Structural Characterization of an S-enantioselective Imine Reductase from Mycobacterium Smegmatis
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression of S-IRED-Ms
2.2. Protein Purification
2.3. Protein Crystallization
2.4. Data Collection, Processing and Structure Determination
3. Results
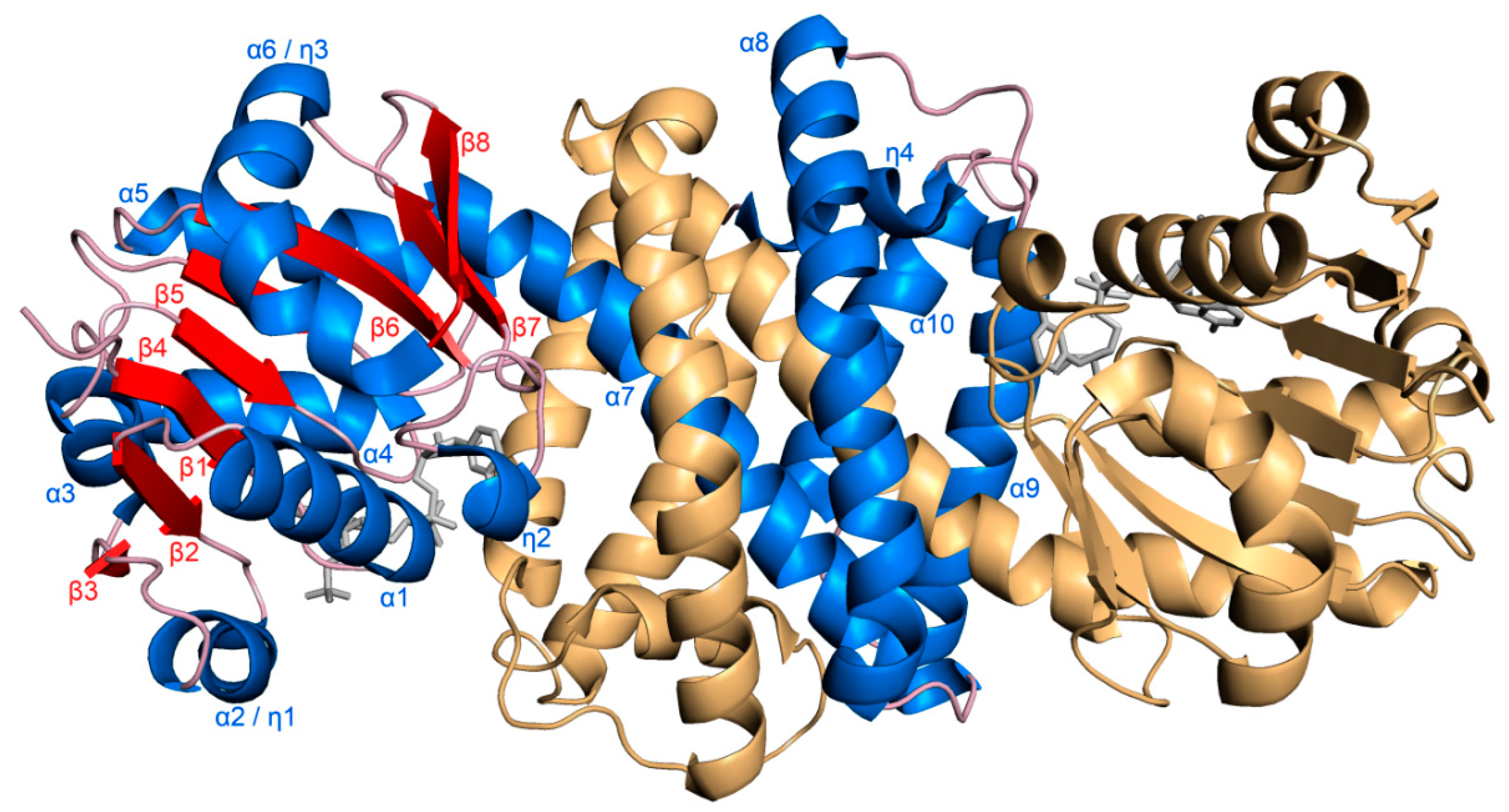
3.1. Overall Structure of S-IRED-Ms and Dimer Formation
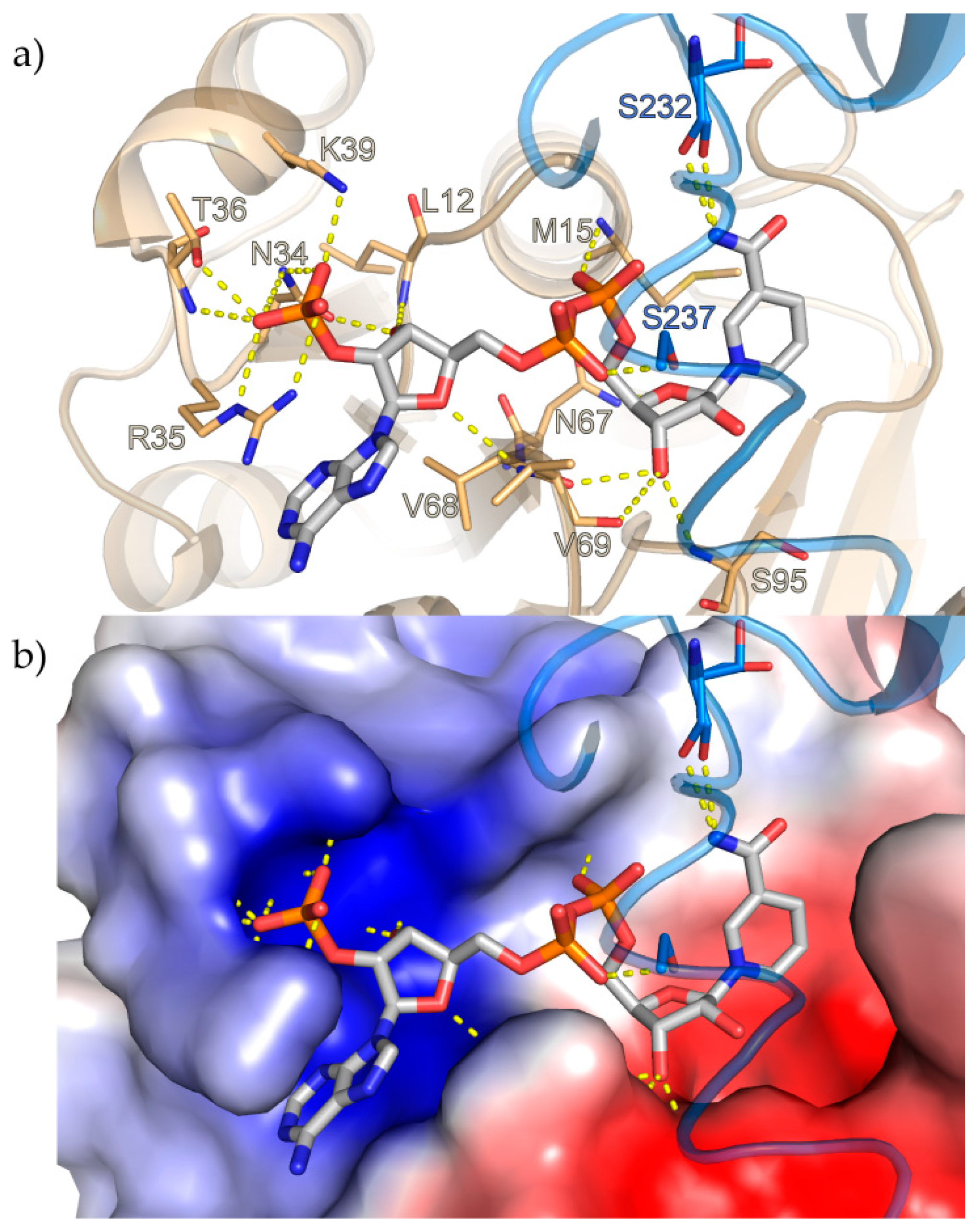
3.2. NADPH-Binding Site
3.3. Substrate-Binding Site
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tomaszewski, J.; Rumore, M.M. Stereoisomeric drugs: FDA’s policy statement and the impact on drug development. Drug Dev. Ind. Pharm. 1994, 20, 119–139. [Google Scholar] [CrossRef]
- Calcaterra, A.; D’Acquarica, I. The market of chiral drugs: Chiral switches versus de novo enantiomerically pure compounds. J. Pharm. Biomed. Anal. 2018, 147, 323–340. [Google Scholar] [CrossRef]
- Elander, R.P. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 2003, 61, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Narang, R.K.; Rawal, R.K. Role of sulphur-heterocycles in medicinal chemistry: An update. Eur. J. Med. Chem. 2019, 180, 486–508. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiao, J. Asymmetric hydrogenation of cyclic imines with an ionic Cp*Rh(III) catalyst. J. Am. Chem. Soc. 2008, 130, 13208–13209. [Google Scholar] [CrossRef] [PubMed]
- Roszkowski, P.; Maurin, J.K.; Czarnocki, Z. Enantioselective synthesis of (R)-(-)-praziquantel (PZQ). Tetrahedron Asymmetry 2006, 17, 1415–1419. [Google Scholar] [CrossRef]
- Reiners, I.; Gröger, H.; Martens, J. A new enantioselective synthetic approach to ß-aminothio-compounds via enantioselective reduction of N,S-heterocyclic imines. J. Prakt. Chem. 1997, 339, 541–546. [Google Scholar] [CrossRef]
- Vu, T.-T.-H.; Kumbhar, P.S.; Figueras, F. Base-catalysed hydrogenation of sulfur-containing aldehydes. Adv. Synth. Catal. 2003, 345, 493–496. [Google Scholar] [CrossRef]
- Baccanari, D.P.; Tansik, R.L.; Joyner, S.S.; Fling, M.E.; Smith, P.L.; Freisheim, J.H. Characterization of Candida albicans dihydrofolate reductase. J. Biol. Chem. 1989, 264, 1100–1107. [Google Scholar]
- Mitsukura, K.; Suzuki, M.; Shinoda, S.; Kuramoto, T.; Yoshida, T.; Nagasawa, T. Purification and characterization of a novel (R)-imine reductase from Streptomyces sp. GF3587. Biosci. Biotechnol. Biochem. 2011, 75, 1778–1782. [Google Scholar] [CrossRef]
- Mitsukura, K.; Kuramoto, T.; Yoshida, T.; Kimoto, N.; Yamamoto, H.; Nagasawa, T. A NADPH-dependent (S)-imine reductase (SIR) from Streptomyces sp. GF3546 for asymmetric synthesis of optically active amines: Purification, characterization, gene cloning, and expression. Appl. Microbiol. Biotechnol. 2013, 97, 8079–8086. [Google Scholar] [CrossRef] [PubMed]
- Scheller, P.N.; Fademrecht, S.; Hofelzer, S.; Pleiss, J.; Leipold, F.; Turner, N.J.; Nestl, B.M.; Hauer, B. Enzyme toolbox: Novel enantiocomplementary imine reductases. ChemBioChem 2014, 15, 2201–2204. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Mata, M.; Frank, A.; Wells, E.; Leipold, F.; Turner, N.J.; Hart, S.; Turkenburg, J.P.; Grogan, G. Structure and activity of NADPH-dependent reductase Q1EQE0 from Streptomyces kanamyceticus, which catalyses the R-selective reduction of an imine substrate. ChemBioChem 2013, 14, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Huber, T.; Schneider, L.; Präg, A.; Gerhardt, S.; Einsle, O.; Müller, M. Direct reductive amination of ketones: Structure and activity of S-selective imine reductases from Streptomyces. ChemCatChem 2014, 6, 2248–2252. [Google Scholar] [CrossRef]
- Man, H.; Wells, E.; Hussain, S.; Leipold, F.; Hart, S.; Turkenburg, J.P.; Turner, N.J.; Grogan, G. Structure, activity and stereoselectivity of NADPH-dependent oxidoreductases catalysing the S-selective reduction of the imine substrate 2-methylpyrroline. ChemBioChem 2015, 16, 1052–1059. [Google Scholar] [CrossRef]
- Aleku, G.A.; Man, H.; France, S.P.; Leipold, F.; Hussain, S.; Toca-Gonzalez, L.; Marchington, R.; Hart, S.; Turkenburg, J.P.; Grogan, G.; et al. Stereoselectivity and structural characterization of an imine reductase (IRED) from Amycolatopsis orientalis. ACS Catal. 2016, 6, 3880–3889. [Google Scholar] [CrossRef]
- Aleku, G.A.; France, S.P.; Man, H.; Mangas-Sanchez, J.; Montgomery, S.L.; Sharma, M.; Leipold, F.; Hussain, S.; Grogan, G.; Turner, N.J. A reductive aminase from Aspergillus oryzae. Nat. Chem. 2017, 9, 961–969. [Google Scholar] [CrossRef]
- France, S.P.; Aleku, G.A.; Sharma, M.; Mangas-Sanchez, J.; Howard, R.M.; Steflik, J.; Kumar, R.; Adams, R.W.; Slabu, I.; Crook, R.; et al. Biocatalytic routes to enantiomerically enriched dibenz[c,e]azepines. Angew. Chem. Int. Ed. Engl. 2017, 129, 15795–15799. [Google Scholar] [CrossRef]
- Lenz, M.; Fademrecht, S.; Sharma, M.; Pleiss, J.; Grogan, G.; Nestl, B.M. New imine-reducing enzymes from β-hydroxyacid dehydrogenases by single amino acid substitutions. Protein Eng. Des. Sel. 2018, 31, 109–120. [Google Scholar] [CrossRef]
- Fademrecht, S.; Scheller, P.N.; Nestl, B.M.; Hauer, B.; Pleiss, J. Identification of imine reductase-specific sequence motifs. Proteins 2016, 84, 600–610. [Google Scholar] [CrossRef]
- Wetzl, D.; Berrera, M.; Sandon, N.; Fishlock, D.; Ebeling, M.; Müller, M.; Hanlon, S.; Wirz, B.; Iding, H. Expanding the imine reductase toolbox by exploring the bacterial protein-sequence space. ChemBioChem 2015, 16, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Zumbrägel, N.; Merten, C.; Huber, S.M.; Gröger, H. Enantioselective reduction of sulfur-containing cyclic imines through biocatalysis. Nat. Commun. 2018, 9, 1949. [Google Scholar] [CrossRef] [PubMed]
- Chaikuad, A.; Knapp, S.; von Delft, F. Defined PEG smears as an alternative approach to enhance the search for crystallization conditions and crystal-quality improvement in reduced screens. Acta Crystallogr. D 2015, 71, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Cianci, M.; Bourenkov, G.; Pompidor, G.; Karpics, I.; Kallio, J.; Bento, I.; Roessle, M.; Cipriani, F.; Fiedler, S.; Schneider, T.R. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat. 2017, 24, 323–332. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 2010, 66, 133–144. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 2011, 67, 235–242. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 2008, 64, 61–69. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adamas, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Klei, H.; Adams, P.D.; Moriarty, N.W.; Cohn, J.D. Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr. D 2006, 62, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.A.; Steiner, R.S.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 dictionary: Organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 2003, 60, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.; Förster, R.; Hellmig, M.; Huschmann, F.U.; Kastner, A.; Malecki, P.; Pühringer, S.; Röwer, M.; Sparta, K.; Steffien, M.; et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 2015, 130, 141–150. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Bellamacina, C.R. The nicotinamide dinucleotide binding motif: A comparison of nucleotide binding proteins. FASEB J. 1996, 10, 1257–1269. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Bope, A.; Haines, S.R.; Hegarty, B.; Weschler, C.J.; Peccia, J.; Dannemiller, K.C. Degradation of phthalate esters in floor dust at elevated relative humidity. Environ. Sci. Process Impacts 2019, 21, 1268–1279. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, P.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk-solvent. Acta Crystallogr. D 2017, 73, 148–157. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Adams, P.D.; Read, R.J.; Zwart, P.H.; Hung, L.-W. Iterative-build OMIT maps: Map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D 2008, 64, 515–524. [Google Scholar] [CrossRef]
- Holm, L. Benchmarking fold detection by DaliLite v.5. Bioinformatics 2019, 35, 5326–5327. [Google Scholar] [CrossRef] [PubMed]
- Meneely, K.M.; Ronnebaum, T.A.; Riley, A.P.; Prisinzano, T.E.; Lamb, A.L. Holo structure and steady state kinetics of the thiazolinyl imine reductases for siderophore biosynthesis. Biochemistry 2016, 55, 5423–5433. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Leipold, F.; Man, H.; Wells, E.; France, S.P.; Mulholland, K.R.; Grogan, G.; Turner, N.J. An (R)-imine reductase biocatalyst for the asymmetric reduction of cyclic imines. ChemCatChem 2015, 7, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Gand, M.; Müller, H.; Wardenga, R.; Höhne, M. Characterization of three novel enzymes with imine reductase activity. J. Mol. Catal. B Enzym. 2014, 110, 126–132. [Google Scholar] [CrossRef]
- Czekster, C.M.; Vandemeulebroucke, A.; Blanchard, J.S. Kinetic and chemical mechanism of the dihydrofolate reductase from Mycobacterium tuberculosis. Biochemistry 2011, 50, 367–375. [Google Scholar] [CrossRef][Green Version]
- Cheng, Q.; Guengerich, F.P. Identification of endogenous substrates of orphan cytochrome P450 enzymes through the use of untargeted metabolomics approaches. Methods Mol. Biol. 2013, 987, 71–77. [Google Scholar]
- Masuo, Y.; Ohba, Y.; Yamada, K.; Al-Shammari, A.H.; Seba, N.; Nakamichi, N.; Ogihara, T.; Kunishima, M.; Kato, Y. Combination metabolomics approach for identifying endogenous substrates of carnitine/organic cation transporter OCTN1. Pharm. Res. 2018, 35, 224. [Google Scholar] [CrossRef]
- Martens, J.; Offermanns, H.; Scherberich, P. Facile synthesis of racemic cysteine. Angew. Chem. Int. Ed. Engl. 1981, 20, 668. [Google Scholar] [CrossRef]
- Battistoni, P.; Bruni, P.; Fava, G. A general method for the synthesis of 3-phenyl-2H-1,4-benzoxazines and 3-phenyl-2H-3,4-dihydro-1,4-benzoxazines. Synthesis 1979, 1979, 220–221. [Google Scholar] [CrossRef]
- Zumbrägel, N.; Machui, P.; Nonnhoff, J.; Gröger, H. Enantioselective biocatalytic reduction of 2H-1,4-benzoxazines using imine reductases. J. Org. Chem. 2019, 84, 1440–1447. [Google Scholar] [CrossRef]
- Stalling, T.; Johannes, K.; Polina, S.; Martens, J. Stereospecific synthesis of β-lactams from heterocyclic imines using the Staudinger reaction. J. Heterocycl. Chem. 2013, 50, 654–659. [Google Scholar] [CrossRef]

| Data Collection Statistics | Holo-S-IRED-Ms |
|---|---|
| Beam line | DESY P13 |
| Detector | DECTRIS PILATUS 6M-F |
| Wavelength (Å) | 0.9762 |
| Temperature (K) | 100 |
| Space Group | I 1 2 1 |
| Unit cell dimensions | a = 117.0 Å |
| b = 77.0 Å | |
| c = 194.5 Å | |
| α = γ = 90.000 ° | |
| β = 90.204 ° | |
| Resolution (highest resolution shell) (Å) | 100.42–1.55 (1.58–1.55) |
| Completeness (%) | 99.1 (96.3) |
| Multiplicity | 6.9 (6.4) |
| Observations | 1,702,995 (76,619) |
| Unique reflections | 248,023 (11,889) |
| I/σ(I) | 13.2 (3.2) |
| Rmerge (%) | 7.3 (38.4) |
| Rmeas (%) | 8.0 (41.7) |
| Rpim (%) | 3.0 (16.2) |
| CC ½ | 99.7 (92.8) |
| Monomers per asymmetric unit | 5 |
| Solvent content (%) | 53.0 |
| Wilson B-factor (A²) | 16.76 |
| Refinement Statistics | |
| Resolution (highest resolution shell) (Å) | 100.42–1.55 (1.59–1.55) |
| Rwork (%) (highest resolution shell) | 13.08 (17.30) |
| Rfree (%) (highest resolution shell) | 15.49 (20.20) |
| Average B-factor (A²) | 21.0 |
| protein | 18.6 |
| ligands | 20.3 |
| solvent | 34.2 |
| Number of non-hydrogen atoms | 13,314 |
| protein | 10,980 |
| ligands | 364 |
| solvent | 1970 |
| r.m.s.d. bondlength (from ideal geometry) | 0.0193 |
| r.m.s.d. angle (from ideal geometry) | 2.2672 |
| Ramachandran favored (%) | 99.86 |
| Ramachandran allowed (%) | 0.14 |
| Ramachandran outlier (%) | 0.00 |
| Rotamer outliers (%) | 0.98 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyer, T.; Zumbrägel, N.; Geerds, C.; Gröger, H.; Niemann, H.H. Structural Characterization of an S-enantioselective Imine Reductase from Mycobacterium Smegmatis. Biomolecules 2020, 10, 1130. https://doi.org/10.3390/biom10081130
Meyer T, Zumbrägel N, Geerds C, Gröger H, Niemann HH. Structural Characterization of an S-enantioselective Imine Reductase from Mycobacterium Smegmatis. Biomolecules. 2020; 10(8):1130. https://doi.org/10.3390/biom10081130
Chicago/Turabian StyleMeyer, Timo, Nadine Zumbrägel, Christina Geerds, Harald Gröger, and Hartmut H. Niemann. 2020. "Structural Characterization of an S-enantioselective Imine Reductase from Mycobacterium Smegmatis" Biomolecules 10, no. 8: 1130. https://doi.org/10.3390/biom10081130
APA StyleMeyer, T., Zumbrägel, N., Geerds, C., Gröger, H., & Niemann, H. H. (2020). Structural Characterization of an S-enantioselective Imine Reductase from Mycobacterium Smegmatis. Biomolecules, 10(8), 1130. https://doi.org/10.3390/biom10081130






