MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics
Abstract
1. Introduction
2. Inhibition or Activation of Translation
3. Suppression or Enhancement of Transcription
Regulation of Alternative Splicing
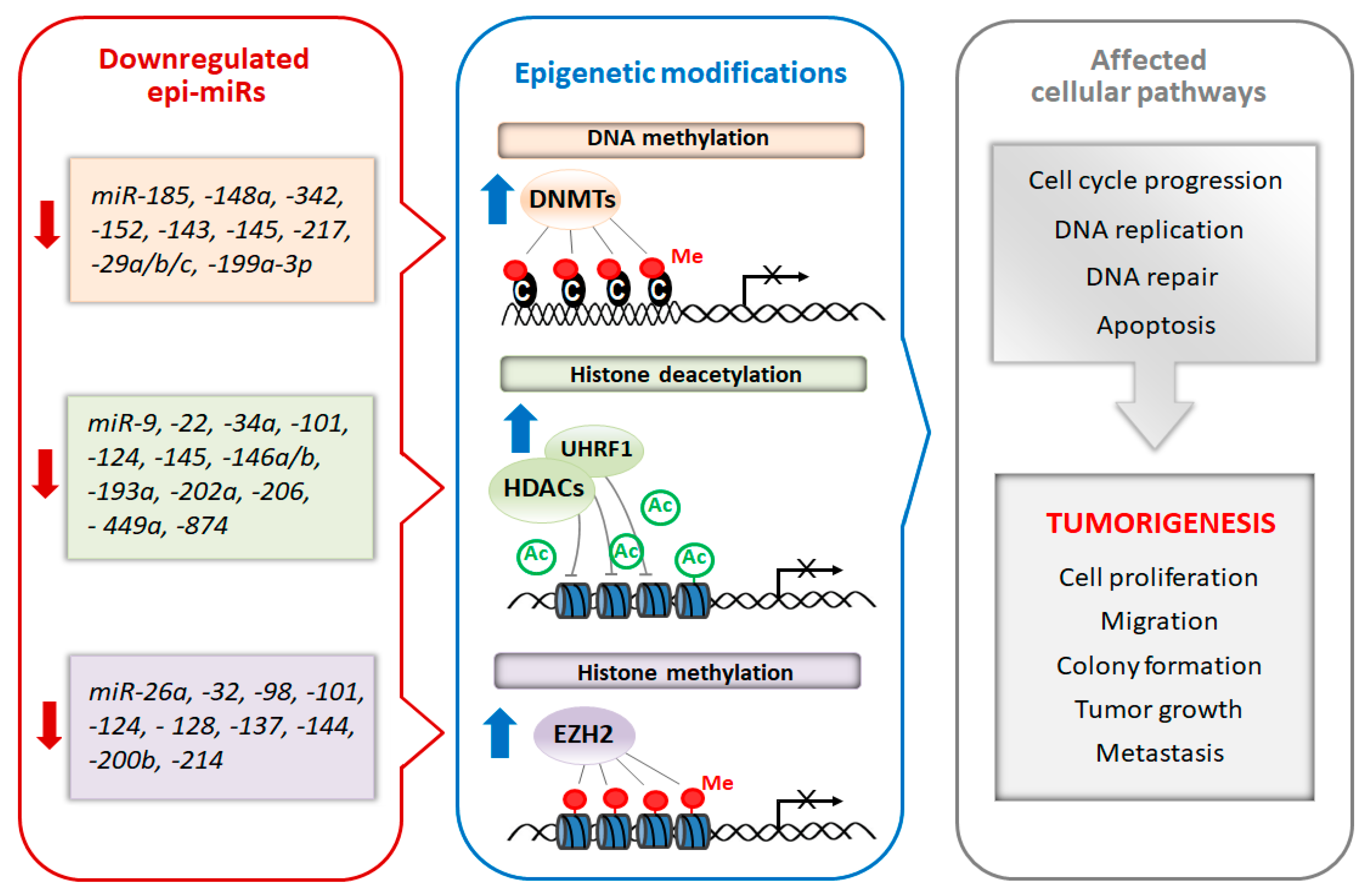
4. miRNAs As Potential Cancer Epi-Therapeutics
| Epi-miRNA | mRNA Target | Cell Lines | Methods | Animal Study | Results | References |
|---|---|---|---|---|---|---|
| Breast cancer | ||||||
| miR-34a | HDAC1 HDAC7 | MCF-7, MDA-MB-231, BT-20, T47-D, PC3, DU-145, LNCaP, NIH:OVCAR, SK-OV-3, HeLa and non-transformed mammary MCF-10A cells | miR-34a mimics; luciferase reporter assay (in MCF-7, PC3, and MDA-MB-231 cells) | ND | miR-34a expression negatively correlates with tumor grades; transfection of miR-34a mimic reduces cell survival and increases the cytotoxicity of chemotherapy drugs; re-expression of miR-34a inhibits the tumorigenic activity of cancer stem cells (CSCs). | [82] |
| miR-101 | EZH2 | SKBR3 | pre-miR-101; luciferase reporter assay (in SKBr3 cells) | ND | miR-101 overexpression in SKBr3 attenuates cell proliferation, migration and inhibits the invasive potential. | [83] |
| miR-128 | BMI1 | SK-3rd, MCF-7 and SKBR3 | lentivirus vector miR-128; luciferase reporter assay (in SK-3rd and MCF-7 cells) | ND | Ectopic expression of miR-128 decreases cell viability and increases apoptosis and DNA damage in the presence of doxorubicin; ectopic miR-128 expression sensitizes BT-ICs (breast tumor–initiating cells) to doxorubicin enhancing the DNA damage and pro-apoptotic effects. | [84] |
| miR-148a | SMAD2 | MDA-MB-231 and Hs-578T | miR-148a mimics; luciferase reporter assay (in MDA-MB-231 cells) | BALB/c nude mice; MDA-MB-231 cells were injected s.c.; glabridin (GLA) was administered intragastrically each day. | GLA enhances the expression of miR-148a; GLA-treated tumors have increased expression of miR-148a and decreased expressions of SMAD2. | [75] |
| miR-185 | DNMT1 | MDA-MB-231, MDA-MB-361, MDA-MB-435, MDA-MB-468, MCF-7, T47D, BT-474, BT-20 and BT-483; normal mammary epithelial cell lines (HBL-100, 184A1 and MCF-10A) | miR-185 mimics | Nude mice; MDA-MB-231 cells were injected s.c.; intratumoral injection of miR-185 mimics. | Ectopic expression of miR-185 inhibits cell proliferation and induces apoptosis; inhibits tumor growth in vivo. | [85] |
| miR200b | SUZ12 | MCF-10A cells containing the ER-Src fusion gene, MCF7, SKBR3, MDAMB-231, MDA-MB-435, NSCCs (non-stem cancer cells) | miR-200b; luciferase reporter assay (in ER-Src cells) | Athymic nude mice; CSCs were pretreated with miR-200b and injected s.c.; ER-Src (treated and untreated with tamoxifen) were injected s.c. and then doxorubicin or combination doxorubicin and miR-200b was administered i.p. | miR-200b overexpression affects CSCs growth and reduces cell invasiveness; pretreatment of CSCs with miR-200b blocked tumor formation in vivo; combinatorial therapy (doxorubicin with miR-200b) causes regression of tumor growth and prevents relapse of the disease. | [72] |
| Bladder cancer | ||||||
| miR-101 | EZH2 | T24, UM-UC-3 and TCCSUP | vector pcDNA3.1 with pre-miR-101; luciferase reporter assay (in UM-UC-3 cells) | ND | Restored miR-101 expression inhibits cell proliferation, suppresses colony formation and hinders EZH2-mediated neoplastic progression. | [86] |
| miR-124 | UHRF1 | J82, T24, HEK 293 and SV-HUC-1 | miR-124 mimics; luciferase reporter assay (in HEK-293cells) | Male BALB/C-A mice; T24 cells were injected s.c. and then intratumoral injection was performed with miR-124 mimics. | miR-124 overexpression attenuates cell proliferation, migration, invasion and vasculogenic mimicry; inhibits tumor growth in vivo. | [87] |
| miR-144 | EZH2 | T24 | vector pcDNA–miR-144; luciferase reporter assay (in HEK293 cells) | ND | miR-144 overexpression inhibits cell proliferation; decreases EZH2 protein levels. | [88] |
| miR-145-5p miR-145-3p | UHRF1 | T24 and BOY | pre-miR-145-5p and pre-miR-145-3p; luciferase reporter assay (in T24 and BOY cells) | ND | Ectopic expression of either miR-145-5p or miR-145-3p suppresses cancer cell growth, migration and invasion and induces apoptosis. | [89] |
| miR-148a | DNMT1 | SV-HUC-1, T24, TCCSUP, J82 and UM-UC-3 | miR-148a mimics; cisplatin or doxorubicin treatment | ND | miR-148a overexpression reduces cell viability by promoting apoptosis; combinatorial therapy (miR-148a/cisplatin or miR-148a/doxorubicin) enhanced apoptosis. | [71] |
| Colorectal cancer | ||||||
| miR-9 | UHRF1 | HCT116 and HT29 | miR-9 oligonucleotides; lentivirus vector miR-9; luciferase reporter assay (in HCT116 and HT29 cells) | ND | miR-9 overexpression attenuates CRC cell proliferation and promotes cell apoptosis; reduces UHRF1 expression. | [90] |
| miR-143 | DNMT3A | 228, CaCO2, Clone A, HCT116, HT-29, MIP101 and SW480 | pre-miR-143; luciferase reporter assay (in 228 and SW480 cells) | ND | Ectopic expression of miR-143 inhibits cell growth, reduces clone formation; restored miR-143 expression decreases tumor cell growth and soft-agar colony formation, and downregulates DNMT3A expression. | [91] |
| miR-342 | DNMT1 | SW480, HT29, HCT116 and HEK293T | miR-342 oligonucleotides; lentivirus vector miR-342; luciferase reporter assay (in SW480 cells) | Female athymic BABL/c nude mice; cell lines stably expressing miR-342 were injected s.c. | Enhanced miR-342 expression inhibits cell proliferation and invasion; miR-342 overexpression leads to demethylation and induction of tumor suppressor genes through blocking DNMT1 expression; miR-342 overexpression inhibits tumor growth and lung metastasis in vivo. | [92] |
| Endometrial cancer | ||||||
| miR-101 | EZH2 | SPAC-1-L and SPAC-1-S; HEC-50 and HOUA-I cell lines were derived from poorly-differentiated endometrioid EC (endometrial carcinoma) | vector with pre-miR-101-3p; luciferase reporter assay (in SPAC-1-L and HOUA-I cells) | ND | Ectopic overexpression of miR-101 suppresses cell proliferation, attenuates the epithelial-mesenchymal transition associated cancer cell migration and invasion, abrogates the sphere-forming capacity and enhances chemosensitivity to paclitaxel. | [93] |
| Esophageal cancer | ||||||
| miR-203 | BMI1 | EC9706 and KYSE150 | lentivirus vector miR-203; luciferase reporter assay (in EC9706 cells) | Female nude mice and nonobese diabetic/severe combined immunodeficient mice; freshly prepared cells were injected s.c. | miR-203 overexpression reduces colony formation, tumorigenicity ability and self-renewal of esophageal cancer stem-like cells; increases sensitivity to cisplatin. | [94] |
| Gastric cancer | ||||||
| miR-29b/c | DNMT3A | AGS and BGC-823 | miR-29b/c mimics; luciferase reporter assay (in BGC-823 cells) | ND | miR-29b/c overexpression decreases migration and reduces invasive ability; miR-29b/c suppresses the expression of DNMT3A. | [95] |
| miR-146a miR-146b | UHRF1 | GC9811, GC9811-P, MKN28NM and MKN28M | pre-miR-146a/b; lentivirus vector miR-146a/b; luciferase reporter assay (in HEK293T and GC9811 cells) | Nude mice; metastasis assay: GC9811-P cells infected with miR-146a/b were injected into the tail vein. | Restored expression of miR-146a/b reduces the expression of UHRF1; upregulation of miR-146a/b suppresses metastasis. | [96] |
| miR-148a | DNMT1 | SGC-7901, BGC-823 and GES-1 (human gastric epithelium-immortalized cell line) | miR-148a mimics | ND | miR-148a mimics suppresses cell proliferation; miR-148a overexpression decreases DNMT1 expression and induces the overexpression of MEG3 (lncRNA). | [97] |
| miR-206 | HDAC4 | SGC-7901, BGC-823, AGS, non-malignant gastric cell line GES-1 and HEK293T | miR-206 mimics; vector with miR-206 | Nude mice; SGC-7901 cells carrying P2GM-miR-206 was injected s.c. | Ectopic expression of miR-206 represses cell proliferation, colony formation, invasion and migration; miR-206 promotes myogenic differentiation and blocks tumor growth in vivo. | [98] |
| Glioblastoma | ||||||
| miR-128 | Bim-1 | U87MG, U251MG and U373MG | pre-miR-128 mimics; lentivirus vector pri-miR-128-1; luciferase reporter assay (in U87, U251, and U373 cells) | Athymic mice; U87 cells stably expressing miR-128 were implanted s.c. | miR-128 expression reduces glioma cell proliferation, self-renewal in vitro and glioma xenograft growth in vivo. | [99] |
| miR-128 | SUZ12 BMI1 | U87 malignant glioma (MG) and U251MG glioblastoma cells | pre-miR-128; lentivirus vector miR-128; luciferase reporter assay (in HEK293 cells) | Mut3 mice (hGFAP-cre; Nf1flox/+; Trp532/+). | miR-128 overexpression reduces proliferative potential and colony formation; reestablishment of miR-128 expression impairs glioma stem-like cells self-renewal and increases their radiosensitivity. | [100] |
| Head and neck squamous cell carcinoma | ||||||
| miR-874 | HDAC1 | SAS, FaDu, HSC3, IMC-3, human fibroblast and MRC-5 | mature miR-874; luciferase reporter assay (in SAS cells) | ND | Restoration of miR-874 inhibits cell proliferation, induces cell cycle arrest and apoptosis. | [101] |
| Hepatobiliary cancer | ||||||
| miR-152 miR-148a | DNMT1 | KMCH-1, Mz-ChA-1, TFK-1 and H69; Mz-IL-6 (KMCH-1 stably transfected with IL-6) | pre-miR-152 and pre-miR-148a; luciferase reporter assay (in Mz-ChA-1 cells) | Male athymic nu/nu mice; Mz-IL-6 cells were injected s.c. | pre-miR-148a and pre-miR-152 decreases DNMT-1 protein expression and reduces cell proliferation; miR-148a and miR-152 expression was reduced in tumor cell xenografts in vivo. | [102] |
| Hepatocellular carcinoma | ||||||
| miR-22 | HDAC4 | Hep3B and SMMC7721 | miR-22 mimics; luciferase reporter assay (in Hep3B cells) | Male BALB/c athymic nude mice; miR-22 mimics transfected Hep3B or SMMC7721 cells were injected s.c. | Restoration of miR-22 expression suppresses cell proliferation and endogenous expression of HDAC4 protein; miR-22 transfection delays tumor formation and reduces tumor size in vivo. | [103] |
| miR-29a miR-185 | DNMT3A DNMT3B | HepG2 and HuH-7 | dendrosomal curcumin (DNC) treatment | ND | Overexpression of miR-29a and miR-185 after dendrosomal curcumin (DNC) treatment, down-regulates the expression of DNMT1, 3A and 3B. | [74] |
| miR-145 | HDAC2 | Hep3B, HepG2, SNU-182, SNU-449 and PLC/PRF/5 | miR-145 mimics; vector with miR-145; luciferase reporter assay (in SNU-449 cells) | Male athymic nude mice; Hep3B cells transfected with miR-145 were injected s.c. | Ectopic expression of miR-145 inhibits cell growth and HDAC2 expression; inhibits tumor growth in vivo. | [104] |
| miR-200a | HDAC4 | SMMC-7721 and HepG2 | miR-200a mimics; lentivirus vector miR-200a; luciferase reporter assay (in SMMC-7721 cells) | Nude mice; HepG2 cells stably transfected with miR-200a were implanted s.c. | miR-200a inhibits cell proliferation and migration both in vivo and in vitro; miR-200a overexpression induces up-regulation of global acetyl-histone H3. | [105] |
| Acute myeloid leukemia | ||||||
| miR-29b | DNMT3A DNMT3B | AML cell lines, Kasumi-1, MV4-11 and K562 | pre-miR-29b; lentivirus vector miR-29b; luciferase reporter assays (in K562 cells) | ND | Enforced expression of miR-29b in AML cells reduces of the expression of DNMT1, DNMT3A, and DNMT3B; pre-miR-29b overexpression induces partial differentiation of AML blasts. | [79] |
| miR-29b | DNMT3B | primary AML blasts, K562 and Kasumi-1 | synthetic miR-29b | Female nude mice; synthetic miR-29b oligonucleotides were injected directly into the tumors. | Restoring miR-29b expression, induces apoptosis and dampens cell growth in AML cells. | [106] |
| miR-193a-3p | DNMT3A HDAC3 | HL60, U937, U937-A/E-HA, Kasumi-1, SKNO-1, SKNO-1-siA/E-RNA and KG1 | miR-193a mimics; lentivirus vector miR-193a; luciferase reporter assay (in 293T cells) | Nude mice; SKNO-1 cells were injected s.c.; intratumor injection of miR-193a. | Enhanced miR-193a levels induce G1 arrest, apoptosis, and restores leukemic cell differentiation; decreases tumor size in vivo. | [107] |
| Chronic myeloid leukemia | ||||||
| miR-217 | DNMT3A | Bcr/Abl-expressing K562 cells | lentivirus vector miR-217; luciferase reporter assay (in K562DR cells) | Female immune deficient BALB/c nude mice; K562 cells were injected s.c.; drug administration: dasatinib or 5-AzadC or a combination of both dasatinib and 5-AzadC. | Forced expression of miR-217 inhibits expression of DNMT3A and sensitizes cells to growth inhibition mediated by the tyrosine kinase inhibitors (prevents drug resistance). | [108] |
| Multiple myeloma | ||||||
| miR-29b | DNMT3A DNMT3B | MM cell lines | pre-miR-29b mimics (formulated with a Neutral Lipid Emulsion (NLE) delivery system); lentivirus vector miR-29b; luciferase reporter assay (in INA-6 cells) | Male CB-17 severe combined immunodeficient (SCID) mice; MM cells were inoculated s.c.; miR-29b mimics were administered intratumorally and systemically via tail vein. | miR-29b mimics impair cell cycle progression and potentiate the growth-inhibitory effects induced by the demethylating agent 5-azacitidine; miR-29b mimics induce anti-tumor effects in vivo. | [70] |
| Leukemia | ||||||
| miR-143 | DNMT3A | AML (HL-60, NB4 and U937), CML (K562), acute erythroleukemia (HEL), T lymphocytic leukemia (Jurkat and CEM), B-cell lymphoma (CA46, Raji cells of Burkitt’s lymphoma) and multiple myeloma (U266) | lentivirus vector miR-143 | ND | miR-143 overexpression decreases DNMT3A mRNA and protein expression, reduces cell proliferation, colony formation and cell cycle progression as well as induces apoptosis. | [109] |
| Lung cancer | ||||||
| miR-29a, -b, -c | DNMT3A DNMT3B | A549 and H1299 | pre-miR-29a, -29b-1, -29c oligonucleotides; luciferase reporter assay (in A549 cells) | Female nude mice; A549 cells transfected with pre-miR-29a, -29b, or -29c, were injected s.c. | Enforced expression of miR-29s restores normal patterns of DNA methylation, induces re-expression of methylation-silenced tumor suppressor genes and inhibits tumorigenicity in vitro and in vivo. | [68] |
| miR-193a-3p miR-193a-5p | UHRF1 | SPC-A-1, SPC-A-1sci, A549, H1299, LC-21, H358 and HEK-293T | miR-193a-3p/5p mimics; lentivirus vector miR-193a-3p/5p; luciferase reporter assay (in HEK293T cells) | BALB/C-nu/nu nude male mice; metastasis assays: SPC-A-1sci cells stably expressing the miR-193a-3p/5p-mimic were injected into the tail vein. | miR-193a-3p/5p overexpression inhibits cell proliferation, migration, invasion and epithelial–mesenchymal transition (EMT); lung metastasis formation in vivo. | [110] |
| Lymphoma | ||||||
| miR-26a | EZH2 | human BL cell lines; murine MYC-induced lymphoma cell lines | vectors with mature miR-26a; luciferase reporter assay (in HEK-293 cells) | ND | miR-26a overexpression reduces cell numbers and results in an anti-proliferative effect. | [111] |
| Melanoma | ||||||
| miR-200c | BMI1 | WM35, WM793, WM115A, M3523A, 1205Lu and 293T | lentivirus vector miR-200c; vector pEZX-miR-200c | Male athymic nu/nu mice; miR-200c–WM115A cells were injected s.c. | miR-200c overexpression decreases cell proliferation, colony formation and migratory capacity as well as drug resistance and increases sensitivity to various chemotherapeutic agents (including cisplatin); inhibits melanoma xenograft growth and metastasis in vivo. | [112] |
| Neuroblastoma | ||||||
| miR-137 | EZH2 | Mouse Neuro-2a (N-2a); human SH-SY5Y | miR-137 mimics; resveratrol (RSV) treatment; luciferase reporter assay (in HEK293 cells) | ND | miR-137 expression is up-regulated after RSV treatment; miR-137 inhibits EZH2 expression after RSV treatment; miR-137 regulates the EZH2-mediated apoptosis after RSV treatment. | [73] |
| miR-124 | EZH2 | Neural Stem Cells (NSCs) and HEK293T | mature miR-124 | ND | miR-124 overexpression down-regulates expression of Ezh2 and up-regulates neuron-specific Ezh2 target genes; promotes neuronal differentiation. | [113] |
| miR-137 | KDM1A | IMR-32, SHEP, SKN-BE and HEK-293 | pre-miR-137; luciferase reporter assay (in SHEP and HEK293 cells) | ND | Re-expression of miR-137 increases apoptosis, decreases cell viability and proliferation, induces neuronal differentiation; downregulates KDM1A. | [114] |
| miR-152 | DNMT1 | SK-N-BE, SH-SY5Y, SK-N-AS and Kelly | pre-miR-152; luciferase reporter assay (in Kelly cells) | ND | Ectopic upregulation of miR-152 declines cell invasiveness and anchorage-independent cell growth, contributing to the differentiated phenotype. | [115] |
| Oral squamous cell carcinoma | ||||||
| miR-32 | EZH2 | SCC-4, SCC-9, SCC-25 and Tca8113; normal oral keratinocyte cell line (hNOK) | mature miR-32 mimics; luciferase reporter assay (in Tca8113 cells) | ND | miR-32 overexpression reduces cell proliferation, migration and invasion, promotes cell apoptosis; miR-32 down-regulates the expression of EZH2. | [116] |
| Ovarian cancer | ||||||
| miR-15a miR-16 | BMI1 | OVCAR-5, OV-167, OV-202, CP-70, A2780 and OSE (ovarian surface epithelial cell) | pre-miR-15a, pre-miR-16; luciferase reporter assay (in OV-202 and CP-70 cells) | ND | miR-15a or miR-16 overexpression decreases cell proliferation and clonal growth; downregulates BMI1 protein levels. | [117] |
| miR-152 miR-185 | DNMT1 | SKOV3, A2780, A2780/DDP (cisplatin-resistant), A549 and HepG2 | miR-152 and miR-185 mimics; luciferase reporter assay (in SKOV3/DDP cells) | CD-1/CD-1 nude mice; SKOV3/DDP cells transfected with miR-152 that were injected i.p. | miR-152 or miR-185 overexpression increases cisplatin sensitivity by inhibiting proliferation and promoting apoptosis; promotes sensitivity to cisplatin through targeting DNMT1 directly. | [118] |
| Pancreatic cancer | ||||||
| miR-148a miR152 | DNMT1 | MIA PaCa-2 and AsPC-1 | pre-miR-148b, pre-miR-152; luciferase reporter assay (in MIA PaCa-2 and AsPC-1 cells) | ND | miR-148b and miR-152 overexpression inhibits cell proliferation and induces apoptosis; decreases DNMT1 expression, returns DNA methylation to normal patterns and induces re-expression of tumor suppressor genes. | [119] |
| Prostate cancer | ||||||
| miR-101 | EZH2 | DU145 | pre-miR-101; luciferase reporter assay (in SKBr3 cells) | Male nude athymic BALB/c nu/nu mice, DU145 stable cells overexpressing miR-101 were injected s.c. | miR-101 overexpression attenuates cell proliferation, migration and invasive potential; reduces colony formation; reduces tumor growth in vivo. | [83] |
| miR-145 | DNMT3B | PC3 | miR-145 mimics; luciferase reporter assay (in PC3 cells) | ND | miR-145 overexpression downregulates the expression of DNMT3B; sensitizes prostate cancer cells to X-ray radiation. | [120] |
| miR-449a | HDAC1 | PC-3, DU-145, BPH-1 and LNCaP | mature miR-449a mimics; a longer, dicer-dependent pre-miR-449a; luciferase reporter assays (in PC-3 cells) | ND | miR-449 expression arrests cell cycle, apoptosis; regulates cell growth and viability in part by repressing the expression of HDAC-1. | [121] |
| Renal cell carcinoma | ||||||
| miR-101 | UHRF1 | 786-O and Caki-1 | pre-miR-101-3p; luciferase reporter assay (in 786-O cells) | ND | Restoration of miR-101 inhibits cell proliferation, migration and decreases invasion activity; suppresses UHRF1 expression. | [122] |
| Rhabdomyosarcoma | ||||||
| miR-29 | Yin Yang 1 (YY1) | C2C12 myoblasts, RH30 and RD2 | pre-miR-29; lentivirus vector miR-29; luciferase reporter assays (in MB cells) | Athymic nu/nu female mice; RH30 cells were injected s.c.; intratumoral injection of lentivirus with miR-29. | miR-29 overexpression reduces cell growth and increases levels of the differentiation markers; intratumoral addition of miR-29 stimulates myogenic differentiation; inhibits tumor growth in vivo. | [123] |
| Testicular cancer | ||||||
| miR-199a-3p | DNMT3A (especially DNMT3A2) | Ntera 2 (NT2) | miR-199a-3p mimics; luciferase reporter assay (in NT2 cells) | ND | miR-199a-3p overexpression restores the expression of tumor-suppressor genes by affecting DNA methylation of their promoter regions. | [124] |
| Waldenström macroglobulinemia | ||||||
| miRNA-9 * | HDAC4 HDAC5 | BCWM.1, WM-WSU, MEC-1 and RL | pre-miRNA-9 * | ND | Restoring miRNA-9 * levels induces toxicity, apoptosis and autophagy; supports down-modulation of HDAC4 and HDAC5 and up-regulation of acetyl-histone-H3 and -H4 | [125] |
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Lemcke, H.; David, R. Potential mechanisms of microRNA mobility. Traffic 2018, 19, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lopez, J.; Brieno-Enriquez, M.A.; Del Mazo, J. MicroRNA biogenesis and variability. Biomol. Concepts 2013, 4, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Jo, M.H.; Shin, S.; Jung, S.R.; Kim, E.; Song, J.J.; Hohng, S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell 2015, 59, 117–124. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Braun, J.E.; Truffault, V.; Boland, A.; Huntzinger, E.; Chang, C.T.; Haas, G.; Weichenrieder, O.; Coles, M.; Izaurralde, E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation. Nat. Struct. Mol. Biol. 2012, 19, 1324–1331. [Google Scholar] [CrossRef]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villen, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef]
- Fukao, A.; Mishima, Y.; Takizawa, N.; Oka, S.; Imataka, H.; Pelletier, J.; Sonenberg, N.; Thoma, C.; Fujiwara, T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol. Cell 2014, 56, 79–89. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Brummer, A.; Hausser, J. MicroRNA binding sites in the coding region of mRNAs: Extending the repertoire of post-transcriptional gene regulation. Bioessays 2014, 36, 617–626. [Google Scholar] [CrossRef]
- Cardinali, B.; Cappella, M.; Provenzano, C.; Garcia-Manteiga, J.M.; Lazarevic, D.; Cittaro, D.; Martelli, F.; Falcone, G. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016, 7, e2086. [Google Scholar] [CrossRef] [PubMed]
- Spengler, R.M.; Zhang, X.; Cheng, C.; McLendon, J.M.; Skeie, J.M.; Johnson, F.L.; Davidson, B.L.; Boudreau, R.L. Elucidation of transcriptome-wide microRNA binding sites in human cardiac tissues by Ago2 HITS-CLIP. Nucleic Acids Res. 2016, 44, 7120–7131. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Steitz, J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; LeTonqueze, O.; Vasudevan, S. MicroRNA-mediated mRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear microRNP. Sci. Rep. 2012, 2, 1–12. [Google Scholar] [CrossRef]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schuttler, C.G.; Fehr, C.; Junemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef]
- Machlin, E.S.; Sarnow, P.; Sagan, S.M. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. USA 2011, 108, 3193–3198. [Google Scholar] [CrossRef]
- Shimakami, T.; Yamane, D.; Jangra, R.K.; Kempf, B.J.; Spaniel, C.; Barton, D.J.; Lemon, S.M. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 941–946. [Google Scholar] [CrossRef]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Tsai, N.P.; Lin, Y.L.; Wei, L.N. MicroRNA mir-346 targets the 5’-untranslated region of receptor-interacting protein 140 (RIP140) mRNA and up-regulates its protein expression. Biochem. J. 2009, 424, 411–418. [Google Scholar] [CrossRef]
- Schraivogel, D.; Meister, G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014, 39, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Schraivogel, D.; Schindler, S.G.; Danner, J.; Kremmer, E.; Pfaff, J.; Hannus, S.; Depping, R.; Meister, G. Importin-β facilitates nuclear import of human GW proteins and balances cytoplasmic gene silencing protein levels. Nucleic Acids Res. 2015, 43, 7447–7461. [Google Scholar] [CrossRef] [PubMed]
- Chaston, J.J.; Stewart, A.G.; Christie, M. Structural characterisation of TNRC6A nuclear localisation signal in complex with importin-alpha. PLoS ONE 2017, 12, e0183587. [Google Scholar] [CrossRef]
- Nishi, K.; Nishi, A.; Nagasawa, T.; Ui-Tei, K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 2013, 19, 17–35. [Google Scholar] [CrossRef]
- Wei, Y.; Li, L.; Wang, D.; Zhang, C.Y.; Zen, K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014, 289, 10270–10275. [Google Scholar] [CrossRef]
- Hwang, H.; Wentzel, E.; Mendell, J. A hexanucleotide element directs microRNA nuclear import. Science 2007, 315. [Google Scholar] [CrossRef]
- Jeffries, C.D.; Fried, H.M.; Perkins, D.O. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA 2011, 17, 675–686. [Google Scholar] [CrossRef]
- Turunen, T.A.; Roberts, T.C.; Laitinen, P.; Väänänen, M.-A.; Korhonen, P.; Malm, T.; Ylä-Herttuala, S.; Turunen, M.P. Changes in nuclear and cytoplasmic microRNA distribution in response to hypoxic stress. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Li, L.C. Small RNA-Guided Transcriptional Gene Activation (RNAa) in Mammalian Cells. Adv. Exp. Med. Biol. 2017, 983, 1–20. [Google Scholar] [CrossRef]
- Roberts, T.C. The MicroRNA Biology of the Mammalian Nucleus. Mol. Ther. Nucleic Acids 2014, 3, e188. [Google Scholar] [CrossRef] [PubMed]
- Benhamed, M.; Herbig, U.; Ye, T.; Dejean, A.; Bischof, O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012, 14, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yao, H.; Li, C.; Pu, M.; Yao, X.; Yang, H.; Qi, X.; Ren, J.; Wang, Y. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim. Biophys. Acta 2016, 1859, 650–662. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, M.; Zhang, X.; Huang, F.; Wu, K.; Zhang, J.; Liu, J.; Huang, Z.; Luo, H.; Tao, L.; et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA 2014, 20, 1878–1889. [Google Scholar] [CrossRef]
- Huang, V.; Place, R.F.; Portnoy, V.; Wang, J.; Qi, Z.; Jia, Z.; Yu, A.; Shuman, M.; Yu, J.; Li, L.C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012, 40, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Saetrom, P.; Snove, O., Jr.; Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef]
- Gonzalez, S.; Pisano, D.G.; Serrano, M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 2008, 7, 2601–2608. [Google Scholar] [CrossRef]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 2012, 119. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbø, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009, 10, 1–9. [Google Scholar] [CrossRef]
- Ramchandran, R.; Chaluvally-Raghavan, P. miRNA-Mediated RNA Activation in Mammalian Cells. Adv. Exp. Med. Biol. 2017, 983, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef]
- Tang, R.; Li, L.; Zhu, D.; Hou, D.; Cao, T.; Gu, H.; Zhang, J.; Chen, J.; Zhang, C.Y.; Zen, K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res. 2012, 22, 504–515. [Google Scholar] [CrossRef]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrom, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef]
- Liang, W.C.; Fu, W.M.; Wang, Y.B.; Sun, Y.X.; Xu, L.L.; Wong, C.W.; Chan, K.M.; Li, G.; Waye, M.M.; Zhang, J.F. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci. Rep. 2016, 6, 20121. [Google Scholar] [CrossRef]
- Fasanaro, P.; Greco, S.; Lorenzi, M.; Pescatori, M.; Brioschi, M.; Kulshreshtha, R.; Banfi, C.; Stubbs, A.; Calin, G.A.; Ivan, M.; et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 2009, 284, 35134–35143. [Google Scholar] [CrossRef]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Allo, M.; Buggiano, V.; Fededa, J.P.; Petrillo, E.; Schor, I.; de la Mata, M.; Agirre, E.; Plass, M.; Eyras, E.; Elela, S.A.; et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 717–724. [Google Scholar] [CrossRef]
- Ameyar-Zazoua, M.; Rachez, C.; Souidi, M.; Robin, P.; Fritsch, L.; Young, R.; Morozova, N.; Fenouil, R.; Descostes, N.; Andrau, J.C.; et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012, 19, 998–1004. [Google Scholar] [CrossRef]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, R.L.; Jiang, P.; Gilmore, B.L.; Spengler, R.M.; Tirabassi, R.; Nelson, J.A.; Ross, C.A.; Xing, Y.; Davidson, B.L. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 2014, 81, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, J.; Hicks, J.A.; Prakash, T.P.; Corey, D.R. Modulation of Splicing by Single-Stranded Silencing RNAs. Nucleic Acid Ther. 2015, 25, 113–120. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Kostrzewa, R.M.; Filip, M. A comprehensive view of the epigenetic landscape part I: DNA methylation, passive and active DNA demethylation pathways and histone variants. Neurotox. Res. 2015, 27, 84–97. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 2020, 301, 89–92. [Google Scholar] [CrossRef]
- Dai, E.; Yu, X.; Zhang, Y.; Meng, F.; Wang, S.; Liu, X.; Liu, D.; Wang, J.; Li, X.; Jiang, W. EpimiR: A database of curated mutual regulation between miRNAs and epigenetic modifications. Database 2014, 2014, bau023. [Google Scholar] [CrossRef]
- Malumbres, M. miRNAs and cancer: An epigenetics view. Mol. Asp. Med. 2013, 34, 863–874. [Google Scholar] [CrossRef]
- Memari, F.; Joneidi, Z.; Taheri, B.; Aval, S.F.; Roointan, A.; Zarghami, N. Epigenetics and Epi-miRNAs: Potential markers/therapeutics in leukemia. Biomed. Pharmacother. 2018, 106, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Corra, F.; Agnoletto, C.; Minotti, L.; Baldassari, F.; Volinia, S. The Network of Non-coding RNAs in Cancer Drug Resistance. Front. Oncol. 2018, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Carvalho, J.; van Grieken, N.C.; Pereira, P.M.; Sousa, S.; Tijssen, M.; Buffart, T.E.; Diosdado, B.; Grabsch, H.; Santos, M.A.; Meijer, G.; et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J. Pathol. 2012, 228, 31–44. [Google Scholar] [CrossRef]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef]
- Amodio, N.; Rossi, M.; Raimondi, L.; Pitari, M.R.; Botta, C.; Tagliaferri, P.; Tassone, P. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015, 6, 12837–12861. [Google Scholar] [CrossRef]
- Lombard, A.P.; Mooso, B.A.; Libertini, S.J.; Lim, R.M.; Nakagawa, R.M.; Vidallo, K.D.; Costanzo, N.C.; Ghosh, P.M.; Mudryj, M. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol. Carcinog. 2016, 55, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Bai, X.; Zhang, X.; Li, Z.; Tang, L.; Zhao, X.; Ren, Y.; Wei, S.; Wang, Q.; Liu, C.; et al. Quantitative nuclear proteomics identifies that miR-137-mediated EZH2 reduction regulates resveratrol-induced apoptosis of neuroblastoma cells. Mol. Cell Proteom. 2015, 14, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Sadeghizadeh, M.; Behmanesh, M.; Najafi, F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine 2015, 22, 961–967. [Google Scholar] [CrossRef]
- Jiang, F.; Li, Y.; Mu, J.; Hu, C.; Zhou, M.; Wang, X.; Si, L.; Ning, S.; Li, Z. Glabridin inhibits cancer stem cell-like properties of human breast cancer cells: An epigenetic regulation of miR-148a/SMAd2 signaling. Mol. Carcinog. 2016, 55, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Baumann, V.; Winkler, J. miRNA-based therapies: Strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014, 6, 1967–1984. [Google Scholar] [CrossRef]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in MicroRNA Delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K.; et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Fu, J.; Xiao, X.; Wu, J.; Wu, R.C. MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7 in breast cancer. Cancer Lett. 2014, 354, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, F.; Jiao, Y.; Feng, J.; Tang, W.; Yao, H.; Gong, C.; Chen, J.; Su, F.; Zhang, Y.; et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 2011, 17, 7105–7115. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Liu, P.; Yang, L.; Xie, X.; Ye, F.; Wu, M.; Liu, X.; Chen, B.; Zhang, L. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 3185–3197. [Google Scholar] [CrossRef]
- Friedman, J.M.; Liang, G.; Liu, C.C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Q.; Xu, B.; Wang, P.; Fan, W.; Cai, Y.; Gu, X.; Meng, F. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015, 282, 4376–4388. [Google Scholar] [CrossRef]
- Guo, Y.; Ying, L.; Tian, Y.; Yang, P.; Zhu, Y.; Wang, Z.; Qiu, F.; Lin, J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013, 280, 4531–4538. [Google Scholar] [CrossRef]
- Matsushita, R.; Yoshino, H.; Enokida, H.; Goto, Y.; Miyamoto, K.; Yonemori, M.; Inoguchi, S.; Nakagawa, M.; Seki, N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget 2016, 7, 28460–28487. [Google Scholar] [CrossRef]
- Zhu, M.; Xu, Y.; Ge, M.; Gui, Z.; Yan, F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015, 106, 833–839. [Google Scholar] [CrossRef]
- Ng, E.K.; Tsang, W.P.; Ng, S.S.; Jin, H.C.; Yu, J.; Li, J.J.; Rocken, C.; Ebert, M.P.; Kwok, T.T.; Sung, J.J. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer 2009, 101, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, J.; Meng, X.; Ying, X.; Zuo, Y.; Liu, R.; Pan, Z.; Kang, T.; Huang, W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis 2011, 32, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Konno, Y.; Dong, P.; Xiong, Y.; Suzuki, F.; Lu, J.; Cai, M.; Watari, H.; Mitamura, T.; Hosaka, M.; Hanley, S.J.; et al. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget 2014, 5, 6049–6062. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jiang, X.; Li, H.; Guo, L.; Jiang, W.; Lu, S.H. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014, 23, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, L.; Gong, P.; Zhao, C.; Zhang, S.; Zhang, K.; Zhou, R.; Zhao, Z.; Fan, H. Deregulation between miR-29b/c and DNMT3A is associated with epigenetic silencing of the CDH1 gene, affecting cell migration and invasion in gastric cancer. PLoS ONE 2015, 10, e0123926. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, X.; Han, Y.; Lu, Y.; Shang, Y.; Liu, C.; Li, T.; Jin, Z.; Fan, D.; Wu, K. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J. 2013, 27, 4929–4939. [Google Scholar] [CrossRef]
- Yan, J.; Guo, X.; Xia, J.; Shan, T.; Gu, C.; Liang, Z.; Zhao, W.; Jin, S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med. Oncol. 2014, 31, 879. [Google Scholar] [CrossRef]
- Ren, J.; Huang, H.J.; Gong, Y.; Yue, S.; Tang, L.M.; Cheng, S.Y. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014, 4, 26. [Google Scholar] [CrossRef]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef]
- Peruzzi, P.; Bronisz, A.; Nowicki, M.O.; Wang, Y.; Ogawa, D.; Price, R.; Nakano, I.; Kwon, C.H.; Hayes, J.; Lawler, S.E.; et al. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro Oncol. 2013, 15, 1212–1224. [Google Scholar] [CrossRef]
- Nohata, N.; Hanazawa, T.; Kinoshita, T.; Inamine, A.; Kikkawa, N.; Itesako, T.; Yoshino, H.; Enokida, H.; Nakagawa, M.; Okamoto, Y.; et al. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br. J. Cancer 2013, 108, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Huang, N.; Patel, T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010, 51, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Yang, T.; Liu, Y.; Li, A.; Fu, S.; Wu, M.; Pan, Z.; Zhou, W. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br. J. Cancer 2010, 103, 1215–1220. [Google Scholar] [CrossRef]
- Noh, J.H.; Chang, Y.G.; Kim, M.G.; Jung, K.H.; Kim, J.K.; Bae, H.J.; Eun, J.W.; Shen, Q.; Kim, S.J.; Kwon, S.H.; et al. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013, 335, 455–462. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Chen, B.F.; Lu, Z.; Huo, X.S.; Zhou, W.P.; Wang, F.; Sun, S.H. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011, 54, 2025–2035. [Google Scholar] [CrossRef]
- Garzon, R.; Heaphy, C.E.; Havelange, V.; Fabbri, M.; Volinia, S.; Tsao, T.; Zanesi, N.; Kornblau, S.M.; Marcucci, G.; Calin, G.A.; et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009, 114, 5331–5341. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, L.; Luo, X.; Wang, L.; Gao, X.; Wang, W.; Sun, J.; Dou, L.; Li, J.; Xu, C.; et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood 2013, 121, 499–509. [Google Scholar] [CrossRef]
- Nishioka, C.; Ikezoe, T.; Yang, J.; Nobumoto, A.; Tsuda, M.; Yokoyama, A. Downregulation of miR-217 correlates with resistance of Ph(+) leukemia cells to ABL tyrosine kinase inhibitors. Cancer Sci. 2014, 105, 297–307. [Google Scholar] [CrossRef]
- Shen, J.Z.; Zhang, Y.Y.; Fu, H.Y.; Wu, D.S.; Zhou, H.R. Overexpression of microRNA-143 inhibits growth and induces apoptosis in human leukemia cells. Oncol. Rep. 2014, 31, 2035–2042. [Google Scholar] [CrossRef]
- Yu, T.; Li, J.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Sun, L.; Zhang, Y.; Cui, Y.; Zhang, F.; et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 2015, 34, 413–423. [Google Scholar] [CrossRef]
- Sander, S.; Bullinger, L.; Klapproth, K.; Fiedler, K.; Kestler, H.A.; Barth, T.F.; Moller, P.; Stilgenbauer, S.; Pollack, J.R.; Wirth, T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008, 112, 4202–4212. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Tetzlaff, M.T.; Cui, R.; Xu, X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am. J. Pathol. 2012, 181, 1823–1835. [Google Scholar] [CrossRef] [PubMed]
- Neo, W.H.; Yap, K.; Lee, S.H.; Looi, L.S.; Khandelia, P.; Neo, S.X.; Makeyev, E.V.; Su, I.H. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J. Biol. Chem. 2014, 289, 20788–20801. [Google Scholar] [CrossRef] [PubMed]
- Althoff, K.; Beckers, A.; Odersky, A.; Mestdagh, P.; Koster, J.; Bray, I.M.; Bryan, K.; Vandesompele, J.; Speleman, F.; Stallings, R.L.; et al. MiR-137 functions as a tumor suppressor in neuroblastoma by downregulating KDM1A. Int. J. Cancer 2013, 133, 1064–1073. [Google Scholar] [CrossRef]
- Das, S.; Foley, N.; Bryan, K.; Watters, K.M.; Bray, I.; Murphy, D.M.; Buckley, P.G.; Stallings, R.L. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer Res. 2010, 70, 7874–7881. [Google Scholar] [CrossRef]
- Zhang, D.; Ni, Z.; Xu, X.; Xiao, J. MiR-32 functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Med. Sci. Monit. 2014, 20, 2527–2535. [Google Scholar] [CrossRef][Green Version]
- Bhattacharya, R.; Nicoloso, M.; Arvizo, R.; Wang, E.; Cortez, A.; Rossi, S.; Calin, G.A.; Mukherjee, P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009, 69, 9090–9095. [Google Scholar] [CrossRef]
- Xiang, Y.; Ma, N.; Wang, D.; Zhang, Y.; Zhou, J.; Wu, G.; Zhao, R.; Huang, H.; Wang, X.; Qiao, Y.; et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: A novel epigenetic therapy independent of decitabine. Oncogene 2014, 33, 378–386. [Google Scholar] [CrossRef]
- Azizi, M.; Teimoori-Toolabi, L.; Arzanani, M.K.; Azadmanesh, K.; Fard-Esfahani, P.; Zeinali, S. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol. Ther. 2014, 15, 419–427. [Google Scholar] [CrossRef]
- Xue, G.; Ren, Z.; Chen, Y.; Zhu, J.; Du, Y.; Pan, D.; Li, X.; Hu, B. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015, 361, 121–127. [Google Scholar] [CrossRef]
- Noonan, E.J.; Place, R.F.; Pookot, D.; Basak, S.; Whitson, J.M.; Hirata, H.; Giardina, C.; Dahiya, R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009, 28, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kurozumi, A.; Nohata, N.; Kojima, S.; Matsushita, R.; Yoshino, H.; Yamazaki, K.; Ishida, Y.; Ichikawa, T.; Naya, Y.; et al. The microRNA signature of patients with sunitinib failure: Regulation of UHRF1 pathways by microRNA-101 in renal cell carcinoma. Oncotarget 2016, 7, 59070–59086. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef]
- Chen, B.F.; Suen, Y.K.; Gu, S.; Li, L.; Chan, W.Y. A miR-199a/miR-214 self-regulatory network via PSMD10, TP53 and DNMT1 in testicular germ cell tumor. Sci. Rep. 2014, 4, 6413. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Sacco, A.; Jia, X.; Azab, A.K.; Maiso, P.; Ngo, H.T.; Azab, F.; Runnels, J.; Quang, P.; Ghobrial, I.M. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood 2010, 116, 1506–1514. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadakierska-Chudy, A. MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules 2020, 10, 1285. https://doi.org/10.3390/biom10091285
Sadakierska-Chudy A. MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules. 2020; 10(9):1285. https://doi.org/10.3390/biom10091285
Chicago/Turabian StyleSadakierska-Chudy, Anna. 2020. "MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics" Biomolecules 10, no. 9: 1285. https://doi.org/10.3390/biom10091285
APA StyleSadakierska-Chudy, A. (2020). MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules, 10(9), 1285. https://doi.org/10.3390/biom10091285





