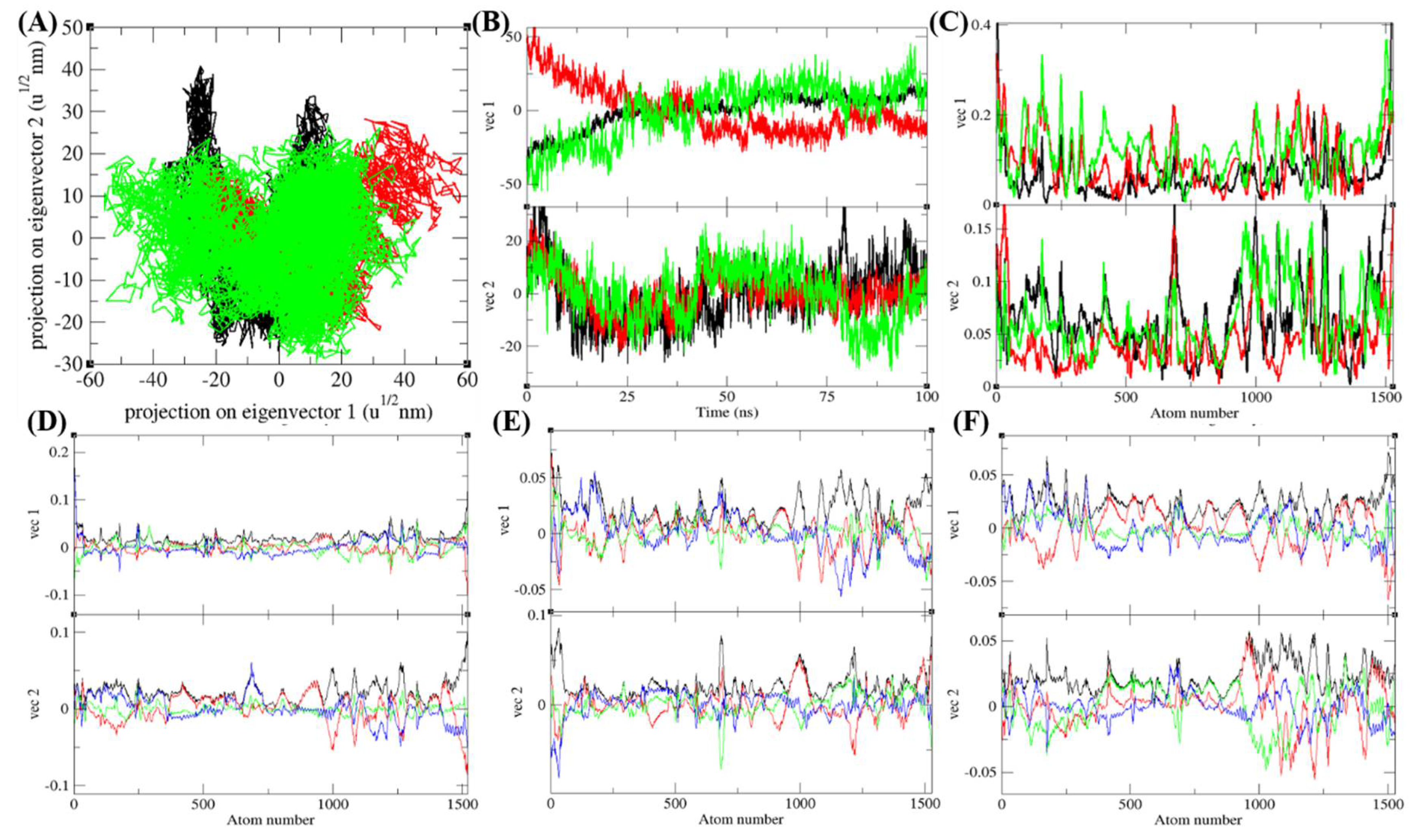
3.7. Principal Component Analysis
3.8. Gibbs Free Energy Landscape
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MD | Molecular dynamics |
| BphPs | Bacteriophytochrome photoreceptors |
| NIR | Near infrared |
| RMSD | Root mean square deviation |
References
- Giraud, E.; Zappa, S.; Vuillet, L.; Adriano, J.-M.; Hannibal, L.; Fardoux, J.; Berthomieu, C.; Bouyer, P.; Pignol, D.; Verméglio, A. A New Type of Bacteriophytochrome Acts in Tandem with a Classical Bacteriophytochrome to Control the Antennae Synthesis inRhodopseudomonas palustris. J. Biol. Chem. 2005, 280, 32389–32397. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kateriya, S.; Singh, V.S.; Tanwar, M.; Agarwal, S.; Singh, H.; Khurana, J.P.; Amla, D.V.; Tripathi, A.K. Bacteriophytochrome controls carotenoid-independent response to photodynamic stress in a non-photosynthetic rhizobacterium, Azospirillum brasilense Sp7. Sci. Rep. 2012, 2, srep00872. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Ntziachristos, V. Shedding light onto live molecular targets. Nat. Med. 2003, 9, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R. A clearer vision for in vivo imaging. Nat. Biotechnol. 2001, 19, 316–317. [Google Scholar] [CrossRef]
- Wagner, J.R.; Zhang, J.; Von Stetten, D.; Günther, M.; Murgida, D.H.; Mroginski, M.A.; Walker, J.M.; Forest, K.T.; Hildebrandt, P.; Vierstra, R.D. Mutational Analysis of Deinococcus radiodurans Bacteriophytochrome Reveals Key Amino Acids Necessary for the Photochromicity and Proton Exchange Cycle of Phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Lagarias, J.C. A Brief History of Phytochromes. ChemPhysChem 2010, 11, 1172–1180. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Malashkevich, V.N.; Morozova, K.S.; Nemkovich, N.A.; Almo, S.C.; Verkhusha, V.V. Extended Stokes Shift in Fluorescent Proteins: Chromophore—Protein Interactions in a Near-Infrared TagRFP675 Variant. Sci. Rep. 2013, 3, srep01847. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Gellman, S.H. Bacterial phytochromes: More than meets the light. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 67–88. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef]
- Baloban, M.; Shcherbakova, D.M.; Pletnev, S.; Pletnev, V.Z.; Lagarias, J.C.; Verkhusha, V.V. Designing brighter near-infrared fluorescent proteins: Insights from structural and biochemical studies. Chem. Sci. 2017, 8, 4546–4557. [Google Scholar] [CrossRef]
- Yu, D.; Dong, Z.; Gustafson, W.C.; Ruiz-Gonzalez, R.; Signor, L.; Marzocca, F.; Borel, F.; Klassen, M.P.; Makhijani, K.; Royant, A.; et al. Rational design of a monomeric and photostable far-red fluorescent protein for fluorescence imaging in vivo. Protein Sci. 2015, 25, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Satyshur, K.A.; Anstrom, D.M.; Forest, K.T. Structure-guided Engineering Enhances a Phytochrome-based Infrared Fluorescent Protein. J. Biol. Chem. 2011, 287, 7000–7009. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.-M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011, 29, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef]
- Tran, M.T.N.; Tanaka, J.; Hamada, M.; Sugiyama, Y.; Sakaguchi, S.; Nakamura, M.; Takahashi, S.; Miwa, Y. In Vivo image Analysis Using iRFP Transgenic Mice. Exp. Anim. 2014, 63, 311–319. [Google Scholar] [CrossRef]
- Yu, D.; Gustafson, W.C.; Han, C.; Lafaye, C.; Noirclerc-Savoye, M.; Ge, W.-P.; Thayer, D.A.; Huang, H.; Kornberg, T.B.; Royant, A.; et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Subach, F.V.; Verkhusha, V.V. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Hassan, F.; Khan, F.I.; Song, H.; Lai, D.; Juan, F. Effects of reverse genetic mutations on the spectral and photochemical behavior of a photoactivatable fluorescent protein PAiRFP1. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117807. [Google Scholar] [CrossRef]
- Lamparter, T.; Krauss, N.; Scheerer, P. Phytochromes from Agrobacterium fabrum. Photochem. Photobiol. 2017, 93, 642–655. [Google Scholar] [CrossRef]
- Karniol, B.; Vierstra, R.D. The Pair of Bacteriophytochromes from Agrobacterium Tumefaciens Are Histidine Kinases with Opposing Photobiological Properties; National Academy of Sciences of the United States of America: Washington, DC, USA, 2003; Volume 100, pp. 2807–2812. [Google Scholar]
- Khan, F.I.; Hassan, F.; Ali, H.; Lai, D. Mechanism of pH-induced conformational changes in MurE ligase obtained from Salmonella enterica serovar Typhi. J. Biomol. Struct. Dyn. 2020, 1–8. [Google Scholar] [CrossRef]
- Kuzmanic, A.; Zagrovic, B. Determination of Ensemble-Average Pairwise Root Mean-Square Deviation from Experimental B-Factors. Biophys. J. 2010, 98, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Lai, D.; Anwer, R.; Azim, I.; Khan, M.K.A. Identifying novel sphingosine kinase 1 inhibitors as therapeutics against breast cancer. J. Enzym. Inhib. Med. Chem. 2020, 35, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.B.; Khan, F.I.; Khan, S.H.; Srivastava, S.; Hasan, G.M.; Lobb, K.; Islam, A.; Ahmad, F.; Hassan, I. Mechanistic insights into the urea-induced denaturation of kinase domain of human integrin linked kinase. Int. J. Biol. Macromol. 2018, 111, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Sauthof, L.; Szczepek, M.; Lopez, M.F.; Escobar, F.V.; Qureshi, B.M.; Michael, N.; Buhrke, D.; Stevens, T.; Kwiatkowski, D.; et al. Structural snapshot of a bacterial phytochrome in its functional intermediate state. Nat. Commun. 2018, 9, 4912. [Google Scholar] [CrossRef]
- Rodriguez, R.; Chinea, G.; Lopez, N.; Pons, T.; Vriend, G. Homology modeling, model and software evaluation: Three related resources. Bioinformatics 1998, 14, 523–528. [Google Scholar] [CrossRef]
- Qausain, S.; Khan, F.I.; Lai, D.; Hassan, M.I.; Basheeruddin, M.; Ahmed, N.; Khan, M.K.A. Mechanistic insights into the urea-induced denaturation of a non-seleno thiol specific antioxidant human peroxiredoxin 6. Int. J. Biol. Macromol. 2020, 161, 1171–1180. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Wang, Q.; Canutescu, A.A.; Dunbrack, R.L., Jr. SCWRL and MolIDE: Computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008, 3, 1832–1847. [Google Scholar] [CrossRef]
- Kaplan, W.; Littlejohn, T.G. Swiss-PDB Viewer (Deep View). Brief. Bioinform. 2001, 2, 195–197. [Google Scholar] [CrossRef]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Tiwari, S.P.; Fuglebakk, E.; Hollup, S.M.; Skjaerven, L.; Cragnolini, T.; Grindhaug, S.H.; Tekle, K.M.; Reuter, N. WEBnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinform. 2014, 15, 427. [Google Scholar] [CrossRef]
- Grant, B.; Rodrigues, A.P.C.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S.D. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Frappier, V.; Najmanovich, R. A Coarse-Grained Elastic Network Atom Contact Model and Its Use in the Simulation of Protein Dynamics and the Prediction of the Effect of Mutations. PLoS Comput. Biol. 2014, 10, e1003569. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Carrillo, O.; Emperador, A.; Orellana, L.; Hospital, A.; Rueda, M.; Cicin-Sain, D.; D’Abramo, M.; Gelpí, J.L.; Orozco, M.; et al. FlexServ: An integrated tool for the analysis of protein flexibility. Bioinformatics 2009, 25, 1709–1710. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z.H. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J. Chem. Inf. Model. 2019, 59, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Potapov, V.Y.; Cohen, M.; Schreiber, G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng. Des. Sel. 2009, 22, 553–560. [Google Scholar] [CrossRef]
- Khan, S.; Vihinen, M. Performance of protein stability predictors. Hum. Mutat. 2010, 31, 675–684. [Google Scholar] [CrossRef]
- Thiltgen, G.; Goldstein, R.A. Assessing Predictors of Changes in Protein Stability upon Mutation Using Self-Consistency. PLoS ONE 2012, 7, e46084. [Google Scholar] [CrossRef]
- Jubb, H.; Blundell, T.L.; Ascher, D.B. Flexibility and small pockets at protein-protein interfaces: New insights into druggability. Prog. Biophys. Mol. Biol. 2015, 119, 2–9. [Google Scholar] [CrossRef]
- Lenngren, N.; Edlund, P.; Takala, H.; Buchli, B.; Rumfeldt, J.; Peshev, I.; Häkkänen, H.; Westenhoff, S.; Ihalainen, J.A. Coordination of the biliverdin D-ring in bacteriophytochromes. Phys. Chem. Chem. Phys. 2018, 20, 18216–18225. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Lehtivuori, H.; Berntsson, O.; Hughes, A.; Nanekar, R.; Niebling, S.; Panman, M.; Henry, L.; Menzel, A.; Westenhoff, S.; et al. On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome. J. Biol. Chem. 2018, 293, 8161–8172. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Flanagan, M.L.; McGillicuddy, R.D.; Zheng, H.; Ginzburg, A.R.; Yang, X.; Moffat, K.; Engel, G.S. Bacteriophytochrome Photoisomerization Proceeds Homogeneously Despite Heterogeneity in Ground State. Biophys. J. 2016, 111, 2125–2134. [Google Scholar] [CrossRef]
- Zhu, K.; Borrelli, K.W.; Greenwood, J.R.; Day, T.; Abel, R.; Farid, R.S.; Harder, E. Docking Covalent Inhibitors: A Parameter Free Approach to Pose Prediction and Scoring. J. Chem. Inf. Model. 2014, 54, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Warshaviak, D.T.; Golan, G.; Borrelli, K.W.; Zhu, K.; Kalid, O. Structure-Based Virtual Screening Approach for Discovery of Covalently Bound Ligands. J. Chem. Inf. Model. 2014, 54, 1941–1950. [Google Scholar] [CrossRef]
- Khan, F.I.; Gupta, P.; Roy, S.; Azum, N.; Alamry, K.A.; Asiri, A.M.; Lai, D.; Hassan, M.I. Mechanistic insights into the urea-induced denaturation of human sphingosine kinase 1. Int. J. Biol. Macromol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Aamir, M.; Wei, D.-Q.; Ahmad, F.; Hassan, I. Molecular mechanism of Ras-related protein Rab-5A and effect of mutations in the catalytically active phosphate-binding loop. J. Biomol. Struct. Dyn. 2016, 35, 105–118. [Google Scholar] [CrossRef]
- Khan, F.I.; Govender, A.; Permaul, K.; Singh, S.; Bisetty, K. Thermostable chitinase II from Thermomyces lanuginosus SSBP: Cloning, structure prediction and molecular dynamics simulations. J. Theor. Biol. 2015, 374, 107–114. [Google Scholar] [CrossRef]
- Stephens, D.E.; Khan, F.I.; Singh, P.; Bisetty, K.; Singh, S.; Permaul, K. Creation of thermostable and alkaline stable xylanase variants by DNA shuffling. J. Biotechnol. 2014, 187, 139–146. [Google Scholar] [CrossRef]
- Wang, Y.-J.; Khan, F.I.; Xu, Q.; Wei, D.-Q. Recent Studies of Mitochondrial SLC25: Integration of Experimental and Computational Approaches. Curr. Protein Pept. Sci. 2018, 19, 507–522. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Beg, A.; Khan, F.I.; Lobb, K.; Islam, A.; Ahmad, F.; Hassan, I. High throughput screening, docking, and molecular dynamics studies to identify potential inhibitors of human calcium/calmodulin-dependent protein kinase IV. J. Biomol. Struct. Dyn. 2018, 37, 2179–2192. [Google Scholar] [CrossRef]
- Khan, F.I.; Nizami, B.; Anwer, R.; Gu, K.-R.; Bisetty, K.; Hassan, I.; Wei, D.-Q. Structure prediction and functional analyses of a thermostable lipase obtained from Shewanella putrefaciens. J. Biomol. Struct. Dyn. 2016, 35, 2123–2135. [Google Scholar] [CrossRef]
- Gupta, P.; Khan, F.I.; Roy, S.; Anwar, S.; Dahiya, R.; Alajmi, M.F.; Hussain, A.; Rehman, T.; Lai, D.; Hassan, I. Functional implications of pH-induced conformational changes in the Sphingosine kinase 1. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 225, 117453. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Durrani, R.; Khan, F.I.; Ali, S.; Wang, Y.; Yang, B. A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study. Biomolecules 2020, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Kneen, M.; Farinas, J.; Li, Y.; Verkman, A. Green Fluorescent Protein as a Noninvasive Intracellular pH Indicator. Biophys. J. 1998, 74, 1591–1599. [Google Scholar] [CrossRef]
- Chapleau, R.R.; Blomberg, R.; Ford, P.C.; Sagermann, M. Design of a highly specific and noninvasive biosensor suitable for real-time in vivo imaging of mercury (II) uptake. Protein Sci. 2008, 17, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, R.; Endoh, T.; Yamamoto, S.; Tainaka, K.; Sugimoto, K.; Fujieda, N.; Kiyonaka, S.; Mori, Y.; Morii, T. A single circularly permuted GFP sensor for inositol-1,3,4,5-tetrakisphosphate based on a split PH domain. Bioorg. Med. Chem. 2009, 17, 7381–7386. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Terwilliger, T.C.; Waldo, G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005, 23, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.G.; Culler, S.J.; Nguyen, P.Q.; McGuire, R.M.; Silberg, J.J.; Smolke, C.D. In Vivo Fluorescent Detection of Fe-S Clusters Coordinated by Human GRX2. Chem. Biol. 2009, 16, 1299–1308. [Google Scholar] [CrossRef]
- Yang, Y.; Linke, M.; Von Haimberger, T.; Hahn, J.; Matute, R.A.; Gonzalez, L.; Schmieder, P.; Heyne, K. Real-Time Tracking of Phytochrome’s Orientational Changes During Pr Photoisomerization. J. Am. Chem. Soc. 2012, 134, 1408–1411. [Google Scholar] [CrossRef] [PubMed]
- Rohmer, T.; Strauss, H.; Hughes, J.; De Groot, H.; Gärtner, W.; Schmieder, P.; Matysik, J. 15N MAS NMR Studies of Cph1 Phytochrome: Chromophore Dynamics and Intramolecular Signal Transduction. J. Phys. Chem. B 2006, 110, 20580–20585. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Brunzelle, J.S.; Forest, K.T.; Vierstra, R.D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 2005, 438, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.R.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schäfer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]


| S. No. | Domains | Agp2 | PAiRFP1 | PAiRFP2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sites | Hydrogen Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | ||
| 1 | PAS domain | K69 | H72 D73 | G67 K68 L70 V71 | – | – | – | R69 | H72 D73 | G67 K68 L70 V71 |
| 2 | R83 | – | T81 G82 T84 T85 | – | – | – | K83 | – | T81 G82 T84 R86 | |
| 3 | G120 | – | S119 D122 S121 | – | – | – | D120 | S121 | S119 D122 S286 | |
| S. No. | Domains | Agp2 | PAiRFP1 | PAiRFP2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sites | Hydrogen Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | ||
| 1 | GAF domain | A123 | – | D122 Q124 P125 P288 | – | – | – | T123 | – | D122 Q124 P125 P288 |
| 2 | G127 | R130 K131 | Q124 P125 L126 T128 A129 | D127 | R130 K131 | Q124 P125 L126 T128 A129 | – | – | – | |
| 3 | S 141 | S144 L145 | A139 D140 V142 E143 L307 | R141 | S144 L145 | A139 D140 V142 E143 L307 | – | – | – | |
| 4 | M163 | Y165 A276 | T150 V162 I164 S177 E178 F187 L274 I275 | L163 | V276 | T150 V162 I164 S177 E178 F187 L274 I275 | L163 | V276 | T150 V162 I164 S177 E178 F187 I275 | |
| 5 | Q168 | R166 G171 A172 | F167 E169 D170 G173 K174 Y191 | L168 | G171 A172 | F167 E169 D170 G173 K174 Q475 W466 R166 | E168 | R166 G171 A172 | F167 E169 D170 G173 K174 Y191 Q475 | |
| 6 | A203 | Q199 Q200 L206 K207 | A201 R202 L204 Y205 | V203 | Q199 Q200 L206 K207 | A201 R202 L204 Y205 | – | – | – | |
| 7 | G218 | D215 | A216 S217 T219 R220 | S218 | D215 | A216 S217 T219 P26 | – | – | – | |
| 8 | R220 | – | I24 P26 G218 T219 I221 S243 V244 P246 R253 | P220 | – | I24 P26 T219 I221 S243 P222 | P220 | – | I24 P26 T219 I221 S243 Y23 | |
| 9 | S 243 | R211 | P21 I24 Y23 Q25 L241 R242 | – | – | – | N243 | Y23 R211 F244 | P21 I24 Q25 L236 L241 R242 | |
| 10 | V244 | – | R211 R242 S243 S245 P246 H248 | F244 | – | R211 S243 S245 P246 H248 C249 | F244 | – | Q25 I212 R242 S245 P246 H248 C249 | |
| 11 | G269 | – | L206 I266 V267 A270 | – | – | – | D269 | – | L206 I266 V267 A270 | |
| 12 | A276 | – | R161 V162 I164 M261 S262 L274 I275 C277 | V276 | – | R161 V162 I164 M261 S262 L274 I275 C277 | V276 | – | R161 V162 I164 M261 S262 I263 L274 I275 | |
| 13 | Y280 | D160 R161 | E185 G256 A258 H279 S281 | C280 | D160 | L184 G256 V257 A258 H279 S281 | C280 | D160 | L184 G256 V257 A258 H279 S281 | |
| 14 | E294 | R290 I291 G297 E298 | T209 L210 A292 A293 M295 F296 | V294 | R290 I291 G297 E298 | T84 T85 T209 A292 A293 M295 F296 | A294 | R290 I291 G297 E298 | T209 L210 A292 A293 M295 F296 | |
| 15 | H303 | F299 F300 V306 L307 | I136 S301 M302 L304 Q305 | R303 | F299 F300 V306 L307 | I136 A139 S301 M302 L304 Q305 | F303 | F299 F300 V306 L307 | R137 S301 M302 L304 Q305 | |
| S. No. | Domains | Agp2 | PAiRFP1 | PAiRFP2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sites | Hydrogen Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | Mutation Sites | H-Bonds | Van der Waals Interactions | ||
| 1 | PHY domain | H333 | A330 | A331 H332 A334 N335 I336 H499 | – | – | – | R333 | A330 | L329 A331 H332 A334 N335 L339 D342 H499 |
| 2 | I336 | L339 L340 | A334 N335 E337 E338 V495 | – | – | – | L336 | L339 L340 | A334 N335 E337 E338 A496 | |
| 3 | D349 | D346 | H318 F347 A348 L350 M351 | – | – | – | R349 | D346 | F347 A348 L350 I351 | |
| 4 | M351 | – | A348 P352 C353 L415 D481 A485 | – | – | – | I 351 | – | A348 L350 P352 C353 R431 W477 D481 | |
| 5 | A386 | F382 V383 | A384 S385 S387 E388 | V386 | F382 V383 | A384 S385 S387 E388 | V386 | F382 V383 | A384 S385 S387 E388 | |
| 6 | G409 | – | Y407 A408 T410 A411 K432 | – | – | – | D409 | – | Y407 A408 T410 A411 K432 E433 Q436 | |
| 7 | L419 | D425 | L,340 L358 V360 I417 S420 Y426 L427 L492 | – | – | – | I419 | D425 | I417 P418 S420 Y426 L427 L492 | |
| 8 | T469 | I465 W466 | P193 K467 E468 V470 R471 Q473 | – | – | – | S469 | I465 W466 | K467 E468 V470 R471 Q473 | |
| 9 | A480 | E483 I484 | S478 E479 D481 R482 | V480 | E483 I484 | I316 S478 E479 D481 R482 | – | – | – | |
| 10 | A487 | E483 I484 I490 A491 | L323 A485 E486 A488 R489 | – | – | – | T487 | E483 I484 A491 | A319 H320 L323 A485 E486 A488 R489 I490 | |
| 11 | E494 | I490 A491 F497 | L327 L492 V493 V495 A496 H499 | – | – | – | G494 | I490 A491 F497 H498. | L327 L492 V493 V495 A496 H498 | |
| 12 | H498 | E494 V495 E501 H502 | L327 A496 F397 H499 S500 | Y498 | E494 Y508 | V495 A496 H499, S500 | – | – | – | |
| S. NO. | Agp2 | PAiRFP1 | ΔΔG ENCoM (kcal/mol) | ΔΔS ENCoM (kcal.mol−1.K−1) | Molecular Flexibility | ΔΔG DynaMut (kcal/mol) | Outcome | DeepDDG (kcal/mol) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | G127 | G127D | 0.04 | −0.05 | Decrease | 0.021 | Stabilizing | 0.220 | Stable |
| 2 | S141 | S141R | −0.073 | 0.092 | Increase | −0.096 | Destabilizing | −0.347 | Unstable |
| 3 | M163 | M163L | −0.158 | 0.197 | Increase | −0.391 | Destabilizing | −1.061 | Unstable |
| 4 | Q168 | Q168L | 0.138 | −0.172 | Decrease | 1.204 | Stabilizing | 0.298 | Stable |
| 5 | A203 | A203V | 0.128 | −0.16 | Decrease | 0.308 | Stabilizing | −0.274 | Unstable |
| 6 | G218 | G218S | 0.209 | −0.261 | Decrease | −0.244 | Destabilizing | −0.661 | Unstable |
| 7 | R220 | R220P | −0.741 | 0.926 | Increase | −0.179 | Destabilizing | 0.361 | Stable |
| 8 | V244 | V244F | 0.519 | −0.649 | Decrease | 1.154 | Stabilizing | −1.383 | Unstable |
| 9 | A276 | A276V | 0.184 | −0.231 | Decrease | 0.791 | Stabilizing | −0.495 | Unstable |
| 10 | Y280 | Y280C | −1.03 | 1.287 | Increase | −0.188 | Destabilizing | −0.786 | Unstable |
| 11 | E294 | E294V | −0.371 | 0.464 | Increase | −0.032 | Destabilizing | −0.614 | Unstable |
| 12 | H303 | H303R | −0.22 | 0.275 | Increase | −0.955 | Destabilizing | −0.072 | Unstable |
| 13 | A386 | A386V | 0.127 | −0.159 | Decrease | 0.09 | Stabilizing | 0.154 | Stable |
| 14 | A480 | A480V | 0.373 | −0.466 | Decrease | 0.964 | Stabilizing | 0.147 | Stable |
| 15 | H498 | H498Y | 0.085 | −0.106 | Decrease | 0.994 | Stabilizing | −1.230 | Unstable |
| S. NO. | Agp2 | PAiRFP2 | ΔΔG ENCoM (kcal/mol) | ΔΔS ENCoM (kcal.mol−1.K−1) | Molecular Flexibility | ΔΔG DynaMut (kcal/mol) | Outcome | DeepDDG (kcal/mol) | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | K69 | K69R | −0.263 | 0.328 | Increase | −0.445 | Destabilizing | −0.148 | Stable |
| 2 | R83 | R83K | −0.077 | 0.097 | Increase | −0.948 | Destabilizing | −0.044 | Unstable |
| 3 | G120 | G120D | 0.078 | −0.098 | Decrease | 0.671 | Stabilizing | 0.043 | Stable |
| 4 | A123 | A123T | −0.034 | 0.042 | Increase | 0.022 | Stabilizing | −0.050 | Unstable |
| 5 | M163 | M163L | −0.158 | 0.197 | Increase | −0.391 | Destabilizing | −1.061 | Unstable |
| 6 | Q168 | Q168E | 0.104 | −0.13 | Decrease | 0.097 | Stabilizing | −0.492 | Unstable |
| 7 | R220 | R220P | −0.741 | 0.926 | Increase | −0.179 | Destabilizing | 0.361 | Stable |
| 8 | S243 | S243N | 0.073 | −0.092 | Decrease | 0.578 | Stabilizing | −2.434 | Unstable |
| 9 | V244 | V244F | 0.519 | −0.649 | Decrease | 1.154 | Stabilizing | −1.383 | Unstable |
| 10 | G269 | G269D | −0.173 | 0.217 | Increase | −1.562 | Destabilizing | −1.040 | Unstable |
| 11 | A276 | A276V | 0.184 | −0.231 | Decrease | 0.791 | Stabilizing | −0.495 | Unstable |
| 12 | Y280 | Y280C | −1.03 | 1.287 | Increase | −0.188 | Destabilizing | −0.786 | Unstable |
| 13 | E294 | E294A | −0.573 | 0.717 | Increase | −0.854 | Destabilizing | −0.667 | Unstable |
| 14 | H303 | H303F | 0.155 | −0.193 | Decrease | 0.479 | Stabilizing | 0.804 | Stable |
| 15 | H333 | H333R | −0.138 | 0.172 | Increase | −0.254 | Destabilizing | −0.402 | Unstable |
| 16 | I336 | I336L | 0.203 | −0.254 | Decrease | 1.153 | Stabilizing | 0.460 | Stable |
| 17 | D349 | D349R | 0.138 | −0.172 | Decrease | 0.824 | Stabilizing | −0.044 | Unstable |
| 18 | M351 | M351I | −0.369 | 0.461 | Increase | −0.516 | Destabilizing | 0.666 | Stable |
| 19 | A386 | A386V | 0.127 | −0.159 | Decrease | 0.09 | Stabilizing | 0.154 | Stable |
| 20 | G409 | G409D | 0.229 | −0.286 | Decrease | 0.803 | Stabilizing | 0.608 | Stable |
| 21 | L419 | L419I | 0.016 | −0.019 | Decrease | 0.173 | Stabilizing | −0.087 | Unstable |
| 22 | T469 | T469S | −0.141 | 0.177 | Increase | −0.67 | Destabilizing | −0.487 | Unstable |
| 23 | A487 | A487T | 0.33 | −0.412 | Decrease | 0.627 | Stabilizing | −0.032 | Unstable |
| 24 | E494 | E494G | −0.313 | 0.391 | Increase | −1.362 | Destabilizing | −1.329 | Unstable |
| Electrostatic Interactions | Van der Waals Interactions | Covalent Bonds | |||
|---|---|---|---|---|---|
| Agp2-BVcis | Agp2-BVtrans | Agp2-BVcis | Agp2-BVtrans | Agp2-BVcis | Agp2-BVtrans |
| Y165 S195 N196 R211 H248 S245 H278 R456 | E16 Y165 D196 Y205 R211 H248 H278 R456 | C13 D14 F187 Q190 F192 I197 P198 Y205 I213 V244 A276 P459 S462 | Y191 F192 S195 I197 P198 F244 S245 I247 Y251 V276 | C13 | C13 |
| PAiRFP1-BVcis | PAiRFP1-BVtrans | PAiRFP1-BVcis | PAiRFP1-BVtrans | PAiRFP1-BVcis | PAiRFP1-BVtrans |
| S195 D196 R211 H248 M255 H278 S260 R456 | Y165 S195 D196 R211 S245 H248 H278 R456 | C13 D14 I18 F192 I197 P198 A201 Y205 F244 I247 Y251 V276 P459 S462 | C13 D14 Q190 F192 I197 P198 Y205 I213 V244 A276 S462 | C13 | C13 |
| PAiRFP2-BVcis | PAiRFP2-BVtrans | PAiRFP2-BVcis | PAiRFP2-BVtrans | PAiRFP2-BVcis | PAiRFP2-BVtrans |
| E16 Y165 Q190 D196 Y205 R211 H248 Y251 H278 R456 S462 | S195 D196 R211 H248 Y251 S260 H278 R456 S462 | F187 F192 S195 I197 P198 F244 S245 I247 M255 V276 P459 | C13 D14 I18 F192 I197 P198 A201 Y205 F244 I247 V276 P459 | C13 | C13 |
| Percentage of Secondary Structure (SS %) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Structure * | Coil | β-Sheet | β-Bridge | Bend | Turn | α-Helix | 310-Helix |
| Agp2 | 65 | 21 | 21 | 1 | 13 | 9 | 34 | 1 |
| PAiRFP1 | 65 | 20 | 21 | 2 | 13 | 10 | 32 | 1 |
| PAiRFP2 | 65 | 21 | 20 | 2 | 13 | 9 | 34 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, F.I.; Hassan, F.; Anwer, R.; Juan, F.; Lai, D. Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2. Biomolecules 2020, 10, 1286. https://doi.org/10.3390/biom10091286
Khan FI, Hassan F, Anwer R, Juan F, Lai D. Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2. Biomolecules. 2020; 10(9):1286. https://doi.org/10.3390/biom10091286
Chicago/Turabian StyleKhan, Faez Iqbal, Fakhrul Hassan, Razique Anwer, Feng Juan, and Dakun Lai. 2020. "Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2" Biomolecules 10, no. 9: 1286. https://doi.org/10.3390/biom10091286
APA StyleKhan, F. I., Hassan, F., Anwer, R., Juan, F., & Lai, D. (2020). Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2. Biomolecules, 10(9), 1286. https://doi.org/10.3390/biom10091286









