TOP mRNPs: Molecular Mechanisms and Principles of Regulation
Abstract
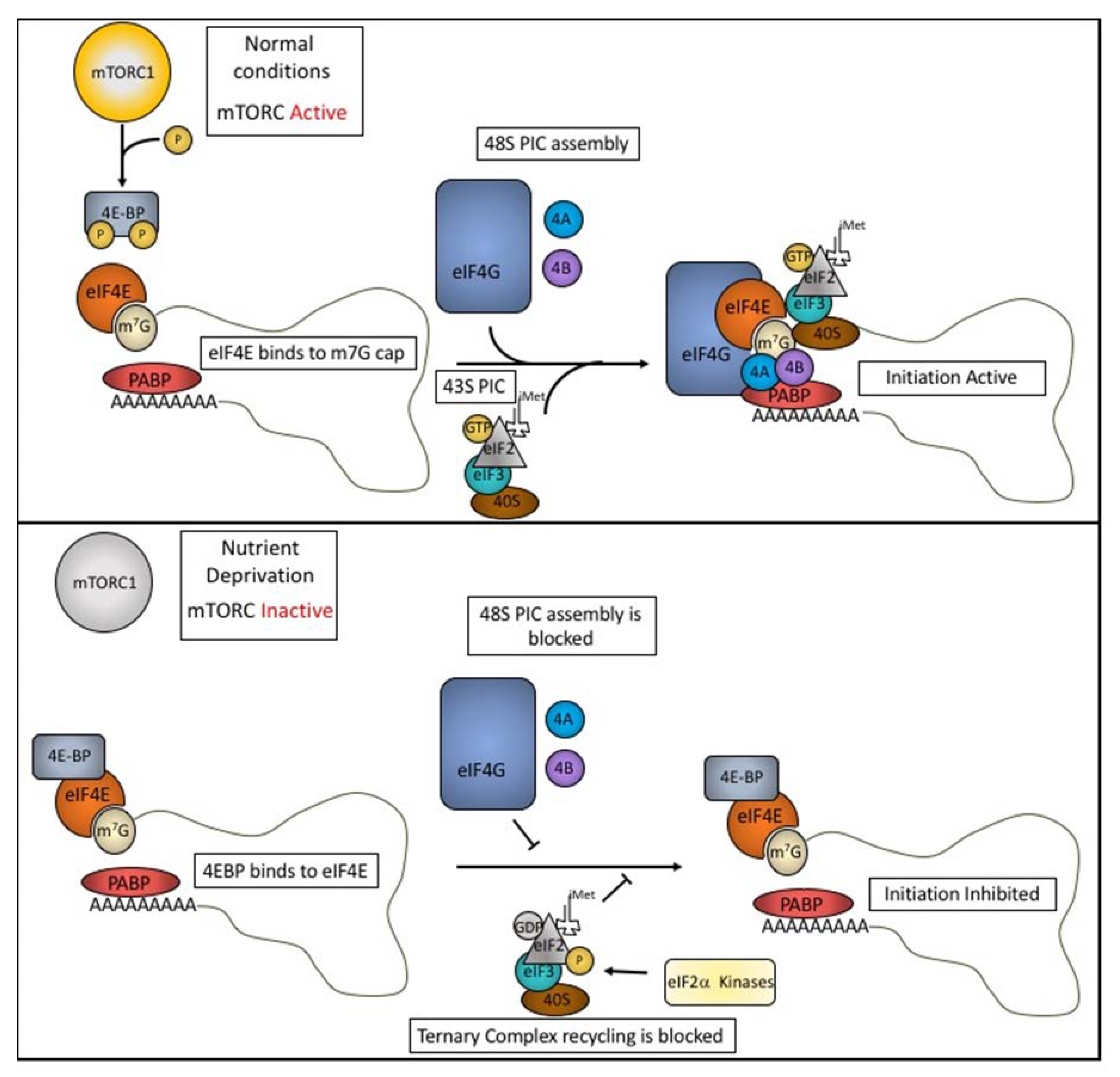
1. Translation Initiation and Regulation
2. The 5’ TOP Motif
3. Regulation of TOP mRNAs
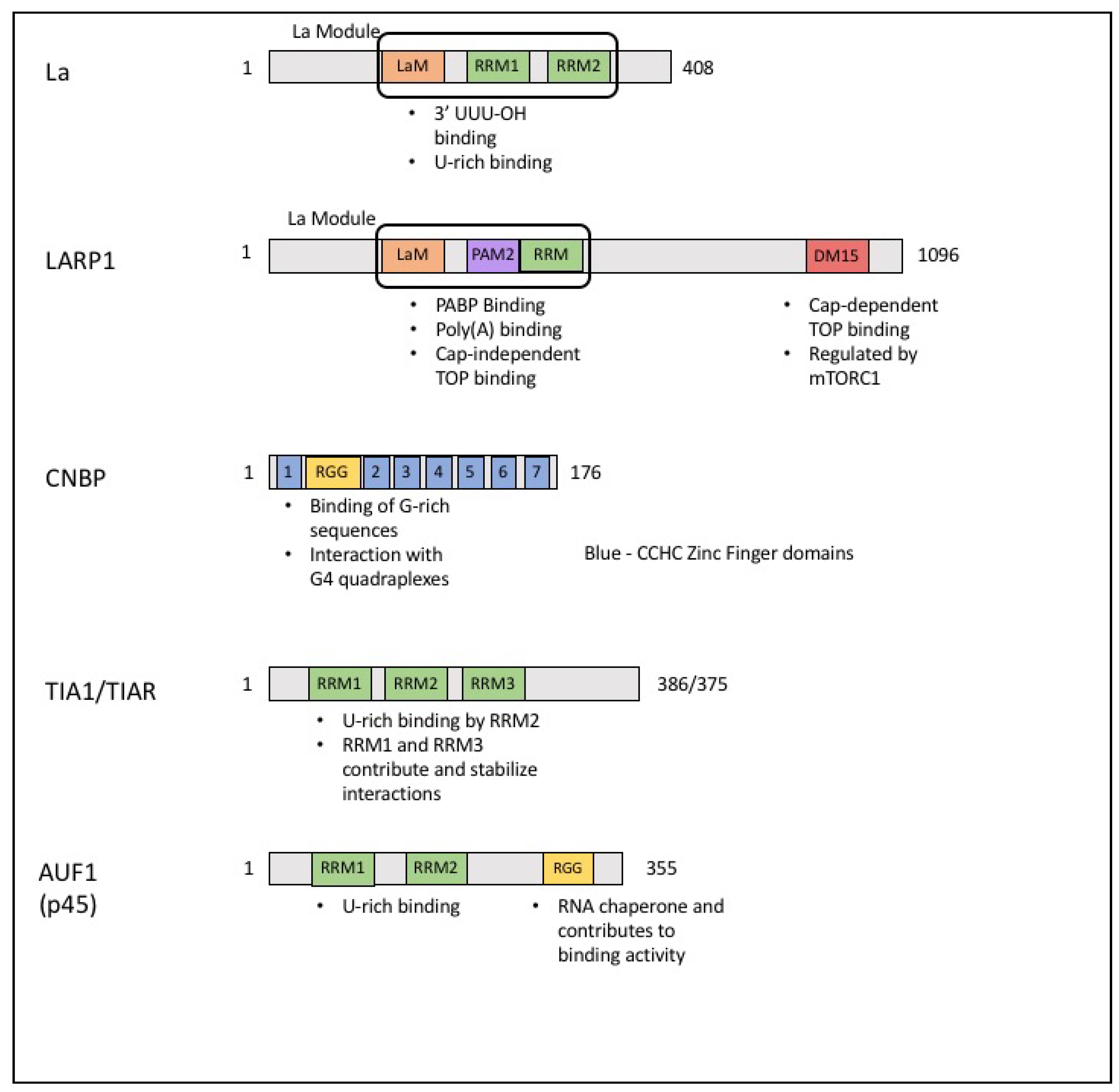
4. The La Family of Proteins
5. La Related Protein 1 (LARP1)
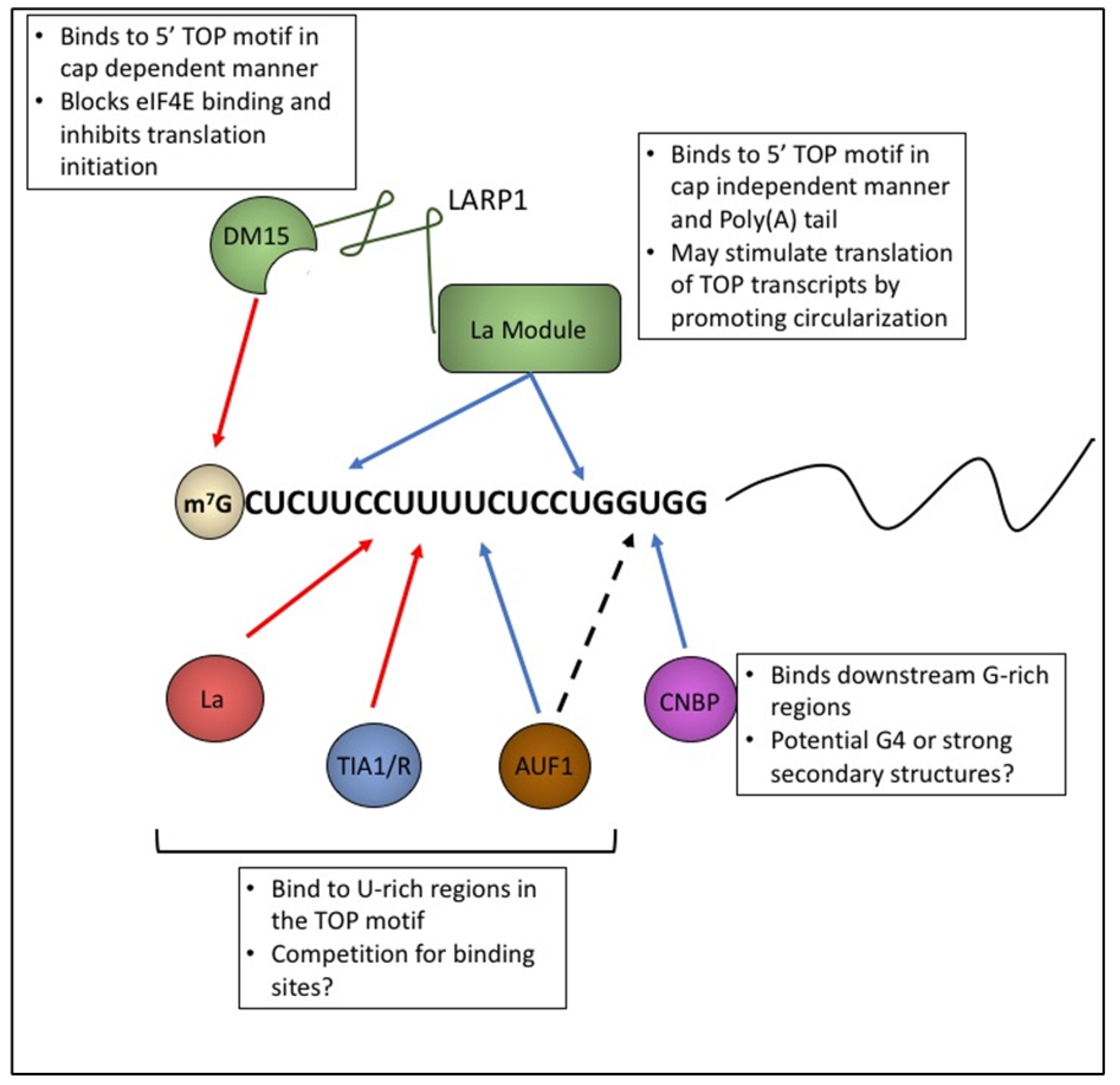
5.1. LARP1 Binds 5’ TOP Motifs in a Sequence Dependent Manner
5.2. LARP1 Represses Translation of TOP Transcripts
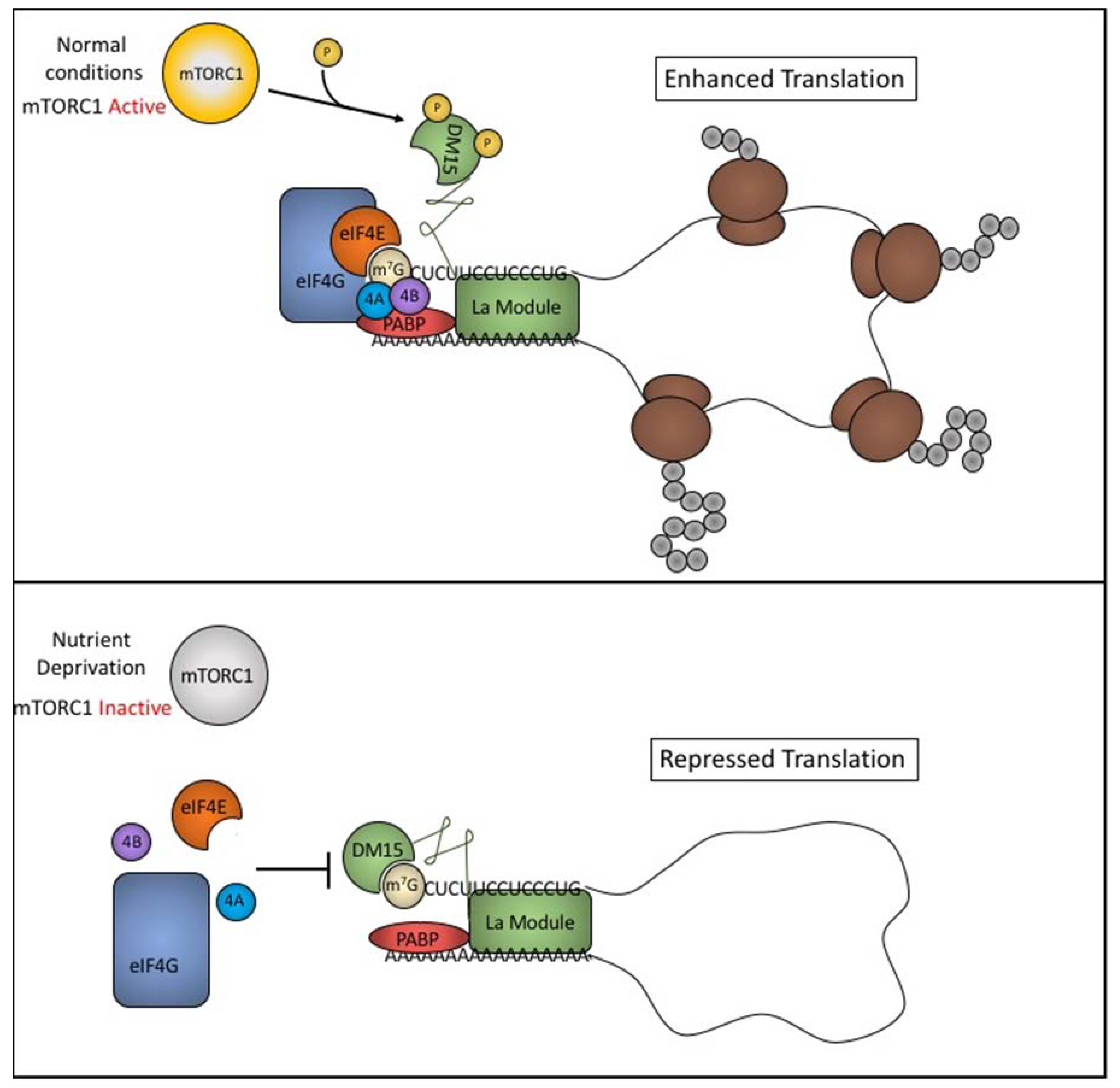
5.3. LARP1 Binding and TOP Repression Is Controlled by mTORC1 Signaling
5.4. LARP1 Inhibits Translation of TOP Transcripts by Competing with Initiation Factors
5.5. LARP1 Regulation of TOP Transcripts is Complex
6. Cellular Nucleic Acid Binding Protein (CNBP)
7. T-Cell Intracellular Antigen (TIA1) and TIA-Related Protein (TIAR)
8. TIA1 and TIAR in TOP Regulation
9. ARE/poly(U)-Binding/Degradation Factor 1 (AUF1)
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Gene | 5’ UTR (First 50 Nucleotides) | Accession No. |
|---|---|---|
| RPSA | CUUUUUCCGUGCUACCUGCAGAGGGGUCCAUACGGCGUUGUUCUGGAUUC | D28372.1 |
| RPS2 | CUUCUUUUCCGACAAAACACCAAAUGGCG | AB055772.1 |
| RPS3 | CCUUUCCUUUCAGCGGAGCGCGGCGGCAAGAUGGCA | D28344.1 |
| RPS3A | CCUUUUGGCUCUCUGACCAGCACCAUGGCG | D28374.1 |
| RPS4X | CUCUUUCCUUGCCUAACGCAGCCAUGGCU | D28359.1 |
| RPS4Y | CUCUUCCGUCGCAGAGUUUCGCCAUGGCC | AB055773.1 |
| RPS5 | CUCUUCCUGUCUGUACCAGGGCGGCGCGUGGUCUACGCCGAGUGACAGAG | D28455.1 |
| RPS6 | CUCUUUUCCGUGGCGCCUCGGAGGCGUUCAGCUGCUUCAAGAUGAAG | D28348.1 |
| RPS7 | CUCUUCCUAAGCCGGCGCUCGGCAAGUUCUCCCAGGAGAAAGCCAUGUUC | AB055774.1 |
| RPS8 | CUCUUUCCAGCCAGCGCCGAGCGAUGGGC | D28361.1 |
| RPS9 | CUCUUUCUCAGUGACCGGGUGGUUUGCUUAGGUGAGGUGCGG | AB061839.1 |
| RPS10 | CUUUUCCAGCCCCGGUACCGGACCCUGCAGCCGCAGAGAUGUUG | D28427.1 |
| RPS11 | CUUUUUUUUUUUCAGGCGGCCGGGAAGAUGGCG | D28407.1 |
| RPS12 | CUUUCCCUGCCGCCGCCGAGUCGCGCGGAGGCGGAGGCUUGGGUGCGUUC | D28378.1 |
| RPS13 | CCUUUCGUUGCCUGAUCGCCGCCAUCAUGGGU | D28429.1 |
| RPS14 | CUUUUUCCGGUGUGGAGUCUGGAGACGACGUGCAGAAAUGGCA | D28352.1 |
| RPS15 | CUUCUGAGGAUCCGGCAAGAUGGCA | AB055776.1 |
| RPS15A | CUCUUUCCGCCAUCUUUCCGCGCCGCCACAAUGGUG | D28347.1 |
| RPS16 | CUUUUCCGGUUGCGGCGCCGCGCGGUGAGGUUGUCUAGUCCACGCUCGGA | D28392.1 |
| RPS17 | CUCUUUUACCAAGGACCCGCCAACAUGGGC | AB055777.1 |
| RPS18 | CUCUUCCACAGGAGGCCUACACGCCGCCGCUUGUGCUGCAGCCAUGUCU | AB055778.1 |
| RPS19 | CUUUCCCCUGGCUGGCAGCGCGGAGGCCGCACGAUGCCU | D28389.1 |
| RPS20 | CUUUUUUUUUGAGGAAGACGCGGUCGUAAGGGCUGAGGAUUUUUGGUCCG | D28358.1 |
| RPS21 | CUUUUCUCUCUCGCGCGCGGUGUGGUGGCAGCAGGCGCAGCCAGCCUCGA | D28422.1 |
| RPS23 | CUCUUUCGCUCAGGCCCGUGGCGCCGACAGGAUGGGC | D28396.1 |
| RPS24 | CUCUUUUCCUCCUUGGCUGUCUGAAGAUAGAUCGCCAUCAUGAAC | D28424.1 |
| RPS25 | CUUCCUUUUUGUCCGACAUCUUGACGAGGCUGCGGUGUCUGCUGCUAUUC | D28369.1 |
| RPS26 | CUCCUCUCUCCGGUCCGUGCCUCCAAGAUGACA | AB056456.1 |
| RPS27 | CUUUCCGGCGGUGACGACCUACGCACACGAGAACAUGCCU | D28454.1 |
| RPS27A | CUUCCUUUUCGAUCCGCCAUCUGCGGUGGAGCCGCCACCAAAAUGCAG | D28404.1 |
| RPS28 | CUCUCCGCCAGACCGCCGCCGCGCCGCCAUCAUGGAC | AB055779.1 |
| RPS29 | CCUUUUACCUCGUUGCACUGCUGAGAGCAAGAUGGGU | AB055780.1 |
| RPS30 | CUCUUUCUCGACUCCAUCUUCGCGGUAGCUGGGACCGCCGUUCAGUCGCC | D28403.1 |
| RPP0 | CUCUCGCCAGGCGUCCUCGUGGAAGUGACAUCGUCUUUAAACCCUGCGUG | D28418.1 |
| RPP1 | CUUUUUUUCCUCAGCUGCCGCCAAGGUGCUCGGUCCUUCCGAGGAAGCUA | D28366.1 |
| RPP2 | CCUUUUCCUCCCUGUCGCCACCGAGGUCGCACGCGUGAGACUUCUCCGCC | D28411.1 |
| RPL3 | CUCUACCGGCGGGAUUUGAUGGCGUGAUGUCU | D28415.1 |
| RPL4 | CUUUUCCUGUGGCAGCAGCCGGGCUGAGAGGAGCGUGGCUGUCUCCUCUC | D23660.1 |
| RPL5 | CUUUUCCCACCCCCUAGCGCCGCUGGGCCUGCAGGUCUCUGUCGAGCAGC | AB055762.1 |
| RPL6 | CUUUCUUUCCCAUCUUGCAAGAUGGCG | D28388.1 |
| RPL7 | CUCUUUUUCCGGCUGGAACCAUGGAG | AB055763.1 |
| RPL7A | CUCUCUCCUCCCGCCGCCCAAGAUGCCG | D28405.1 |
| RPL8 | CUCUUUCGGCCGCGCUGGUGAACAGGACCCGUCGCCAUGGGC | D28421.1 |
| RPL9 | CUUUUUUUGCUGCGUCUACUGCGAGAAUGAAG | D28399.1 |
| RPL10 | CUCUUUCCCUUCGGUGUGCCACUGAAGAUCCUGGUGUCGCCAUGGGC | D28410.1 |
| RPL11 | CUCUUCCUGCUCUCCAUC | NM_000975.5 |
| RPL10A | CUCUUUUCCGGUUAGCGCGGCGUGAGAAGCCAUGAGC | AB055764.1 |
| RPL12 | CUCUCGGCUUUCGGCUCGGAGGAGGCCAAGGUGCAACUUCCUUCGGUCGU | D28443.1 |
| RPL13 | CUUUCCGCUCGGCUGUUUUCCUGCGCAGGAGCCGCAGGGCCGUAGGCAGC | NM_000977.4 |
| RPL13A | CUUUUCCAAGCGGCUGCCGAAGAUGGCG | D28409.1 |
| RPL14 | CUUCUUCCUUCUCGCCUAACGCUGCCAACAUGGUG | AB055765.1 |
| RPL15 | CCUUUCCGUCUGGCGGCAGCAUCAGGUAAGCCAAGAUGGGU | D28417.1 |
| RPL17 | CUCUUUCCCUAAGCAGCCUGAGGUGAUCUGUGAAAAUGGUU | D28373.1 |
| RPL18 | CUUUCCGGACCUGGCCGAGCAGGAGGCGCAAUCAUGGGA | D28461.1 |
| RPL18A | CUUUUGCGGGUGGCGGCGAACGCGGAGAGCACGCCAUGAAG | D28393.1 |
| RPL19 | CUUUCCUUUCGCUGCUGCGGCCGCAGCCAUGAGU | AB055766.1 |
| RPL21 | CCUUUCGGCCGGAACCGCCAUCUUCCAGUAAUUCGCCAAA | D28406.1 |
| RPL22 | CUUUUUUUUUCUAACUCCGCUGCCGCC | D28346.1 |
| RPL23 | CCUUUUUUCUUUUUUCCGGCGUUCAAG | D28349.1 |
| RPL23A | CCUUUUCACAAG | D28401.1 |
| RPL24 | CUUUUCUUUCGCCAUCUUUUGUCUUUCCGUGGAGCUGUCGCC | D28400.1 |
| RPL26 | CUCUUCCCUUUUGCGGCCAUCACCGAAGCGGGAGCGCCAAA | D28413.1 |
| RPL27 | CCUUUUUGCUGUAGGCCCGGGUGGUUGCUGCCGAA | D28453.1 |
| RPL27A | CUUUUUCGUCUGGGCUGCCAAC | AB055767.1 |
| RPL30 | CUUUUCUCGUUCCCCGGCCAUCUUAGCGGCUGCUGUUGGUUGGGGGCCGU | D28438.1 |
| RPL31 | CUUCCUUUCCAACUUAGACGCUGCAGA | D28386.1 |
| RPL32 | CUUCCUCGGCGCUGCCUACGGAGGUGGCAGCCAUCUCCUUCUCGGCAUCA | D28385.1 |
| RPL34 | CUUCCUCUUCCGGGGACGUUGUCUGCAGGCACUCAGAAUGGUC | D28420.1 |
| RPL35 | CUCUUUCCCUCGGAGCGGGCGGCGGCGUUGGCGGCUUGUGCAGCAAUGGC | D28448.1 |
| RPL36 | CUUUGCGCCACGGCCGUCUCUGGAGAGCAGCAGCCAUGGCC | AB055769.1 |
| RPL36A | CUUUUUCCGCGCCGAUAGCGCUCACGCAAGCAUGGUU | D28414.1 |
| RPL37 | CUUUCUGGUCUCGGCCGCAGAAGCGAGAUGACG | AB055770.1 |
| RPL39 | CUUCCGCCAGCUUCCCUCCUCUUCCUUUCUCCGCCAUCGUGGUGUGUUCU | D28397.1 |
| RPL40 | CUUCUUUUUCUUCAGCGAGGCGGCCGAGCUGGUUGGUGGCGGCGGUCGUG | NM_001321022 |
| RPL41 | CUCUCGGCCUUAGCGCCAUUUUUUUGGAAACCUCUGCGCCAUGAGA | D28462.1 |
| eIF3A | CUCCUUCCUUUCCGUCUCUGGCCGGCUGGGCGCGGGCGACUGCUGGCGAG | JX312508.1 |
| eIF3E | CUUUUCUUUGGCAAGAUGGCGGAGUACGACUUGACUACUCGCAUCGCGCA | NM_001568.3 |
| eiF3F | CUUCUUUCUCGACAAGAUGG | NM_003754.3 |
| eIF3H | CUCUUUCUUCCUGUCUGCUUGGAAAGAUG | NM_003756.3 |
| eIF4B | CUUUUGCGUUCUCUUUCCCUCUCCCAACAUG | NM_001300821 |
| eEF1A | CUUUUUCGCAACGGGUUUGCCGCCAGAACACAGGUGUCGUGAAAACUACC | NM_001402.6 |
| eEF1B2 | CCUUUUUCCUCUCUUCAGCGUGGGGCGCCCACAAUUUGCGCGCUCUCUUU | NM_001959.4 |
| eEF1D | CUCCCUUUCAUCAGUCUUCCCGCGUCCGCCGAUUCCUCCUCCUUGGUCGC | NM_032378.6 |
| eEF1G | CCUUUCUUUGCGGAAUCACCAUG | NM_001404.5 |
| eEF2 | CUCUUCCGCCGUCGUCGCCGCCAUCCUCGGCGCGACUCGCUUCUUUCGGU | NM_001961.4 |
| Rack1 | CUCUCUUUCACUGCAAGGCGGCGGCAGGAGAGGUUGUGGUGCUAGUUUCU | NM_006098.5 |
| PABP | CCCCUUCUCCCCGGCGGUUAGUGCUGAGAGUGCGGAGUGUGUGCUCCGGG | NM_002568.4 |
| hnRNP A1 | CCUUUCUGCCCGUGGACGCCGCCGAAGAAGCAUCGUUAAAGUCUCUCUU | NM_002136.4 |
| Nucleophosmin | CUUUCCCUGGUGUGAUUCCGUCCUGCGCGGUUGUUCUCUGGAGCAGCGUU | NM_002520.6 |
| NAP1L1 | CUUUUUUAGCGCCAUCUGCUCGCGGCGCCGCCUCCUGCUCCUCCCGCUGC | NM_139207.5 |
| TCTP | CUUUUCCGCCCGCUCCCCCCUCCCCCCGAGCGCCGCUCC | NM_00128627 |
| Vimentin | CCUCUGCCACUCUCGCUCCGAGGUCCCCGCGCCAGAGACGCAGCCGCGCU | NM_003380.5 |
References
- Harding, H.P.; Zhang, Y.; Bertolotti, A.; Zeng, H.; Ron, D. Perk Is Essential for Translational Regulation and Cell Survival during the Unfolded Protein Response. Mol. Cell 2000, 5, 897–904. [Google Scholar] [CrossRef]
- Lu, L.; Han, A.-P.; Chen, J.-J. Translation Initiation Control by Heme-Regulated Eukaryotic Initiation Factor 2α Kinase in Erythroid Cells under Cytoplasmic Stresses. Mol. Cell. Biol. 2001, 21, 7971–7980. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Kumar, K.U.; Kaufman, R.J. Phosphorylation of Eukaryotic Translation Initiation Factor 2 Mediates Apoptosis in Response to Activation of the Double-stranded RNA-dependent Protein Kinase. J. Biol. Chem. 1998, 273, 2416–2423. [Google Scholar] [CrossRef] [PubMed]
- Wek, S.A.; Zhu, S.; Wek, R.C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 1995, 15, 4497–4506. [Google Scholar] [CrossRef] [PubMed]
- Wek, R.C. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol. 2018, 10, a032870. [Google Scholar] [CrossRef]
- Advani, V.M.; Ivanov, P. Translational Control under Stress: Reshaping the Translatome. Bioessays 2019, 41, e1900009. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Khoutorsky, A.; Mathews, M.B.; Sonenberg, N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 791–807. [Google Scholar] [CrossRef]
- Mader, S.; Lee, H.; Pause, A.; Sonenberg, N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995, 15, 4990–4997. [Google Scholar] [CrossRef]
- Yoshihama, M.; Uechi, T.; Asakawa, S.; Kawasaki, K.; Kato, S.; Higa, S.; Maeda, N.; Minoshima, S.; Tanaka, T.; Shimizu, N.; et al. The Human Ribosomal Protein Genes: Sequencing and Comparative Analysis of 73 Genes. Genome Res. 2002, 12, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Avni, D.; Shama, S.; Loreni, F.; Meyuhas, O. Vertebrate mRNAs with a 5’-terminal pyrimidine tract are candidates for translational repression in quiescent cells: Characterization of the translational cis-regulatory element. Mol. Cell Biol. 1994, 14, 3822–3833. [Google Scholar] [CrossRef]
- Meyuhas, O.; Kahan, T. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Bioenerg. 2015, 1849, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Iadevaia, V.; Caldarola, S.; Tino, E.; Amaldi, F.; Loreni, F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 2008, 14, 1730–1736. [Google Scholar] [CrossRef]
- Fonseca, B.D.; Zakaria, C.; Jia, J.-J.; Graber, T.E.; Svitkin, Y.; Tahmasebi, S.; Healy, D.; Hoang, H.-D.; Jensen, J.M.; Diao, I.T.; et al. La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1). J. Biol. Chem. 2015, 290, 15996–16020. [Google Scholar] [CrossRef] [PubMed]
- Miloslavski, R.; Cohen, E.; Avraham, A.; Iluz, Y.; Hayouka, Z.; Kasir, J.; Mudhasani, R.; Jones, S.N.; Cybulski, N.; Rüegg, M.A.; et al. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J. Mol. Cell Biol. 2014, 6, 255–266. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Sabatini, D.M. Rapamycin inhibits mTORC1, but not completely. Autophagy 2009, 5, 725–726. [Google Scholar] [CrossRef] [PubMed]
- Philippe, L.; Vasseur, J.-J.; Debart, F.; Thoreen, C.C. La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 2018, 46, 1457–1469. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Chantranupong, L.; Keys, H.R.; Wang, T.C.; Gray, N.S.; Sabatini, D.M. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012, 485, 109–113. [Google Scholar] [CrossRef]
- Li, B.B.; Qian, C.; Gameiro, P.A.; Liu, C.-C.; Jiang, T.; Roberts, T.M.; Struhl, K.; Zhao, J.J. Targeted profiling of RNA translation reveals mTOR-4EBP1/2-independent translation regulation of mRNAs encoding ribosomal proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E9325–E9332. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Mattijssen, S.; Cruz-Gallardo, I.; Conte, M.R. The La and related RNA-binding proteins (LARPs): Structures, functions, and evolving perspectives. Wiley Interdiscip. Rev. RNA 2017, 8, e1430. [Google Scholar] [CrossRef] [PubMed]
- Kotik-Kogan, O.; Valentine, E.R.; Sanfelice, D.; Conte, M.R.; Curry, S. Structural Analysis Reveals Conformational Plasticity in the Recognition of RNA 3′ Ends by the Human La Protein. Structure 2008, 16, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Martino, L.; Pennell, S.; Kelly, G.; Bui, T.T.T.; Kotik-Kogan, O.; Smerdon, S.; Drake, A.F.; Curry, S.; Conte, M.R. Analysis of the interaction with the hepatitis C virus mRNA reveals an alternative mode of RNA recognition by the human La protein. Nucleic Acids Res. 2011, 40, 1381–1394. [Google Scholar] [CrossRef] [PubMed]
- Merret, R.; Martino, L.; Bousquet-Antonelli, C.; Fneich, S.; Descombin, J.; Élodie, B.; Conte, M.R.; Deragon, J.-M. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA 2012, 19, 36–50. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Jahchan, N.S.; Hong, E.; Li, Q.; Bayfield, M.A.; Maraia, R.J.; Luo, K.; Zhou, Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 2008, 29, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Alfano, C.; Sanfelice, D.; Babon, J.; Kelly, G.; Jacks, A.; Curry, S.; Conte, M.R.; Babon, J.J. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004, 11, 323–329. [Google Scholar] [CrossRef]
- Martino, L.; Pennell, S.; Kelly, G.; Busi, B.; Brown, P.; Atkinson, R.A.; Salisbury, N.J.; Ooi, Z.-H.; See, K.-W.; Smerdon, S.; et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2014, 43, 645–660. [Google Scholar] [CrossRef]
- Boelens, W.C.; Palacios, I.; Mattaj, I.W. Nuclear retention of RNA as a mechanism for localization. RNA 1995, 1, 273–283. [Google Scholar]
- Huang, Y.; Intine, R.V.; Mozlin, A.; Hasson, S.A.; Maraia, R.J. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3′ Cleavage Activity Increase 3′-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol. Cell. Biol. 2005, 25, 621–636. [Google Scholar] [CrossRef]
- Stefano, J. Purified lupus antigen la recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 1984, 36, 145–154. [Google Scholar] [CrossRef]
- Copela, L.A.; Chakshusmathi, G.; Sherrer, R.; Wolin, S.L. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 2006, 12, 644–654. [Google Scholar] [CrossRef]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J.T. Nuclear surveillance, and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genome Res. 2004, 18, 1227–1240. [Google Scholar] [CrossRef]
- Intine, R.V.; Sakulich, A.L.; Koduru, S.B.; Huang, Y.; Pierstorff, E.; Goodier, J.L.; Phan, L.; Maraia, R.J. Control of Transfer RNA Maturation by Phosphorylation of the Human La Antigen on Serine 366. Mol. Cell 2000, 6, 339–348. [Google Scholar] [CrossRef]
- Wolin, S.L.; Matera, A.G. The trials and travels of tRNA. Genome Res. 1999, 13, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pellizzoni, L.; Lotti, F.; Maras, B.; Pierandrei-Amaldi, P. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J. Mol. Biol. 1997, 267, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Crosio, C. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000, 28, 2927–2934. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, B.; Carissimi, C.; Gravina, P.; Pierandrei-Amaldi, P. La Protein Is Associated with Terminal Oligopyrimidine mRNAs in Actively Translating Polysomes. J. Biol. Chem. 2003, 278, 35145–35151. [Google Scholar] [CrossRef]
- Intine, R.V.; Tenenbaum, S.A.; Sakulich, A.L.; Keene, J.D.; Maraia, R.J. Differential Phosphorylation and Subcellular Localization of La RNPs Associated with Precursor tRNAs and Translation-Related mRNAs. Mol. Cell 2003, 12, 1301–1307. [Google Scholar] [CrossRef]
- Schwartz, E.I.; Intine, R.V.; Maraia, R.J. CK2 Is Responsible for Phosphorylation of Human La Protein Serine-366 and Can Modulate rpL37 5′-Terminal Oligopyrimidine mRNA Metabolism. Mol. Cell. Biol. 2004, 24, 9580–9591. [Google Scholar] [CrossRef]
- Zhu, J.; Hayakawa, A.; Kakegawa, T.; Kaspar, R.L. Binding of the La autoantigen to the 5′ untranslated region of a chimeric human translation elongation factor 1A reporter mRNA inhibits translation in vitro. Biochim. Biophys. Acta Gene Struct. Expr. 2001, 1521, 19–29. [Google Scholar] [CrossRef]
- Caldarola, S.; de Stefano, M.C.; Amaldi, F.; Loreni, F. Synthesis and function of ribosomal proteins—Fading models and new perspectives. FEBS J. 2009, 276, 3199–3210. [Google Scholar] [CrossRef]
- Tcherkezian, J.; Cargnello, M.; Romeo, Y.; Huttlin, E.L.; Lavoie, G.; Gygi, S.P.; Roux, P.P. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014, 28, 357–371. [Google Scholar] [CrossRef]
- Lahr, R.; Mack, S.M.; Héroux, A.; Blagden, S.P.; Bousquet-Antonelli, C.; Deragon, J.-M.; Berman, A.J. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5′TOP sequence. Nucleic Acids Res. 2015, 43, 8077–8088. [Google Scholar] [CrossRef]
- Lahr, R.; Fonseca, B.D.; Ciotti, G.E.; A Al-Ashtal, H.; Jia, J.-J.; Niklaus, M.R.; Blagden, S.P.; Alain, T.; Berman, A. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Blagden, S.P.; Gatt, M.K.; Archambault, V.; Lada, K.; Ichihara, K.; Lilley, K.S.; Inoue, Y.H.; Glover, D.M. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 2009, 334, 186–197. [Google Scholar] [CrossRef]
- Burrows, C.; Latip, N.A.; Lam, S.-J.; Carpenter, L.; Sawicka, K.; Tzolovsky, G.; Gabra, H.; Bushell, M.; Glover, D.M.; Willis, A.E.; et al. The RNA binding protein Larp1 regulates cell division, apoptosis, and cell migration. Nucleic Acids Res. 2010, 38, 5542–5553. [Google Scholar] [CrossRef]
- Xie, C.; Huang, L.; Xie, S.-B.; Xie, D.-Y.; Zhang, G.-L.; Wang, P.; Peng, L.; Gao, Z.-L. LARP1 predict the prognosis for early-stage and AFP-normal hepatocellular carcinoma. J. Transl. Med. 2013, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, J.; Lu, H.; Lin, B.; Cai, S.; Guo, J.; Zang, F.; Chen, R. LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma. Oncol. Rep. 2017, 38, 3659–3667. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Lin, S.-T.; Mi, Y.-S.; Liu, Y.; Ma, Y.; Sun, H.-M.; Peng, Z.-H.; Fan, J.-W. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumor Biol. 2016, 37, 14585–14594. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Adachi, S.; Homoto, M.; Kusano, H.; Koike, K.; Natsume, T. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013, 587, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Gentilella, A.; Morón-Duran, F.D.; Fuentes, P.; Rocha, G.; Riaño-Canalias, F.; Pelletier, J.; Ruiz, M.; Turón, G.; Castaño, J.; Tauler, A.; et al. Autogenous Control of 5′TOP mRNA Stability by 40S Ribosomes. Mol. Cell 2017, 67, 55–70. [Google Scholar] [CrossRef]
- Hong, S.; Freeberg, M.; Han, T.; Kamath, A.; Yao, Y.; Fukuda, T.; Suzuki, T.; Kim, J.K.; Inoki, K. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife 2017, 6. [Google Scholar] [CrossRef]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.A.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-Regulated Phosphoproteome Reveals a Mechanism of mTORC1-Mediated Inhibition of Growth Factor Signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Ma, X.M.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic Analysis Identifies Grb10 as an mTORC1 Substrate That Negatively Regulates Insulin Signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.A.; Pacold, M.E.; Cervantes, C.L.; Lim, D.; Lou, H.J.; Ottina, K.; Gray, N.S.; Turk, B.E.; Yaffe, M.B.; Sabatini, D.M. mTORC1 Phosphorylation Sites Encode Their Sensitivity to Starvation and Rapamycin. Science 2013, 341, 1236566. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.; Hopkins, T.G.; Michael, T.; Abd-Latip, N.; Weir, J.; Aboagye, E.; Mauri, F.; Jameson, C.; Sturge, J.; Gabra, H.; et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 2014, 34, 5025–5036. [Google Scholar] [CrossRef]
- Quiocho, F.A.; Hu, G.; Gershon, P.D. Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol. 2000, 10, 78–86. [Google Scholar] [CrossRef]
- Tomoo, K.; Shen, X.; Okabe, K.; Nozoe, Y.; Fukuhara, S.; Morino, S.; Sasaki, M.; Taniguchi, T.; Miyagawa, H.; Kitamura, K.; et al. Structural features of human initiation factor 4E, studied by X-ray crystal analyses and molecular dynamics simulations. J. Mol. Biol. 2003, 328, 365–383. [Google Scholar] [CrossRef]
- Shama, S.; Avni, D.; Frederickson, R.M.; Sonenberg, N.; Meyuhas, O. Overexpression of Initiation Factor eIF-4E Does Not Relieve the Translational Repression of Ribosomal Protein mRNAs in Quiescent Cells. Gene Expr. 2018, 4, 241–252. [Google Scholar]
- Al-Ashtal, H.A.; Rubottom, C.M.; Leeper, T.C.; Berman, A. The LARP1 La-Module recognizes both ends of TOP mRNAs. RNA Biol. 2019, 1–11. [Google Scholar] [CrossRef]
- Huichalaf, C.; Schoser, B.; Schneider-Gold, C.; Jin, B.; Sarkar, P.; Timchenko, L.T. Reduction of the rate of protein translation in patients with myotonic dystrophy 2. J. Neurosci. 2009, 29, 9042–9049. [Google Scholar] [CrossRef]
- Pellizzoni, L.; Lotti, F.; Rutjes, S.A.; Pierandrei-Amaldi, P. Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′UTR of L4 ribosomal protein mRNA. J. Mol. Biol. 1998, 281, 593–608. [Google Scholar] [CrossRef]
- Liquori, C.L. Myotonic Dystrophy Type 2 Caused by a CCTG Expansion in Intron 1 of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef]
- Schoser, B.; Timchenko, L.T. Myotonic Dystrophies 1 and 2: Complex Diseases with Complex Mechanisms. Curr. Genom. 2010, 11, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Stock, L.; Schneider-Gold, C.; Sommer, C.; Timchenko, N.A.; Timchenko, L. Reduction of Cellular Nucleic Acid Binding Protein Encoded by a Myotonic Dystrophy Type 2 Gene Causes Muscle Atrophy. Mol. Cell. Biol. 2018, 38, mcb–00649. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, R.; Hamel, F.; Beaulieu, D.; Patry, L.; Haineault, C.; Tarnopolsky, M.; Schoser, B.; Puymirat, J. Absence of a differentiation defect in muscle satellite cells from DM2 patients. Neurobiol. Dis. 2009, 36, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Gold, C.; Timchenko, L.T. CCUG Repeats Reduce the Rate of Global Protein Synthesis in Myotonic Dystrophy Type 2. Rev. Neurosci. 2010, 21, 19–28. [Google Scholar] [CrossRef]
- Benhalevy, D.; Gupta, S.K.; Danan, C.H.; Ghosal, S.; Sun, H.-W.; Kazemier, H.G.; Paeschke, K.; Hafner, M.; Juranek, S.A. The Human CCHC-type Zinc Finger Nucleic Acid-Binding Protein Binds G-Rich Elements in Target mRNA Coding Sequences and Promotes Translation. Cell Rep. 2017, 18, 2979–2990. [Google Scholar] [CrossRef]
- Fay, M.M.; Lyons, S.M.; Ivanov, P. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017, 429, 2127–2147. [Google Scholar] [CrossRef]
- Sauer, M.; Juranek, S.A.; Marks, J.; de Magis, A.; Kazemier, H.G.; Hilbig, D.; Benhalevy, D.; Wang, X.; Hafner, M.; Paeschke, K. DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat. Commun. 2019, 10, 2421. [Google Scholar] [CrossRef]
- Ghosal, G.; Muniyappa, K. Hoogsteen base-pairing revisited: Resolving a role in normal biological processes and human diseases. Biochem. Biophys. Res. Commun. 2006, 343, 1–7. [Google Scholar] [CrossRef]
- Dember, L.M.; Kim, N.D.; Liu, K.-Q.; Anderson, P. Individual RNA Recognition Motifs of TIA-1 and TIAR Have Different RNA Binding Specificities. J. Biol. Chem. 1996, 271, 2783–2788. [Google Scholar] [CrossRef]
- Gueydan, C.; Droogmans, L.; Chalon, P.; Huez, G.; Caput, D.; Kruys, V. Identification of TIAR as a Protein Binding to the Translational Regulatory AU-rich Element of Tumor Necrosis Factor α mRNA. J. Biol. Chem. 1999, 274, 2322–2326. [Google Scholar] [CrossRef]
- de Silanes, I.L.; Galbán, S.; Martindale, J.L.; Yang, X.; Mazan-Mamczarz, K.; Indig, F.E.; Falco, G.; Zhan, M.; Gorospe, M. Identification and Functional Outcome of mRNAs Associated with RNA-Binding Protein TIA-1. Mol. Cell. Biol. 2005, 25, 9520–9531. [Google Scholar] [CrossRef]
- Mazan-Mamczarz, K.; Lal, A.; Martindale, J.L.; Kawai, T.; Gorospe, M. Translational Repression by RNA-Binding Protein TIAR. Mol. Cell. Biol. 2006, 26, 2716–2727. [Google Scholar] [CrossRef]
- Piecyk, M.; Wax, S.; Beck, A.R.; Kedersha, N.; Gupta, M.; Maritim, B.; Chen, S.; Gueydan, C.; Kruys, V.; Streuli, M.; et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000, 19, 4154–4163. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence That Ternary Complex (eIF2-GTP-tRNAiMet)–Deficient Preinitiation Complexes Are Core Constituents of Mammalian Stress Granules. Mol. Biol. Cell 2002, 13, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604. [Google Scholar] [CrossRef]
- Kedersha, N.; Cho, M.R.; Li, W.; Yacono, P.W.; Chen, S.; Gilks, N.; Golan, D.E.; Anderson, P. Dynamic Shuttling of Tia-1 Accompanies the Recruitment of mRNA to Mammalian Stress Granules. J. Cell Biol. 2000, 151, 1257–1268. [Google Scholar] [CrossRef]
- Damgaard, C.K.; Lykke-Andersen, J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011, 25, 2057–2068. [Google Scholar] [CrossRef]
- Kim, H.S.; Kuwano, Y.; Zhan, M.; Pullmann, R.; Mazan-Mamczarz, K.; Li, H.; Kedersha, N.; Anderson, P.; Wilce, M.C.J.; Gorospe, M.; et al. Elucidation of a C-Rich Signature Motif in Target mRNAs of RNA-Binding Protein TIAR. Mol. Cell. Biol. 2007, 27, 6806–6817. [Google Scholar] [CrossRef] [PubMed]
- Wagner, B.J.; de Maria, C.T.; Sun, Y.; Wilson, G.; Brewer, G. Structure and Genomic Organization of the Human AUF1 Gene: Alternative Pre-mRNA Splicing Generates Four Protein Isoforms. Genomics 1998, 48, 195–202. [Google Scholar] [CrossRef] [PubMed]
- de Maria, C.T.; Sun, Y.; Long, L.; Wagner, B.J.; Brewer, G. Structural Determinants in AUF1 Required for High Affinity Binding to A + U-rich Elements. J. Biol. Chem. 1997, 272, 27635–27643. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.; Sun, Y.; Lu, H.; Brewer, G. Assembly of AUF1 Oligomers on U-rich RNA Targets by Sequential Dimer Association. J. Biol. Chem. 1999, 274, 33374–33381. [Google Scholar] [CrossRef]
- Meyer, A.; Golbik, R.P.; Sänger, L.; Schmidt, T.; Behrens, S.-E.; Friedrich, S. The RGG/RG motif of AUF1 isoform p45 is a key modulator of the protein’s RNA chaperone and RNA annealing activities. RNA Biol. 2019, 16, 960–971. [Google Scholar] [CrossRef]
- Zucconi, B.E.; Ballin, J.D.; Brewer, B.Y.; Ross, C.R.; Huang, J.; Toth, E.A.; Wilson, G. Alternatively Expressed Domains of AU-rich Element RNA-binding Protein 1 (AUF1) Regulate RNA-binding Affinity, RNA-induced Protein Oligomerization, and the Local Conformation of Bound RNA Ligands. J. Biol. Chem. 2010, 285, 39127–39139. [Google Scholar] [CrossRef]
- White, E.J.; Brewer, G.; Wilson, G. Post-transcriptional control of gene expression by AUF1: Mechanisms, physiological targets, and regulation. Biochim. Biophys. Acta Bioenerg. 2012, 1829, 680–688. [Google Scholar] [CrossRef]
- Hendrayani, S.-F.; Al-Harbi, B.; Al-Ansari, M.; Silva, G.; Aboussekhra, A. The inflammatory/cancer-related IL-6/STAT3/NF-κB positive feedback loop includes AUF1 and maintains the active state of breast myofibroblasts. Oncotarget 2016, 7, 41974–41985. [Google Scholar] [CrossRef]
- Hendrayani, S.-F.; Al-Khalaf, H.H.; Aboussekhra, A. The Cytokine IL-6 Reactivates Breast Stromal Fibroblasts through Transcription Factor STAT3-dependent Up-regulation of the RNA-binding Protein AUF1. J. Biol. Chem. 2014, 289, 30962–30976. [Google Scholar] [CrossRef]
- Al-Khalaf, H.H.; Aboussekhra, A. AUF1 positively controls angiogenesis through mRNA stabilization-dependent up-regulation of HIF-1α and VEGF-A in human osteosarcoma. Oncotarget 2019, 10, 4868–4879. [Google Scholar] [CrossRef]
- Liao, B.; Hu, Y.; Brewer, G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007, 14, 511–518. [Google Scholar] [CrossRef]
- Yoon, J.-H.; de, S.; Srikantan, S.; Abdelmohsen, K.; Grammatikakis, I.; Kim, J.; Kim, K.M.; Noh, J.H.; White, E.J.F.; Martindale, J.L.; et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014, 5, 5248. [Google Scholar] [CrossRef] [PubMed]
- Kakegawa, T.; Ohuchi, N.; Hayakawa, A.; Hirata, S.; Matsuda, M.; Kogure, K.; Kobayashi, H.; Inoue, A.; Kaspar, R.L. Identification of AUF1 as a rapamycin-responsive binding protein to the 5′-terminal oligopyrimidine element of mRNAs. Arch. Biochem. Biophys. 2007, 465, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, T.G.; Mura, M.; Al-Ashtal, H.A.; Lahr, R.M.; Abd-Latip, N.; Sweeney, K.; Lu, H.; Weir, J.; El-Bahrawy, M.; Steel, J.H.; et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 2015, 44, 1227–1246. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Farr, G.W.; Fernandez, C.F.; Lauden, L.; McCormack, J.C.; Wolin, S.L. Yeast Gis2 and Its Human Ortholog CNBP Are Novel Components of Stress-Induced RNP Granules. PLoS ONE 2012, 7, e52824. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lin, L.; Zhao, W.; Li, X.; Wang, Y.; Si, X.; Wang, T.; Wu, H.; Zhai, X.; Zhong, X.; et al. AUF1 is recruited to the stress granules induced by coxsackievirus B3. Virus Res. 2014, 192, 52–61. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cockman, E.; Anderson, P.; Ivanov, P. TOP mRNPs: Molecular Mechanisms and Principles of Regulation. Biomolecules 2020, 10, 969. https://doi.org/10.3390/biom10070969
Cockman E, Anderson P, Ivanov P. TOP mRNPs: Molecular Mechanisms and Principles of Regulation. Biomolecules. 2020; 10(7):969. https://doi.org/10.3390/biom10070969
Chicago/Turabian StyleCockman, Eric, Paul Anderson, and Pavel Ivanov. 2020. "TOP mRNPs: Molecular Mechanisms and Principles of Regulation" Biomolecules 10, no. 7: 969. https://doi.org/10.3390/biom10070969
APA StyleCockman, E., Anderson, P., & Ivanov, P. (2020). TOP mRNPs: Molecular Mechanisms and Principles of Regulation. Biomolecules, 10(7), 969. https://doi.org/10.3390/biom10070969





