Biological Evaluation of Oxindole Derivative as a Novel Anticancer Agent against Human Kidney Carcinoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
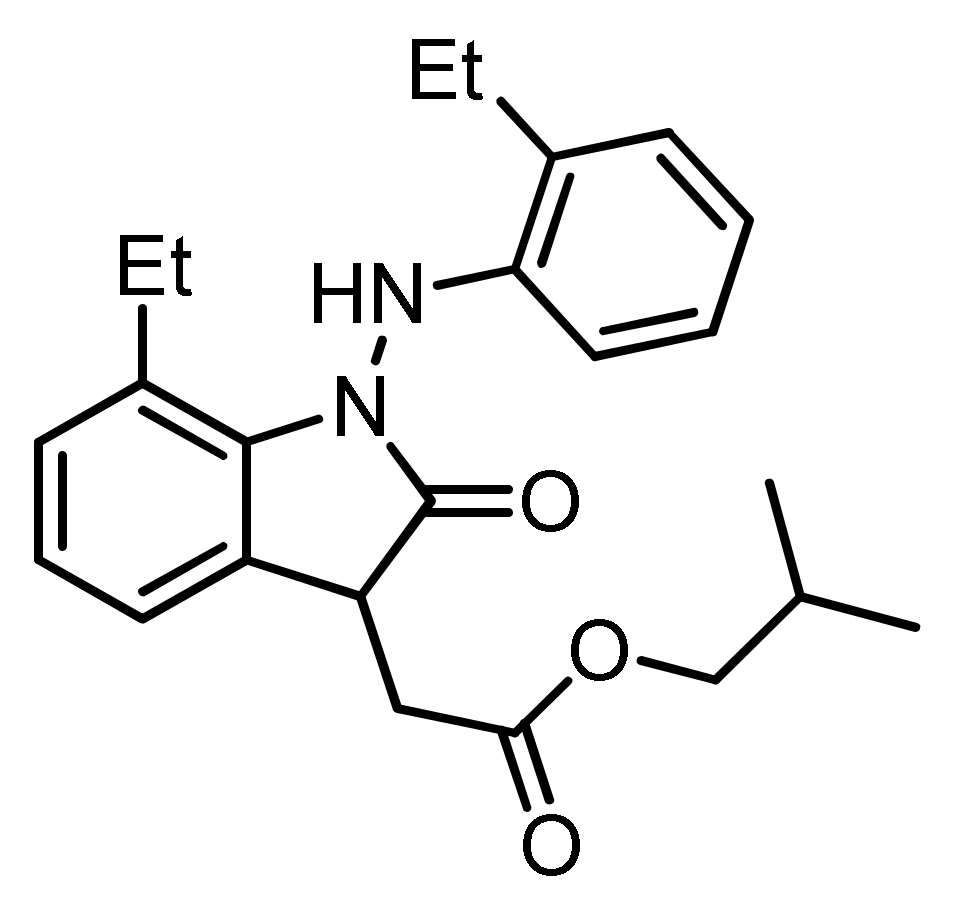
2.2. Preparation of the Oxindole Derivative (SH-859)
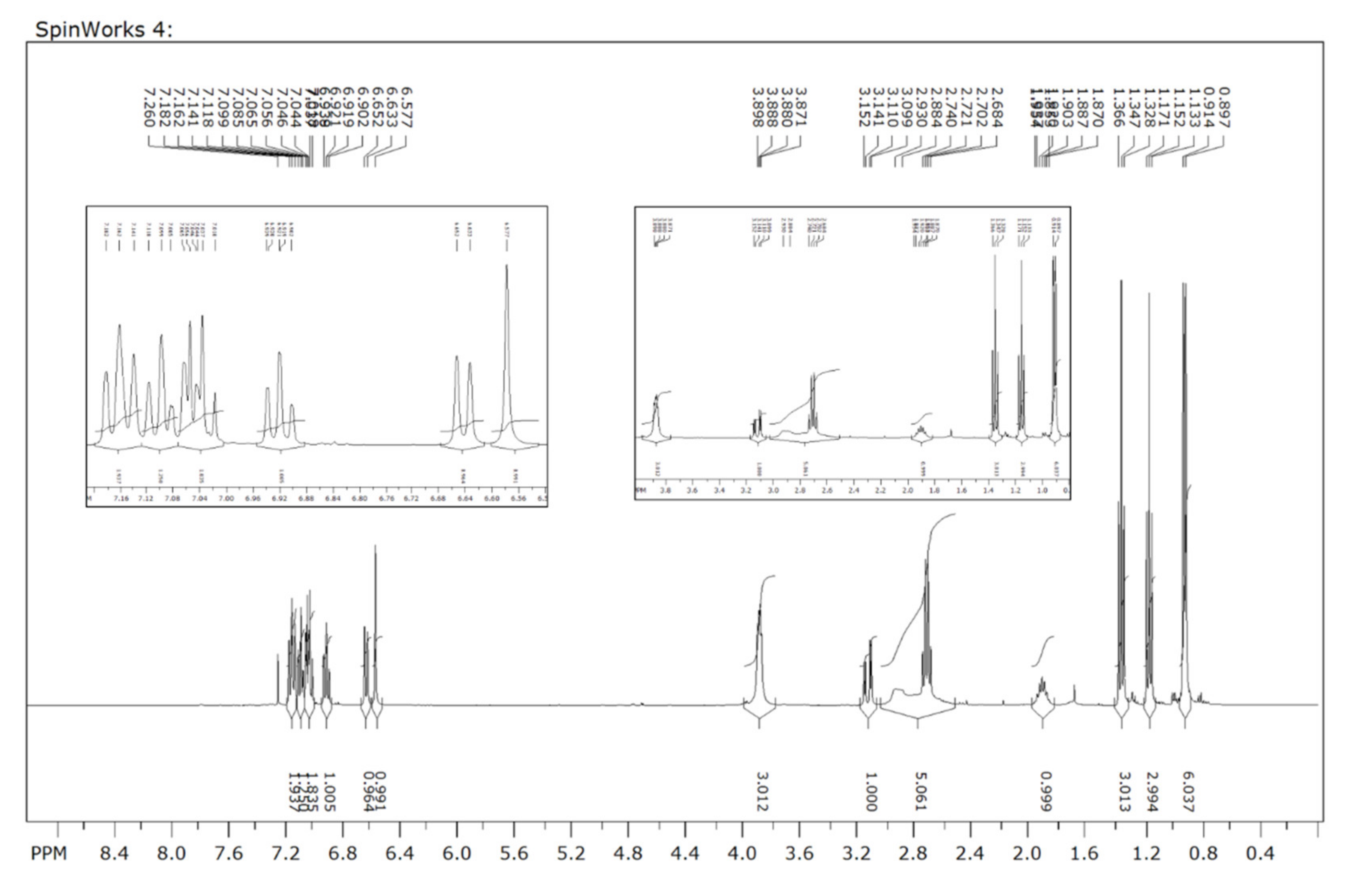
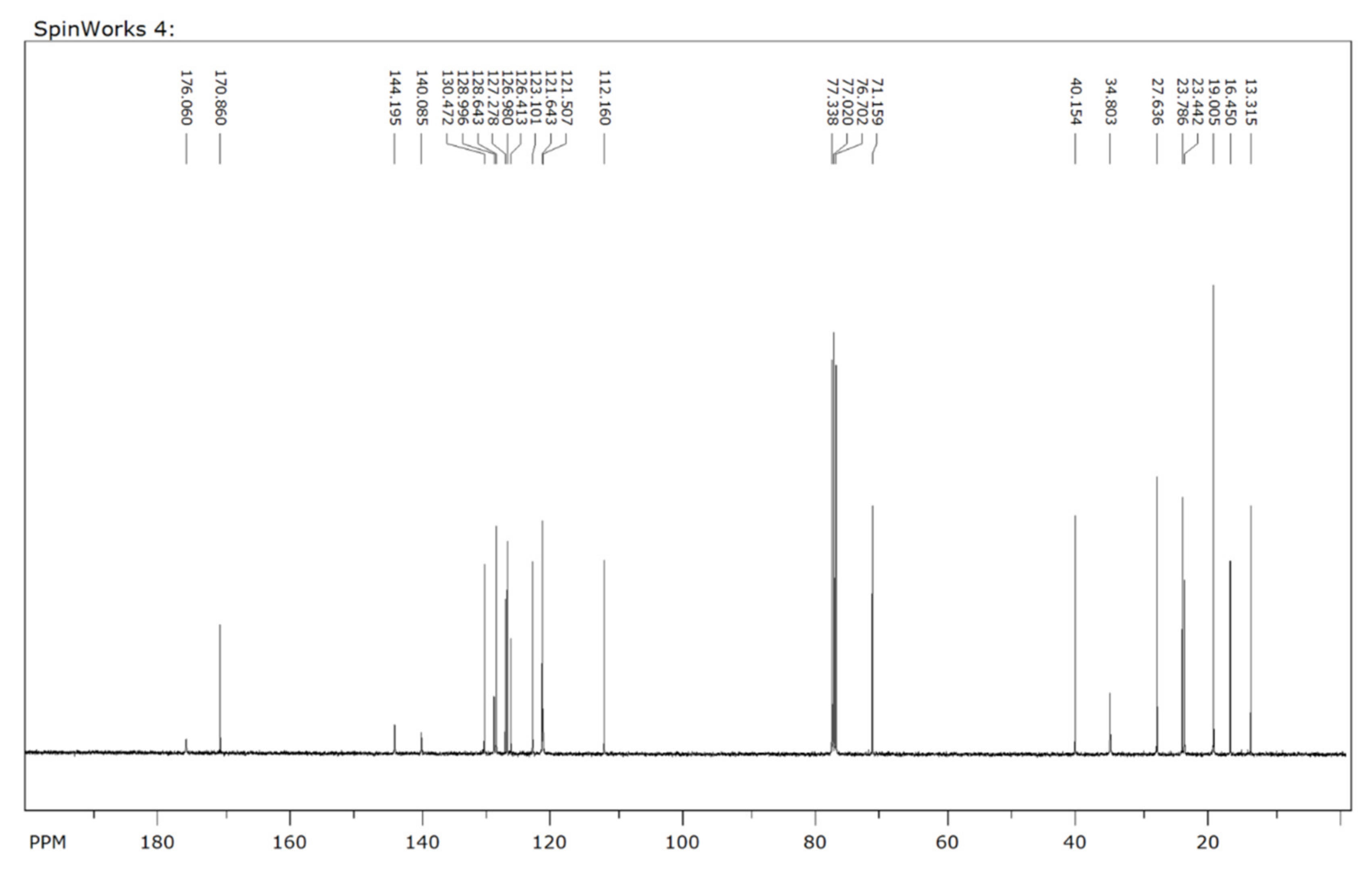
2.3. Characterization Data for SH-859
2.4. Cell Lines and Cell Culture
2.5. Cytotoxicity Assay
2.6. Analysis of Cell Cycle Progression
2.7. Annexin V/PI Double Staining
2.8. Western Blot Analysis
2.9. Acridine Orange Staining
2.10. MDC Staining
2.11. Immunofluorescence Analysis
2.12. Pyruvate Kinase Activity Assay
2.13. Analysis of Glycolysis, Glycolytic Intermediates, and Mitochondrial Activity
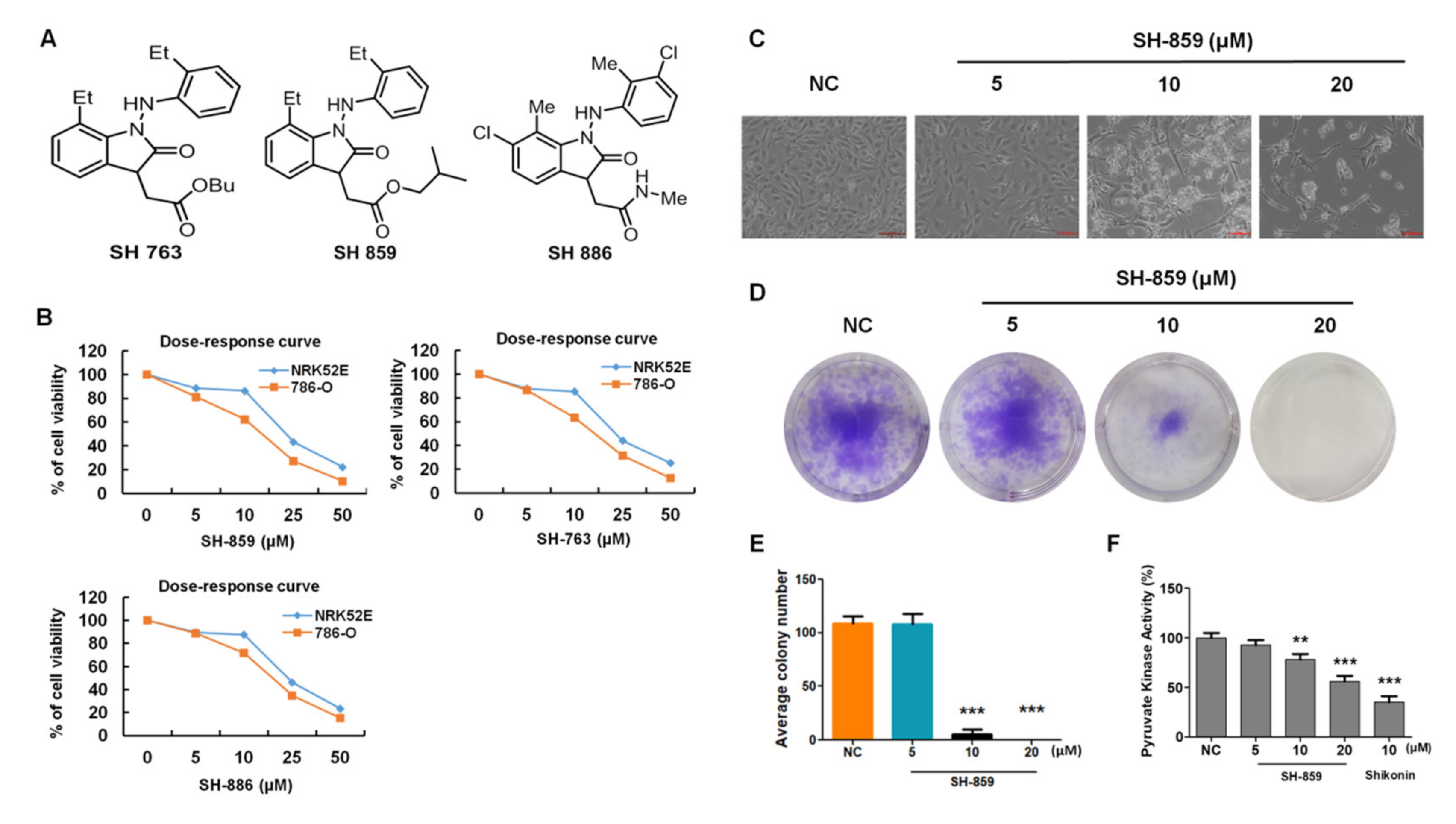
2.14. Colony Formation Assay
2.15. Tumor Xenograft Study
2.16. Statistical Analysis
3. Results
3.1. SH-859 Prevented 786-O Cell Progression
3.2. Analysis of Cell Cycle Progression
3.3. Induction of Cellular Apoptosis by SH-859
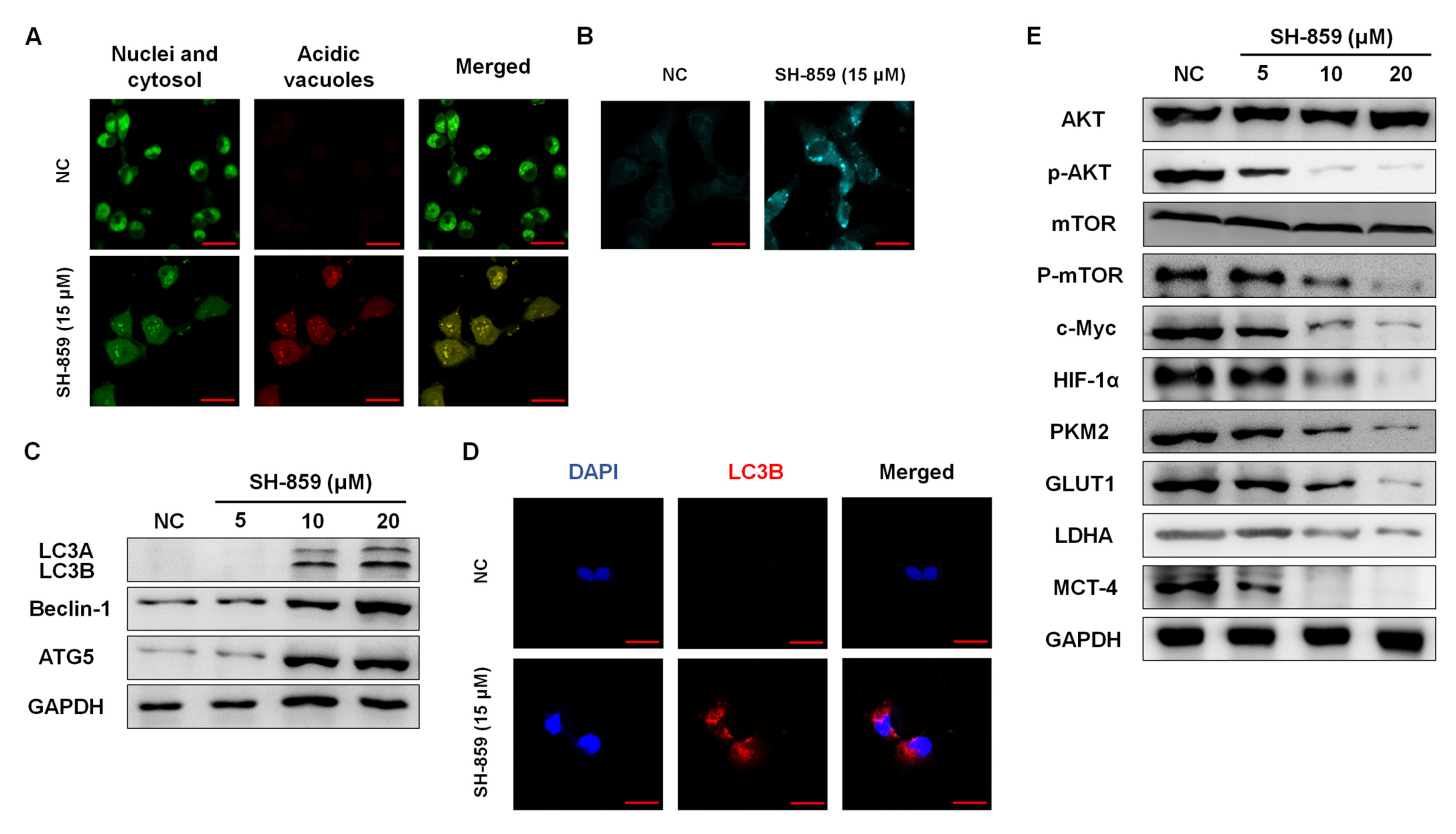
3.4. SH-859 Induced Autophagy
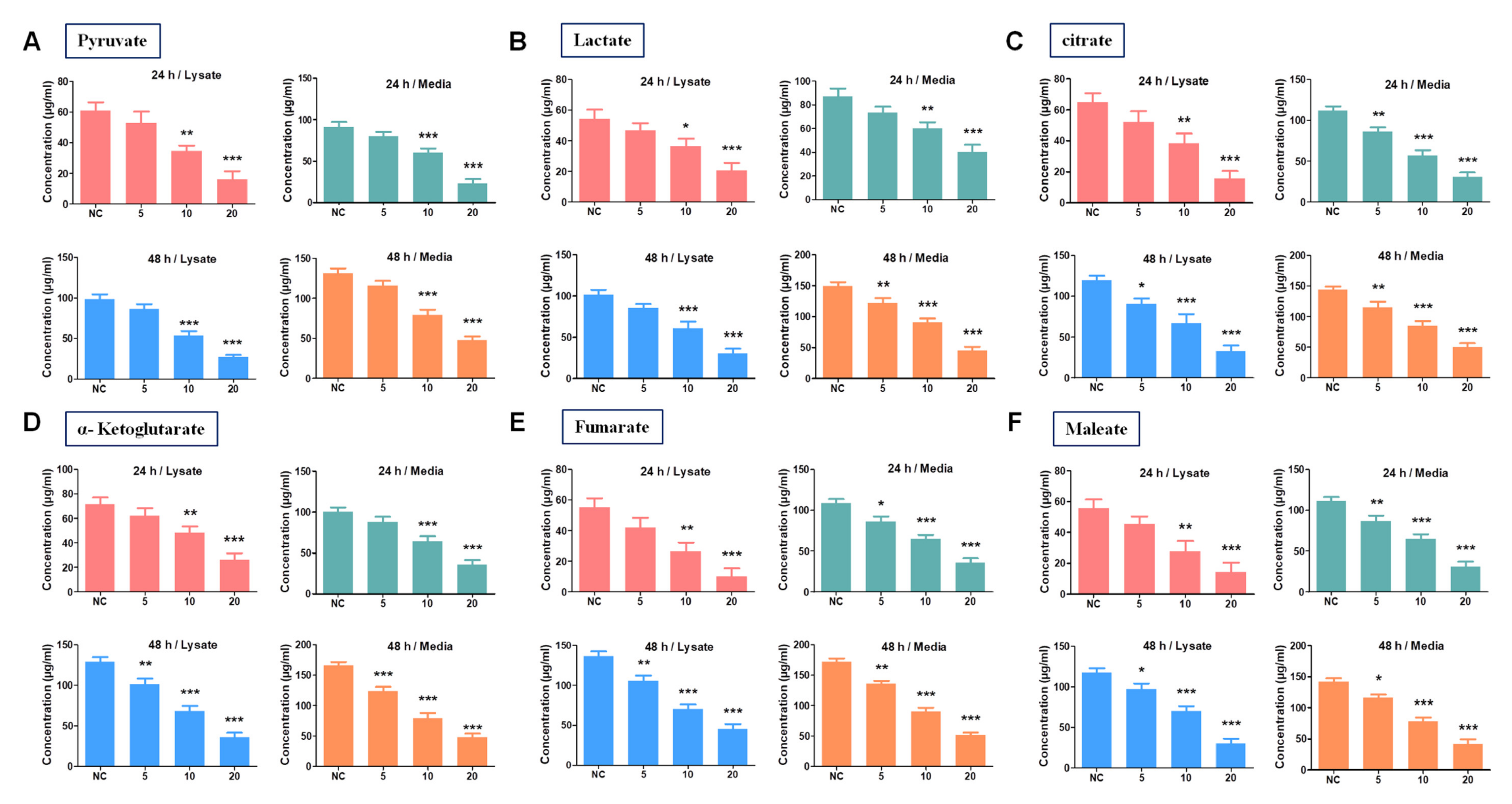
3.5. SH-859 Attenuated Glycolysis, Cellular Metabolite Level, and Mitochondrial Activity
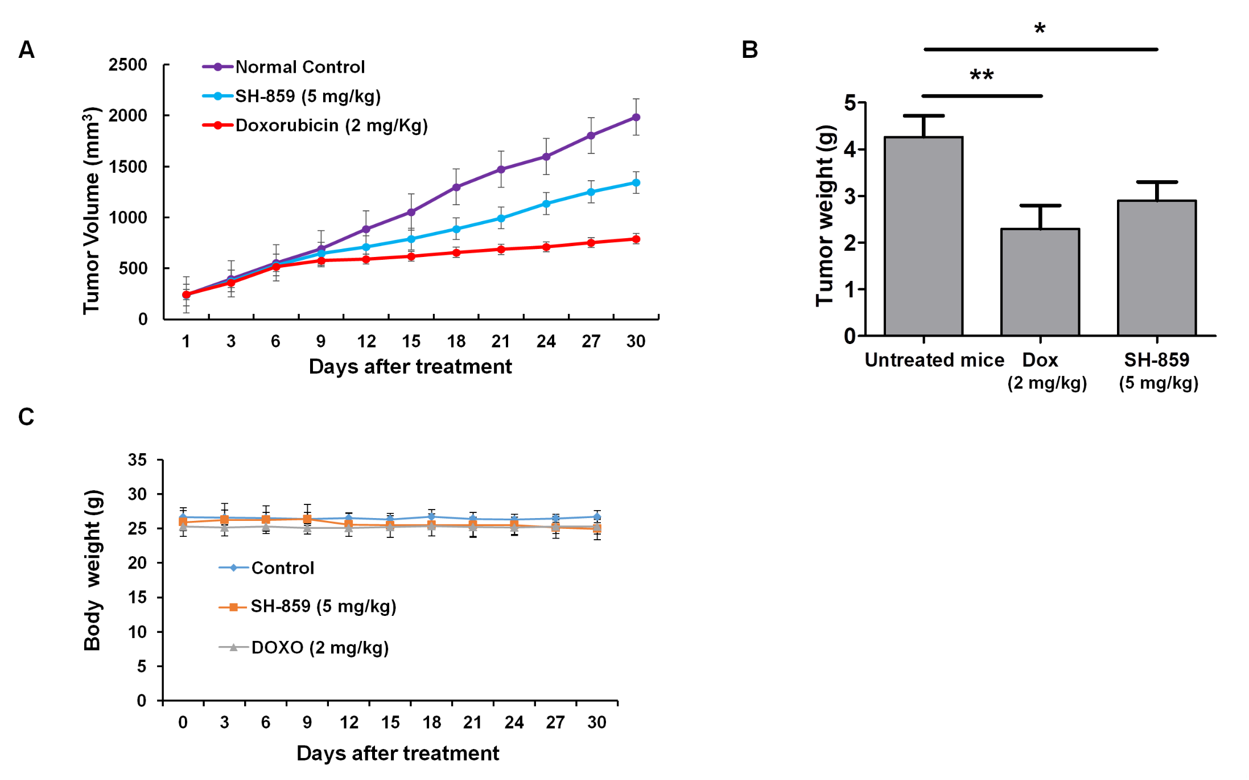
3.6. SH-859 Inhibited 786-O Cell Growth in a Xenograft Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, L.; Zhang, S.; MacLennan, G.T.; Lopez-Beltran, A.; Montironi, R. Molecular and cytogenetic insights into the pathogenesis, classification, differential diagnosis, and prognosis of renal epithelial neoplasms. Hum. Pathol. 2009, 40, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wu, Y.; Zhang, J.; Fang, Z.; Liu, Z.; Xu, Z.; Fan, Y. Nomograms for predicting long-term overall survival and disease-specific survival of patients with clear cell renal cell carcinoma. OncoTargets Ther. 2018, 11, 5535. [Google Scholar] [CrossRef] [PubMed]
- Xing, T.; He, H. Epigenomics of clear cell renal cell carcinoma: Mechanisms and potential use in molecular pathology. Chin. J. Cancer Res. 2016, 28, 80. [Google Scholar]
- Atkins, M.B.; Tannir, N.M. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat. Rev. 2018, 70, 127–137. [Google Scholar] [CrossRef]
- Cao, Q.; Ruan, H.; Wang, K.; Song, Z.; Bao, L.; Xu, T.; Xiao, H.; Wang, C.; Cheng, G.; Tong, J. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int. J. Oncol. 2018, 53, 137–147. [Google Scholar] [CrossRef]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.G.; Kolenko, V.M. Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar] [CrossRef]
- Son, J.Y.; Yoon, S.; Tae, I.H.; Park, Y.J.; De, U.; Jeon, Y.; Park, Y.J.; Rhyu, I.J.; Lee, B.M.; Chung, K.H.; et al. Novel therapeutic roles of MC—4 in combination with everolimus against advanced renal cell carcinoma by dual targeting of Akt/pyruvate kinase muscle isozyme M2 and mechanistic target of rapamycin complex 1 pathways. Cancer Med. 2018, 7, 5083–5095. [Google Scholar] [CrossRef]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Ferri, K.F.; Kroemer, G. Organelle-specific initiation of cell death pathways. Nat. Cell Biol. 2001, 3, E255. [Google Scholar] [CrossRef]
- Su, M.; Mei, Y.; Sinha, S. Role of the crosstalk between autophagy and apoptosis in cancer. J. Oncol. 2013, 2013, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Garber, K. Energy deregulation: Licensing tumors to grow. Science 2006, 312, 1158–1159. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Singh, M.; Chadha, N.; Silakari, O. Oxindole: A chemical prism carrying plethora of therapeutic benefits. Eur. J. Med. Chem. 2016, 10, 858–894. [Google Scholar] [CrossRef]
- Saraswat, P.; Jeyabalan, G.; Hassan, M.Z.; Rahman, M.U.; Nyola, N.K. Review of synthesis and various biological activities of spiro heterocyclic compounds comprising oxindole and pyrrolidine moieties. Synth. Commun. 2016, 46, 1643–1664. [Google Scholar] [CrossRef]
- Romagnoli, R.; Baraldi, P.G.; Prencipe, F.; Oliva, P.; Baraldi, S.; Salvador, M.K.; Lopez-Cara, L.C.; Bortolozzi, R.; Mattiuzzo, E.; Basso, G.; et al. Design, synthesis and biological evaluation of 3-substituted-2-oxindole hybrid derivatives as novel anticancer agents. Eur. J. Med. Chem. 2017, 7, 258–270. [Google Scholar] [CrossRef]
- Nesi, G.; Sestito, S.; Mey, V.; Ricciardi, S.; Falasca, M.; Danesi, R.; Lapucci, A.; Breschi, M.C.; Fogli, S.; Rapposelli, S. Synthesis of novel 3,5-disubstituted-2-oxindole derivatives as antitumor agents against human non small cell lung cancer. ACS Med. Chem. Lett. 2013, 4, 1137–1141. [Google Scholar] [CrossRef]
- Bort, A.; Quesada, S.; Ramos-Torres, Á.; Gargantilla, M.; Priego, E.M.; Raynal, S.; Lepifre, F.; Gasalla, J.M.; Rodriguez-Henche, N.; Castro, A.; et al. Identification of a novel 2-oxindole fluorinated derivative as in vivo antitumor agent for prostate cancer acting via AMPK activation. Sci. Rep. 2018, 12, 4370. [Google Scholar] [CrossRef]
- Hong, L.; Wang, R. Recent advances in asymmetric organocatalytic construction of 3,3′—Spirocyclic oxindoles. Adv. Synth. Catal. 2013, 355, 1023–1052. [Google Scholar] [CrossRef]
- Cao, Z.Y.; Eng, X.P.; Zhou, J. Catalytic asymmetric synthesis of 3,3-disubstituted oxindoles: Diazooxindole joins the field. Tetrahedron Lett. 2014, 55, 2571–2584. [Google Scholar] [CrossRef]
- Han, S.H.; Mishra, N.K.; Jo, H.; Oh, Y.; Jeon, M.; Kim, S.; Kim, W.J.; Lee, J.S.; Kim, H.S.; Kim, I.S. One-pot Synthesis of Oxindoles through C−H Alkylation and Intramolecular Cyclization of Azobenzenes with Internal Olefins. Adv. Synth. Catal. 2017, 359, 2396–2401. [Google Scholar] [CrossRef]
- Jeon, M.; Park, J.; Dey, P.; Oh, Y.; Oh, H.; Han, S.; Um, S.H.; Kim, H.S.; Mishra, N.K.; Kim, I.S. Site-selective rhodium (III)-catalyzed C−H amination of 7-azaindoles with anthranils: Synthesis and anticancer evaluation. Adv. Synth. Catal. 2017, 359, 3471–3478. [Google Scholar] [CrossRef]
- Dey, P.; Son, J.Y.; Kundu, A.; Kim, K.S.; Lee, Y.; Yoon, K.; Yoon, S.; Lee, B.M.; Nam, K.T.; Kim, H.S.; et al. Knockdown of pyruvate kinase M2 inhibits cell proliferation, metabolism, and migration in Renal CELL carcinoma. Int. J. Mol. Sci. 2019, 20, 5622. [Google Scholar] [CrossRef]
- Tae, I.H.; Park, E.Y.; Dey, P.; Son, J.Y.; Lee, S.-Y.; Jung, J.H.; Saloni, S.; Kim, M.-H.; Kim, H.S. Novel SIRT1 inhibitor 15-deoxy-Δ12, 14-prostaglandin J2 and its derivatives exhibit anticancer activity through apoptotic or autophagic cell death pathways in SKOV3 cells. Int. J. Oncol. 2018, 53, 2518–2530. [Google Scholar]
- Iansante, V.; Choy, P.M.; Fung, S.W.; Liu, Y.; Chai, J.-G.; Dyson, J.; Del Rio, A.; D’Santos, C.; Williams, R.; Chokshi, S.; et al. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat. Commun. 2015, 6, 7882. [Google Scholar] [CrossRef]
- Nair, H.K.; Rao, K.V.; Aalinkeel, R.; Mahajan, S.; Chawda, R.; Schwartz, S.A. Inhibition of prostate cancer cell colony formation by the flavonoid quercetin correlates with modulation of specific regulatory genes. Clin. Diagn. Lab. Immunol. 2004, 11, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; Van Bree, C. Clonogenic assay of cells in vitro. Nat. Prot. 2006, 1, 2315. [Google Scholar] [CrossRef]
- Mascetti, G.; Carrara, S.; Vergani, L. Relationship between chromatin compactness and dye uptake for in situ chromatin stained with DAPI. Cytometry 2001, 44, 113–119. [Google Scholar] [CrossRef]
- Omran, M.A.A.; Fabb, S.A.; Dickson, G. Biochemical and morphological analysis of cell death induced by Egyptian cobra (Naja haje) venom on cultured cells. J. Venom. Anim. Toxins Incl. Trop. Dis. 2004, 10, 219–241. [Google Scholar] [CrossRef]
- Su, X.; Wang, P.; Wang, X.; Cao, B.; Li, L.; Liu, Q. Apoptosis of U937 cells induced by hematoporphyrin monomethyl ether-mediated sonodynamic action. Cancer Bioth. Radiopharm. 2013, 28, 207–217. [Google Scholar] [CrossRef]
- Mandelkow, R.; Guembel, D.; Ahrend, H.; Kaul, A.; Zimmermann, U.; Burchardt, M.; Stope, M.B. Detection and quantification of nuclear morphology changes in apoptotic cells by fluorescence microscopy and subsequent analysis of visualized fluorescent signals. Anticancer Res. 2017, 37, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- He, C.-L.; Bian, Y.-Y.; Xue, Y.; Liu, Z.-X.; Zhou, K.-Q.; Yao, C.-F.; Lin, Y.; Zou, H.-F.; Luo, F.-X.; Qu, Y.-Y.; et al. Pyruvate kinase M2 activates mTORC1 by phosphorylating AKT1S1. Sci. Rep. 2016, 6, 21524. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Jiang, J.; Ji, J.; Cai, Q.; Chen, X.; Yu, Y.; Zhu, Z.; Zhang, J. PKM2 promotes cell migration and inhibits autophagy by mediating PI3K/AKT activation and contributes to the malignant development of gastric cancer. Scie. Rep. 2017, 7, 2886. [Google Scholar] [CrossRef]
- Chu, B.; Wang, J.; Wang, Y.; Yang, G. Knockdown of PKM2 induces apoptosis and autophagy in human A549 alveolar adenocarcinoma cells. Mol. Med. Rep. 2015, 12, 4358–4363. [Google Scholar] [CrossRef]
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of renal cell carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef]
- Dutcher, J.P. Recent developments in the treatment of renal cell carcinoma. Ther. Adv. Urol. 2013, 5, 338–353. [Google Scholar] [CrossRef]
- Kanesvaran, R.; Tan, M.-H. Targeted therapy for renal cell carcinoma: The next lap. J. Carcinog. 2014, 13, 3. [Google Scholar]
- Ghatalia, P.; Koenigsberg, R.; Pisarcik, D.; Handorf, E.A.; Geynisman, D.M.; Zibelman, M. The evolution of clinical trials in renal cell carcinoma: A status report for 2013–2016 from the ClinicalTrials.gov website. Kidney Cancer 2017, 1, 151–159. [Google Scholar] [CrossRef]
- Sathianathen, N.J.; Krishna, S.; Anderson, J.K.; Weight, C.J.; Gupta, S.; Konety, B.R.; Griffith, T.S. The current status of immunobased therapies for metastatic renal-cell carcinoma. ImmunoTargets Ther. 2017, 6, 83. [Google Scholar] [CrossRef]
- Jeong, T.; Lee, S.H.; Mishra, N.K.; De, U.; Park, J.; Dey, P.; Kwak, J.H.; Jung, Y.H.; Kim, H.S.; Kim, I.S. Synthesis and cytotoxic evaluation of N-aroylureas through rhodium (III)-Catalyzed C−H functionalization of indolines with isocyanates. Adv. Synth. Catal. 2017, 359, 2329–2336. [Google Scholar] [CrossRef]
- Jeong, T.; Mishra, N.K.; Dey, P.; Oh, H.; Han, S.; Lee, S.H.; Kim, H.S.; Park, J.; Kim, I.S. C (sp 3)–H amination of 8-methylquinolines with azodicarboxylates under Rh (iii) catalysis: Cytotoxic evaluation of quinolin-8-ylmethanamines. Chem. Commun. 2017, 53, 11197–11200. [Google Scholar] [CrossRef] [PubMed]
- Richa, S.; Dey, P.; Park, C.; Yang, J.; Son, J.Y.; Park, J.H.; Lee, S.H.; Ahn, M.-Y.; Kim, I.S.; Moon, H.R.; et al. A new histone deacetylase inhibitor, MHY4381, induces apoptosis via generation of reactive oxygen species in human prostate cancer cells. Biomol. Ther. 2019, 28, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, K.; Jeon, Y.U.; Kundu, A.; Dey, P.; Hwang, J.Y.; Mishra, N.K.; Kim, H.S.; Kim, I.S. Lewis acid-mediated cross-coupling reaction of 7-azaindoles and aldehydes: Cytotoxic evaluation of C3-linked bis-7-azaindoles. Tetrahedron Lett. 2019, 60, 150974. [Google Scholar] [CrossRef]
- Otto, T.; Sicinski, P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 2017, 17, 93. [Google Scholar] [CrossRef] [PubMed]
- Meeran, S.M.; Katiyar, S.K. Cell cycle control as a basis for cancer chemoprevention through dietary agents. Front. Biosci. 2008, 13, 2191. [Google Scholar] [CrossRef]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316. [Google Scholar] [CrossRef] [PubMed]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 2008, 10, 10. [Google Scholar] [CrossRef]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef]
- Onen-Bayram, F.E.; Durmaz, I.; Scherman, D.; Herscovici, J.; Cetin-Atalay, R. A novel thiazolidine compound induces caspase-9 dependent apoptosis in cancer cells. Bioorgan. Med. Chem. 2012, 20, 5094–5102. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Li, X.; Zhang, J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef] [PubMed]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/mTOR signaling in cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular survival: A play in three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef]
- Facompre, N.D.; Sinha, I.; El-Bayoumy, K.; Pinto, J.T.; Sinha, R. Remarkable inhibition of mTOR signaling by the combination of rapamycin and 1, 4-phenylenebis (methylene) selenocyanate in human prostate cancer cells. Int. J. Cancer 2012, 131, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Robb, V.A.; Karbowniczek, M.; Klein-Szanto, A.J.; Henske, E.P. Activation of the mTOR signaling pathway in renal clear cell carcinoma. J. Urol. 2007, 177, 346–352. [Google Scholar] [CrossRef]
- He, D.; Sun, X.; Yang, H.; Li, X.; Yang, D. TOFA induces cell cycle arrest and apoptosis in ACHN and 786-O cells through inhibiting PI3K/Akt/mTOR pathway. J. Cancer 2018, 9, 2734. [Google Scholar] [CrossRef]
- Guo, H.; German, P.; Bai, S.; Barnes, S.; Guo, W.; Qi, X.; Lou, H.; Liang, J.; Jonasch, E.; Mills, G.B.; et al. The PI3K/AKT pathway and renal cell carcinoma. J. Genet. Genom. 2015, 42, 343–353. [Google Scholar] [CrossRef]
- Xiang, C.; Cui, S.-p.; Ke, Y. MiR-144 inhibits cell proliferation of renal cell carcinoma by targeting MTOR. J. Huazhong Univ. Sci. Technol. Med. Sci. 2016, 36, 186–192. [Google Scholar] [CrossRef]
- Yousefi, S.; Perozzo, R.; Schmid, I.; Ziemiecki, A.; Schaffner, T.; Scapozza, L.; Brunner, T.; Simon, H.U. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006, 8, 1124–1132. [Google Scholar] [CrossRef]
- Djavaheri-Mergny, M.; Maiuri, M.C.; Kroemer, G. Crosstalk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010, 29, 1717–1719. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.H.; Jo, Y.K.; Hwang, J.J.; Lee, Y.M.; Roh, S.A.; Kim, J.C. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009, 274, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Lerner, A.; Bialik, S.; Simon, H.U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Moretti, L.; Cha, Y.I.; Niermann, K.J.; Lu, B. Switch between apoptosis and autophagy: Radiation-induced endoplasmic reticulum stress? Cell Cycle 2007, 6, 793–798. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, P.; Kundu, A.; Han, S.H.; Kim, K.-S.; Park, J.H.; Yoon, S.; Kim, I.S.; Kim, H.S. Biological Evaluation of Oxindole Derivative as a Novel Anticancer Agent against Human Kidney Carcinoma Cells. Biomolecules 2020, 10, 1260. https://doi.org/10.3390/biom10091260
Dey P, Kundu A, Han SH, Kim K-S, Park JH, Yoon S, Kim IS, Kim HS. Biological Evaluation of Oxindole Derivative as a Novel Anticancer Agent against Human Kidney Carcinoma Cells. Biomolecules. 2020; 10(9):1260. https://doi.org/10.3390/biom10091260
Chicago/Turabian StyleDey, Prasanta, Amit Kundu, Sang Hoon Han, Kyeong-Seok Kim, Jae Hyeon Park, Sungpil Yoon, In Su Kim, and Hyung Sik Kim. 2020. "Biological Evaluation of Oxindole Derivative as a Novel Anticancer Agent against Human Kidney Carcinoma Cells" Biomolecules 10, no. 9: 1260. https://doi.org/10.3390/biom10091260
APA StyleDey, P., Kundu, A., Han, S. H., Kim, K.-S., Park, J. H., Yoon, S., Kim, I. S., & Kim, H. S. (2020). Biological Evaluation of Oxindole Derivative as a Novel Anticancer Agent against Human Kidney Carcinoma Cells. Biomolecules, 10(9), 1260. https://doi.org/10.3390/biom10091260






