Metabolic Roles of Plant Mitochondrial Carriers
Abstract
1. Introduction
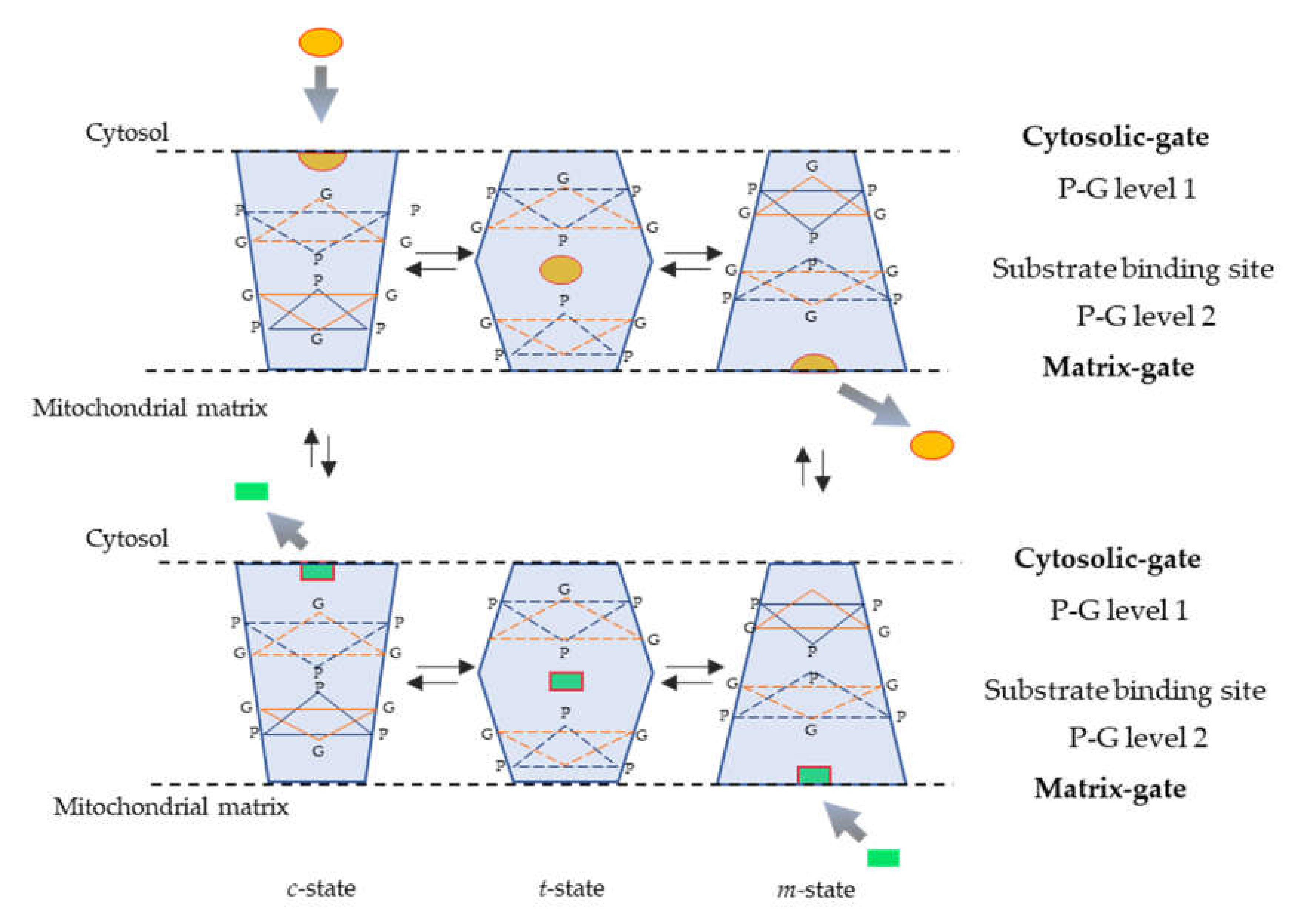
2. Structure and Transport Mechanisms
3. Extension of the MCF
4. Biochemical Characterization of Plant MCF Members
4.1. Coenzyme A Carriers
4.2. Nicotinamide Adenine Dinucleotide (NAD) Carriers
4.3. Adenylate Carriers
4.4. Amino Acid Carriers
4.5. Uncoupling Proteins
4.6. Dicarboxylate Carriers
4.7. Dicarboxylate/Tricarboxylate Carrier
4.8. Succinate/Fumarate Carriers
4.9. Phosphate Carriers
4.10. Pyruvate Carriers
4.11. Iron Transporters (Mitoferrins)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. CB 2017, 27, 1177–1192. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Eastmond, P.J.; Astley, H.M.; Parsley, K.; Aubry, S.; Williams, B.P.; Menard, G.N.; Craddock, C.P.; Nunes-Nesi, A.; Fernie, A.R.; Hibberd, J.M. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Giege, P.; Heazlewood, J.L.; Roessner-Tunali, U.; Millar, A.H.; Fernie, A.R.; Leaver, C.J.; Sweetlove, L.J. Enzymes of glycolysis are functionally associated with the mitochondrion in arabidopsis cells. Plant Cell 2003, 15, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Picault, N.; Hodges, M.; Paimieri, L.; Palmieri, F. The growing family of mitochondrial carriers in arabidopsis. Trends Plant Sci. 2004, 9, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Mintz-Oron, S.; Meir, S.; Malitsky, S.; Ruppin, E.; Aharoni, A.; Shlomi, T. Reconstruction of arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 339–344. [Google Scholar] [CrossRef]
- Smith, A.C.; Robinson, A.J. A metabolic model of the mitochondrion and its use in modelling diseases of the tricarboxylic acid cycle. BMC Syst. Biol. 2011, 5, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Millar, A.H. The plant mitochondrial transportome: Balancing metabolic demands with energetic constraints. Trends Plant Sci. 2016, 21, 662–676. [Google Scholar] [CrossRef]
- Colombini, M. Candidate for the permeability pathway of the outer mitochondrial-membrane. Nature 1979, 279, 643–645. [Google Scholar] [CrossRef]
- Benz, R.; Kottke, M.; Brdiczka, D. The cationically selective state of the mitochondrial outer-membrane pore—A study with intact mitochondria and reconstituted mitochondrial porin. Biochim. Biophys. Acta 1990, 1022, 311–318. [Google Scholar] [CrossRef]
- Pfaff, E.; Klingenberg, M.; Ritt, E.; Vogell, W. Correlation of unspecific permeable mitochondrial spaces with intermembrane spaces. Eur. J. Biochem. 1968, 5, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Pierri, C.L.; De Grassi, A.; Nunes-Nesi, A.; Fernie, A.R. Evolution, structure and function of mitochondrial carriers: A review with new insights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Pierri, C.L. Mitochondrial metabolite transport. Essays in Biochem. 2010, 47, 37–52. [Google Scholar]
- Palmieri, F.; Pierri, C.L. Structure and function of mitochondrial carriers—Role of the transmembrane helix p and g residues in the gating and transport mechanism. Febs Lett. 2010, 584, 1931–1939. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef] [PubMed]
- Monne, M.; Miniero, D.V.; Obata, T.; Daddabbo, L.; Palmieri, L.; Vozza, A.; Nicolardi, M.C.; Fernie, A.R.; Palmieri, F. Functional characterization and organ distribution of three mitochondrial ATP-Mg/Pi carriers in Arabidopsis thaliana. Biochim. Biophys. Acta-Bioenerg. 2015, 1847, 1220–1230. [Google Scholar] [CrossRef]
- Kunji, E.R.S.; Robinson, A.J. Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers. Curr. Opin. Struct. Biol. 2010, 20, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family (slc25): Physiological and pathological implications. Pflug. Arch.-Eur. J. Physiol. 2004, 447, 689–709. [Google Scholar] [CrossRef]
- Klingenberg, M. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 1978–2021. [Google Scholar] [CrossRef]
- Satrustegui, J.; Pardo, B.; Del Arco, A. Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol. Rev. 2007, 87, 29–67. [Google Scholar] [CrossRef]
- Monne, M.; Miniero, D.V.; Daddabbo, L.; Palmieri, L.; Porcelli, V.; Palmieri, F. Mitochondrial transporters for ornithine and related amino acids: A review. Amino Acids 2015, 47, 1763–1777. [Google Scholar] [CrossRef]
- Monne, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial carriers for aspartate, glutamate and other amino acids: A review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. Mitochondrial transporters of the slc25 family and associated diseases: A review. J. Inherit. Metab. Dis. 2014, 37, 565–575. [Google Scholar] [CrossRef]
- Robinson, A.J.; Kunji, E.R.; Gross, A. Mitochondrial carrier homolog 2 (MTCH2): The recruitment and evolution of a mitochondrial carrier protein to a critical player in apoptosis. Exp. Cell Res. 2012, 318, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, J.J.; Kunji, E.R.S. The SLC25 mitochondrial carrier family: Structure and mechanism. Trends Biochem. Sci. 2020, 45, 244–258. [Google Scholar] [CrossRef]
- Taylor, E.B. Functional properties of the mitochondrial carrier system. Trends Cell Biol. 2017, 27, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Colasante, C.; Peña Diaz, P.; Clayton, C.; Voncken, F. Mitochondrial carrier family inventory of Trypanosoma brucei brucei: Identification, expression and subcellular localisation. Mol. Biochem. Parasitol. 2009, 167, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Cavalcanti, J.H.; Fernie, A.R. Characterization of in vivo function(s) of members of the plant mitochondrial carrier family. Biomolecules. under review.
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trezeguet, V.; Lauquin, G.J.M.; Brandolin, R. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Saraste, M.; Walker, J.E. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. Febs Lett. 1982, 144, 250–254. [Google Scholar] [CrossRef]
- Kunji, E.R.S.; Harding, M. Projection structure of the atractyloside-inhibited mitochondrial ADP/ATP carrier of Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 36985–36988. [Google Scholar] [CrossRef]
- Walters, D.E.; Kaplan, R.S. Homology-modeled structure of the yeast mitochondrial citrate transport protein. Biophys. J. 2004, 87, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Tonazzi, A.; Giangregorio, N.; Indiveri, C.; Palmieri, F. Identification by site-directed mutagenesis and chemical modification of three vicinal cysteine residues in rat mitochondrial carnitine/acylcarnitine transporter. J. Biol. Chem. 2005, 280, 19607–19612. [Google Scholar] [CrossRef]
- Wohlrab, H. Novel inter- and intrasubunit contacts between transport-relevant residues of the homodimeric mitochondrial phosphate transport protein. Biochem. Biophys. Res. Commun. 2004, 320, 685–688. [Google Scholar] [CrossRef]
- Cappello, A.R.; Curcio, R.; Miniero, D.V.; Stipani, I.; Robinson, A.J.; Kunji, E.R.S.; Palmieri, F. Functional and structural role of amino acid residues in the even-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J. Mol. Biol. 2006, 363, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Cappello, A.R.; Miniero, D.V.; Curcio, R.; Ludovico, A.; Daddabbo, L.; Stipani, I.; Robinson, A.J.; Kunji, E.R.S.; Palmieri, F. Functional and structural role of amino acid residues in the odd-numbered transmembrane alpha-helices of the bovine mitochondrial oxoglutarate carrier. J. Mol. Biol. 2007, 369, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.J.; Kunji, E.R.S. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. USA 2006, 103, 2617–2622. [Google Scholar] [CrossRef]
- Robinson, A.J.; Overy, C.; Kunji, E.R.S. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. USA 2008, 105, 17766–17771. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; Hellawell, A.M.; Harding, M.; Crichton, P.G.; McCoy, A.J.; Kunji, E.R. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. USA 2014, 111, 426–434. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; King, M.S.; Zogg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R.S. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 2019, 176, 435–447. [Google Scholar] [CrossRef]
- Indiveri, C.; Iacobazzi, V.; Giangregorio, N.; Palmieri, F. The mitochondrial carnitine carrier protein: cDNA cloning, primary structure and comparison with other mitochondrial transport proteins. Biochem. J. 1997, 321, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Palmieri, F.; Runswick, M.J.; Walker, J.E. Identification by bacterial expression and functional reconstitution of the yeast genomic sequence encoding the mitochondrial dicarboxylate carrier protein. Febs Lett. 1996, 399, 299–302. [Google Scholar] [CrossRef]
- Fiermonte, G.; Walker, J.E.; Palmieri, F. Abundant bacterial expression and reconstitution of an intrinsic membrane-transport protein from bovine mitochondria. Biochem. J. 1993, 294, 293–299. [Google Scholar] [CrossRef]
- Palmieri, L.; Lasorsa, F.M.; Iacobazzi, V.; Runswick, M.J.; Palmieri, F.; Walker, J.E. Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. Febs Lett. 1999, 462, 472–476. [Google Scholar] [CrossRef]
- Palmieri, L.; Pardo, B.; Lasorsa, F.M.; del Arco, A.; Kobayashi, K.; Iijima, M.; Runswick, M.J.; Walker, J.E.; Saheki, T.; Satrustegui, J.; et al. Citrin and aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. Embo J. 2001, 20, 5060–5069. [Google Scholar] [CrossRef]
- Palmieri, L.; Rottensteiner, H.; Girzalsky, W.; Scarcia, P.; Palmieri, F.; Erdmann, R. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. Embo J. 2001, 20, 5049–5059. [Google Scholar] [CrossRef]
- Marobbio, C.M.T.; Giannuzzi, G.; Paradies, E.; Pierri, C.L.; Palmieri, F. Alpha-isopropylmalate, a leucine biosynthesis intermediate in yeast, is transported by the mitochondrial oxalacetate carrier. J. Biol. Chem. 2008, 283, 28445–28453. [Google Scholar] [CrossRef]
- Marobbio, C.M.T.; Di Noia, M.A.; Palmieri, F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: Bacterial expression, reconstitution and functional characterization. Biochem. J. 2006, 393, 441–446. [Google Scholar] [CrossRef][Green Version]
- Todisco, S.; Agrimi, G.; Castegna, A.; Palmieri, F. Identification of the mitochondrial NAD(+) transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 1524–1531. [Google Scholar] [CrossRef]
- Picault, N.; Palmieri, L.; Pisano, I.; Hodges, M.; Palmieri, F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria—Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2002, 277, 24204–24211. [Google Scholar] [CrossRef]
- Monne, M.; Daddabbo, L.; Gagneul, D.; Obata, T.; Hielscher, B.; Palmieri, L.; Miniero, D.V.; Fernie, A.R.; Weber, A.P.M.; Palmieri, F. Uncoupling proteins 1 and 2 (UCP1 and UCP2) from Arabidopsis thaliana are mitochondrial transporters of aspartate, glutamate, and dicarboxylates. J. Biol. Chem. 2018, 293, 4213–4227. [Google Scholar] [CrossRef]
- Toleco, M.R.; Naake, T.; Zhang, Y.; Heazlewood, J.L.; Fernie, A.R. Plant mitochondrial carriers: Molecular gatekeepers that help to regulate plant central carbon metabolism. Plants 2020, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. Orthofinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Mutwil, M.; Klie, S.; Tohge, T.; Giorgi, F.M.; Wilkins, O.; Campbell, M.M.; Fernie, A.R.; Usadel, B.; Nikoloski, Z.; Persson, S. Planet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 2011, 23, 895–910. [Google Scholar] [CrossRef]
- Ruprecht, C.; Mendrinna, A.; Tohge, T.; Sampathkumar, A.; Klie, S.; Fernie, A.R.; Nikoloski, Z.; Persson, S.; Mutwil, M. Famnet: A framework to identify multiplied modules driving pathway expansion in plants. Plant Physiol. 2016, 170, 1878–1894. [Google Scholar] [CrossRef] [PubMed]
- Iacobazzi, V.; Lauria, G.; Palmieri, F. Organization and sequence of the human gene for the mitochondrial citrate transport protein. DNA Seq. 1997, 7, 127–139. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Naglieri, M.A.; Stanley, C.A.; Wanders, R.J.A.; Palmieri, F. The structure and organization of the human carnitine/acylcarnitine translocase (CACT1) gene. Biochem. Biophys. Res. Commun. 1998, 252, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Fiermonte, G.; Dolce, V.; Arrigoni, R.; Runswick, M.J.; Walker, J.E.; Palmieri, F. Organization and sequence of the gene for the human mitochondrial dicarboxylate carrier: Evolution of the carrier family. Biochem. J. 1999, 344, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouze, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The tiny eukaryote ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef]
- Soltis, D.E.; Bell, C.D.; Kim, S.; Soltis, P.S. Origin and early evolution of angiosperms. Ann. N. Y. Acad. Sci. 2008, 1133, 3–25. [Google Scholar] [CrossRef]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Ann. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Vandepoele, K.; Simillion, C.; Van de Peer, Y. Detecting the undetectable: Uncovering duplicated segments in arabidopsis by comparison with rice. Trends Genet. 2002, 18, 606–608. [Google Scholar] [CrossRef]
- Thomas, B.C.; Pedersen, B.; Freeling, M. Following tetraploidy in an arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006, 16, 934–946. [Google Scholar] [CrossRef]
- Zallot, R.; Agrimi, G.; Lerma-Ortiz, C.; Teresinski, H.J.; Frelin, O.; Ellens, K.W.; Castegna, A.; Russo, A.; de Crecy-Lagard, V.; Mullen, R.T.; et al. Identification of mitochondrial coenzyme a transporters from maize and arabidopsis. Plant Physiol. 2013, 162, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Prohl, C.; Pelzer, W.; Diekert, K.; Kmita, H.; Bedekovics, T.; Kispal, G.; Lill, R. The yeast mitochondrial carrier leu5p and its human homologue graves’ disease protein are required for accumulation of coenzyme A in the matrix. Mol. Cell. Biol. 2001, 21, 1089–1097. [Google Scholar] [CrossRef]
- Agrimi, G.; Russo, A.; Scarcia, P.; Palmieri, F. The human gene SLC25A17 encodes a peroxisomal transporter of coenzyme a, FAD and NAD+. Biochem. J. 2012, 443, 241–247. [Google Scholar] [CrossRef] [PubMed]
- van Roermund, C.W.; Schroers, M.G.; Wiese, J.; Facchinelli, F.; Kurz, S.; Wilkinson, S.; Charton, L.; Wanders, R.J.; Waterham, H.R.; Weber, A.P.; et al. The peroxisomal NAD carrier from arabidopsis imports NAD in exchange with AMP. Plant Physiol. 2016, 171, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F.; Rieder, B.; Ventrella, A.; Blanco, E.; Do, P.T.; Nunes-Nesi, A.; Trauth, A.U.; Fiermonte, G.; Tjaden, J.; Agrimi, G.; et al. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 2009, 284, 31249–31259. [Google Scholar] [CrossRef]
- Senkler, J.; Senkler, M.; Eubel, H.; Hildebrandt, T.; Lengwenus, C.; Schertl, P.; Schwarzlander, M.; Wagner, S.; Wittig, I.; Braun, H.P. The mitochondrial complexome of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2017, 89, 1079–1092. [Google Scholar] [CrossRef]
- Bedhomme, M.; Hoffmann, M.; McCarthy, E.A.; Gambonnet, B.; Moran, R.G.; Rebeille, F.; Ravanel, S. Folate metabolism in plants—An arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J. Biol. Chem. 2005, 280, 34823–34831. [Google Scholar] [CrossRef]
- de Souza Chaves, I.; Feitosa-Araujo, E.; Florian, A.; Medeiros, D.B.; da Fonseca-Pereira, P.; Charton, L.; Heyneke, E.; Apfata, J.A.C.; Pires, M.V.; Mettler-Altmann, T.; et al. The mitochondrial nad(+) transporter (ndt1) plays important roles in cellular NAD(+) homeostasis in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2019, 100, 487–504. [Google Scholar] [CrossRef] [PubMed]
- da Fonseca-Pereira, P.; Neri-Silva, R.; Cavalcanti, J.H.F.; Brito, D.S.; Weber, A.P.M.; Araujo, W.L.; Nunes-Nesi, A. Data-mining bioinformatics: Connecting adenylate transport and metabolic responses to stress. Trends Plant Sci. 2018, 23, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Pena-Diaz, P.; Pelosi, L.; Ebikeme, C.; Colasante, C.; Gao, F.; Bringaud, F.; Voncken, F. Functional characterization of tbMCP5, a conserved and essential ADP/ATP carrier present in the mitochondrion of the human pathogen Trypanosoma brucei. J. Biol. Chem. 2012, 287, 41861–41874. [Google Scholar] [CrossRef]
- Gnipová, A.; Šubrtová, K.; Panicucci, B.; Horváth, A.; Lukeš, J.; Zíková, A. The ADP/ATP carrier and its relationship to oxidative phosphorylation in ancestral protist Trypanosoma brucei. Eukaryot. Cell 2015, 14, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsasser, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kuhn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from arabidopsis mitochondrial proteomics. Plant J. Cell Mol. Biol. 2020, 101, 420–441. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Santoro, A.; Carrari, F.; Blanco, E.; Nunes-Nesi, A.; Arrigoni, R.; Genchi, F.; Fernie, A.R.; Palmieri, F. Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from arabidopsis. Plant Physiol. 2008, 148, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Rocha, A.G.; Robinson, A.J.; Kmiecik, P.; Vothknecht, U.C.; Teige, M. Arabidopsis calcium-binding mitochondrial carrier proteins as potential facilitators of mitochondrial ATP-import and plastid SAM-import. FEBS Lett. 2011, 585, 3935–3940. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, A.; Lorenz, M.; Vothknecht, U.C.; Niopek-Witz, S.; Neuhaus, H.E.; Haferkamp, I. In vitro analyses of mitochondrial atp/phosphate carriers from Arabidopsis thaliana revealed unexpected Ca2+-effects. BMC Plant Biol. 2015, 15, 238–254. [Google Scholar] [CrossRef]
- Monne, M.; Daddabbo, L.; Giannossa, L.C.; Nicolardi, M.C.; Palmieri, L.; Miniero, D.V.; Mangone, A.; Palmieri, F. Mitochondrial ATP-Mg/phosphate carriers transport divalent inorganic cations in complex with ATP. J. Bioenerg. Biomembr. 2017, 49, 369–380. [Google Scholar] [CrossRef]
- Fiore, C.; Trezeguet, V.; Le Saux, A.; Roux, P.; Schwimmer, C.; Dianoux, A.C.; Noel, F.; Lauquin, G.J.-M.; Brandolin, G.; Vignais, P.V. The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochemie. 1998, 80, 137–150. [Google Scholar] [CrossRef]
- Fiermonte, G.; Paradies, E.; Todisco, S.; Marobbio, C.M.T.; Palmieri, F. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme a and adenosine 3‘,5‘-diphosphate in human mitochondria. J. Biol. Chem. 2009, 284, 18152–18159. [Google Scholar] [CrossRef] [PubMed]
- Fiermonte, G.; De Leonardis, F.; Todisco, S.; Palmieri, L.; Lasorsa, F.M.; Palmieri, F. Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. J. Biol. Chem. 2004, 279, 30722–30730. [Google Scholar] [CrossRef] [PubMed]
- Traba, J.; Froschauer, E.; Wiesenberger, G.; Satrustegui, J.; del Arco, A. Yeast mitochondria import ATP through the calcium-dependent ATP-Mg/Pi carrier Sal1p, and are ATP consumers during aerobis growth in glucose. Mol. Microbiol. 2008, 69, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Traba, J.; Satrustegui, J.; del Arco, A. Characterization of SCaMC-3-like/slc25a41, a novel calcium-independent mitochondrial ATP-Mg/Pi carrier. Biochem. J. 2009, 418, 125–133. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dolce, V.; Fiermonte, F.; Runswick, M.J.; Palmieri, F.; Walker, J.E. The human mitochondrial deoxynucleotide carrier and its role in toxicity of nucleoside antivirals. Proc. Natl Acad. Sci. USA. 2001, 98, 2284–2288. [Google Scholar] [CrossRef] [PubMed]
- Marobbio, C.M.T.; Vozza, A.; Harding, M.; Bisaccia, F.; Palmieri, F.; Walker, J.E. Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 2002, 21, 5653–5661. [Google Scholar] [CrossRef]
- Lindhurst, M.J.; Fiermonte, G.; Song, S. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc. Natl Acad. Sci. USA 2006, 103, 15927–15932. [Google Scholar] [CrossRef]
- Floyd, S.; Favre, C.; Lasorsa, F.M. The IGF-I–mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell. 2007, 18, 3545–3555. [Google Scholar] [CrossRef]
- Tzagoloff, A.; Jang, J.; Glerum, D.M.; Wu, M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 1996, 271, 7392–7397. [Google Scholar] [CrossRef]
- Titus, S.A.; Moran, R.G. Retrovirally mediated complementation of the glyB phenotype: cloning of a human gene encoding the carrier for entry of folates into mitochondria. J. Biol. Chem. 2000, 275, 36811–36817. [Google Scholar] [CrossRef]
- Vozza, A.; Blanco, E.; Palmieri, L.; Palmieri, F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20850–20857. [Google Scholar] [CrossRef]
- Fiermonte, G.; Palmieri, L.; Dolce, V.; Lasorsa, F.M.; Palmieri, F.; Runswick, M.J.; Walker, J.E. The sequence, bacterial expression and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. J. Biol. Chem. 1998, 273, 24754–24759. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Picault, N.; Arrigoni, R.; Besin, E.; Palmieri, F.; Hodges, M. Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: Organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 2008, 410, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Brito, D.S.; Agrimi, G.; Charton, L.; Brilhaus, D.; Bitetto, M.G.; Lana-Costa, J.; Messina, E.; Nascimento, C.P.; Araujo, E.F.; Viana Pires, M.; et al. Biochemical and functional characterization of a mitochondrial citrate carrier in Arabidopsis thaliana. Biochem. J. 2020, 477, 1759–1777. [Google Scholar] [CrossRef] [PubMed]
- Catoni, E.; Schwab, R.; Hilpert, M.; Desimone, M.; Schwacke, R.; Flugge, U.I.; Schumacher, K.; Frommer, W.B. Identification of an arabidopsis mitochondrial succinate-fumarate translocator. Febs Lett. 2003, 534, 87–92. [Google Scholar] [CrossRef]
- Palmieri, L.; Lasorsa, F.M.; DePalma, A.; Palmieri, F.; Runswick, M.J.; Walker, J.E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. Febs Lett. 1997, 417, 114–118. [Google Scholar] [CrossRef]
- Kaplan, R.S.; Mayor, J.A.; Wood, D.O. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J. Biol. Chem. 1993, 268, 13682–13690. [Google Scholar] [PubMed]
- Kaplan, R.S.; Mayor, J.A.; Gremse, D.A.; Wood, D.O. High level expression and characterization of the mitochondrial citrate transport protein from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 4108–4114. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, C.; Palmieri, F.; Bisaccia, F.; Kramer, R. Kinetics of the reconstituted 2-oxoglutarate carrier from bovine heart mitochondria. Biochim. Biophys. Acta 1987, 890, 310–318. [Google Scholar] [CrossRef]
- Palmieri, L.; Agrimi, G.; Runswick, M.J.; Fearnley, I.M.; Palmieri, F.; Walker, J.E. Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 2001, 276, 1916–1922. [Google Scholar] [CrossRef]
- Fiermonte, G.; Dolce, V.; Palmieri, L.; Ventura, M.; Runswick, M.J.; Palmieri, F.; Walker, J.E. Identification of the human mitochondrial oxodicarboxylate carrier: bacterial expression, reconstitution, functional characterization, tissue distribution, and chromosomal location. J. Biol. Chem. 2001, 276, 8225–8230. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Vozza, A.; Agrimi, G.; De Marco, V.; Runswick, M.J.; Palmieri, F.; Walker, J.E. Identification of the yeast mitochondrial transporter for oxaloacetate and sulfate. J. Biol. Chem. 1999, 274, 22184–22190. [Google Scholar] [CrossRef]
- Fiermonte, G.; Palmieri, L.; Todisco, S.; Agrimi, G.; Palmieri, F.; Walker, J.E. Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 2002, 277, 19289–19294. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, V.; Vozza, A.; Calcagnile, V.; Gorgoglione, R.; Arrigoni, R.; Fontanesi, F.; Marobbio, C.M.T.; Castegna, A.; Palmieri, F.; Palmieri, L. Molecular identification and functional characterization of a novel glutamate transporter in yeast and plant mitochondria. Biochim. Biophys. Acta-Bioenerg. 2018, 1859, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Cavero, S.; Vozza, A.; del Arco, A. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 2003, 50, 1257–1269. [Google Scholar] [CrossRef]
- Hoyos, M.E.; Palmieri, L.; Wertin, T.; Arrigoni, R.; Polacco, J.C.; Palmieri, F. Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J. 2003, 33, 1027–1035. [Google Scholar] [CrossRef]
- Palmieri, L.; De Marco, V.; Iacobazzi, V.; Palmieri, F.; Runswick, M.J.; Walker, J.E. Identification of the yeast ARG-11 gene as a mitochondrial ornithine carrier involved in arginine biosynthesis. FEBS Lett. 1997, 410, 447–451. [Google Scholar] [CrossRef]
- Fiermonte, G.; Dolce, V.; David, L.; Santorelli, F.M.; Dionisi-Vici, C.; Palmieri, F.; Walker, J.E. The mitochondrial ornithine transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J. Biol. Chem. 2003, 278, 32778–32783. [Google Scholar] [CrossRef]
- Indiveri, C.; Tonazzi, A.; Palmieri, F. Identification and purification of the carnitine carrier from rat liver mitochondria. Biochim. Biophys. Acta 1990, 1020, 81–86. [Google Scholar] [CrossRef]
- Marobbio, C.M.T.; Agrimi, G.; Lasorsa, F.M.; Palmieri, F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003, 22, 5975–5982. [Google Scholar] [CrossRef]
- Agrimi, G.; Di Noia, M.A.; Marobbio, C.M.T.; Fiermonte, G.; Lasorsa, F.M.; Palmieri, F. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 2004, 379, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Arrigoni, R.; Blanco, E.; Carrari, F.; Zanor, M.I.; Studart-Guimarães, C.; Fernie, A.R.; Palmieri, F. Molecular identification of an Arabidopsis thaliana S-adenosylmethionine transporter: analysis of organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Plant Physiol. 2006, 142, 855–865. [Google Scholar] [CrossRef]
- Bouvier, F.; Linka, N.; Isner, J.C.; Mutterer, J.; Weber, A.P.M.; Camara, B. Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 2006, 18, 3088–3105. [Google Scholar] [CrossRef] [PubMed]
- Wohlrab, H.; Briggs, C. Yeast mitochondrial phosphate transport protein expressed in Escherichia coli. Site-directed mutations at threonine-43 and at a similar location in the second tandem repeat (isoleucine-141). Biochemistry 1994, 33, 9371–9375. [Google Scholar] [CrossRef]
- Fiermonte, G.; Dolce, V.; Palmieri, F. Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem. 1998, 273, 22782–22787. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Dashner, Z.S.; Connolly, E.L. Mitochondrial iron transporters (mit1 and mit2) are essential for iron homeostasis and embryogenesis in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1449. [Google Scholar] [CrossRef] [PubMed]
- Lawand, S.; Dorne, A.J.; Long, D.; Coupland, G.; Mache, R.; Carol, P. Arabidopsis a bout de souffle, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light. Plant Cell 2002, 14, 2161–2173. [Google Scholar] [CrossRef] [PubMed]
- Eisenhut, M.; Planchais, S.; Cabassa, C.; Guivarc’h, A.; Justin, A.M.; Taconnat, L.; Renou, J.P.; Linka, M.; Gagneul, D.; Timm, S.; et al. Arabidopsis a bout de souffle is a putative mitochondrial transporter involved in photorespiratory metabolism and is required for meristem growth at ambient CO2 levels. Plant J. Cell Mol. Biol. 2013, 73, 836–849. [Google Scholar] [CrossRef]
- Catoni, E.; Desimone, M.; Hilpert, M.; Wipf, D.; Kunze, R.; Schneider, A.; Fluegge, U.-I.; Schumacher, K.; Frommer, W.B. Expression pattern of a nuclear encoded mitochondrial arginine-ornithine translocator gene from arabidopsis. BMC Plant Biol. 2003, 3, 1–10. [Google Scholar] [CrossRef]
- Palmieri, L.; Todd, C.D.; Arrigoni, R.; Hoyos, M.E.; Santoro, A.; Polacco, J.C.; Palmieri, F. Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim. Biophys. Acta-Bioenerg. 2006, 1757, 1277–1283. [Google Scholar] [CrossRef]
- Taylor, N.L.; Howell, K.A.; Heazlewood, J.L.; Tan, T.Y.; Narsai, R.; Huang, S.; Whelan, J.; Millar, A.H. Analysis of the rice mitochondrial carrier family reveals anaerobic accumulation of a basic amino acid carrier involved in arginine metabolism during seed germination. Plant Physiol. 2010, 154, 691–704. [Google Scholar] [CrossRef]
- Toka, I.; Planchais, S.; Cabassa, C.; Justin, A.M.; De Vos, D.; Richard, L.; Savoure, A.; Carol, P. Mutations in the hyperosmotic stress-responsive mitochondrial basic amino acid carrier2 enhance proline accumulation in arabidopsis. Plant Physiol. 2010, 152, 1851–1862. [Google Scholar] [CrossRef]
- Planchais, S.; Cabassa, C.; Toka, I.; Justin, A.M.; Renou, J.P.; Savoure, A.; Carol, P. Basic amino acid carrier 2 gene expression modulates arginine and urea content and stress recovery in arabidopsis leaves. Front. Plant Sci. 2014, 5, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Borecky, J.; Maia, I.G.; Costa, A.D.T.; Jezek, P.; Chaimovich, H.; de Andrade, P.B.M.; Vercesi, A.E.; Arruda, P. Functional reconstitution of Arabidopsis thaliana plant uncoupling mitochondrial protein (AtPUMP1) expressed in Escherichia coli. Febs Lett. 2001, 505, 240–244. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Lytovchenko, A.; Morgan, M.; Nunes-Nesi, A.; Taylor, N.L.; Baxter, C.J.; Eickmeier, I.; Fernie, A.R. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA. 2006, 103, 19587–19592. [Google Scholar] [CrossRef] [PubMed]
- Vercesi, A.E.; Borecky, J.; Maia Ide, G.; Arruda, P.; Cuccovia, I.M.; Chaimovich, H. Plant uncoupling mitochondrial proteins. Ann. Rev. Plant Biol. 2006, 57, 383–404. [Google Scholar] [CrossRef]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.M.; Morrison, S.; McInerney, P.; Hadi, M.Z.; et al. Isolation and proteomic characterization of the arabidopsis golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef]
- Nikolovski, N.; Rubtsov, D.; Segura, M.P.; Miles, G.P.; Stevens, T.J.; Dunkley, T.P.; Munro, S.; Lilley, K.S.; Dupree, P. Putative glycosyltransferases and other plant golgi apparatus proteins are revealed by lopit proteomics. Plant Physiol. 2012, 160, 1037–1051. [Google Scholar] [CrossRef]
- Colasante, C.; Zheng, F.; Kemp, C.; Voncken, F. A plant-like mitochondrial carrier family protein facilitates mitochondrial transport of di- and tricarboxylates in Trypanosoma brucei. Mol. Biochem. Parasitol. 2018, 221, 36–51. [Google Scholar] [CrossRef]
- Regalado, A.; Pierri, C.L.; Bitetto, M.; Laera, V.L.; Pimentel, C.; Francisco, R.; Passarinho, J.; Chaves, M.M.; Agrimi, G. Characterization of mitochondrial dicarboxylate/tricarboxylate transporters from grape berries. Planta 2013, 237, 693–703. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, Z.; Yang, Y. Molecular cloning and characterization of a mitochondrial dicarboxylate/tricarboxylate transporter gene in Citrus junos response to aluminum stress. Mitochondrial DNA 2008, 19, 376–384. [Google Scholar] [PubMed]
- Spagnoletta, A.; De Santis, A.; Tampieri, E.; Baraldi, E.; Bachi, A.; Genchi, G. Identification and kinetic characterization of HtDTC, the mitochondrial dicarboxylate-tricarboxylate carrier of jerusalem artichoke tubers. J. Bioenerg. Biomembr. 2006, 38, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Spagnoletta, A.; De Santis, A.; Stefanizzi, L.; Palmieri, F. Purification and characterization of the reconstitutively active citrate carrier from maize mitochondria. Plant Physiol. 1999, 120, 841–848. [Google Scholar] [CrossRef]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the arabidopsis endomembrane system. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Haferkamp, I.; Schmitz-Esser, S. The plant mitochondrial carrier family: Functional and evolutionary aspects. Front. Plant Sci. 2012, 3, 2–21. [Google Scholar] [CrossRef]
- Pratt, R.D.; Ferreira, G.C.; Pedersen, P.L. Mitochondrial phosphate-transport—Import of the H+/Pi symporter and role of the presequence. J. Biol. Chem. 1991, 266, 1276–1280. [Google Scholar]
- Stappen, R.; Kramer, R. Kinetic mechanism of phosphate phosphate and phosphate OH- antiports catalyzed by reconstituted phosphate carrier from beef-heart mitochondria. J. Biol. Chem. 1994, 269, 11240–11246. [Google Scholar]
- Zhang, Y.; Swart, C.; Alseekh, S.; Scossa, F.; Jiang, L.; Obata, T.; Graf, A.; Fernie, A.R. The extra-pathway interactome of the TCA cycle: Expected and unexpected metabolic interactions. Plant Physiol. 2018, 177, 966–979. [Google Scholar] [CrossRef]
- Zhang, Y.; Beard, K.F.M.; Swart, C.; Bergmann, S.; Krahnert, I.; Nikoloski, Z.; Graf, A.; Ratcliffe, R.G.; Sweetlove, L.J.; Fernie, A.R.; et al. Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat. Commun. 2017, 8, 15212–15223. [Google Scholar] [CrossRef] [PubMed]
- Bricker, D.K.; Taylor, E.B.; Schell, J.C.; Orsak, T.; Boutron, A.; Chen, Y.C.; Cox, J.E.; Cardon, C.M.; Van Vranken, J.G.; Dephoure, N.; et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, drosophila, and humans. Science 2012, 337, 96–100. [Google Scholar] [CrossRef]
- Herzig, S.; Raemy, E.; Montessuit, S.; Veuthey, J.L.; Zamboni, N.; Westermann, B.; Kunji, E.R.; Martinou, J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science 2012, 337, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Štáfková, J.; Mach, J.; Biran, M.; Verner, Z.; Bringaud, F.; Tachezy, J. Mitochondrial pyruvate carrier in Trypanosoma brucei. Mol. Microbiol. 2016, 100, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Vanderperre, B.; Bender, T.; Kunji, E.R.S.; Martinou, J.C. Mitochondrial pyruvate import and its effects on homeostasis. Curr. Opin. Cell Biol. 2015, 33, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Furumoto, T. Pyruvate transport systems in organelles: Future directions in C4 biology research. Curr. Opin. Plant Biol. 2016, 31, 143–148. [Google Scholar] [CrossRef]
- McCommis, K.S.; Finck, B.N. Mitochondrial pyruvate transport: A historical perspective and future research directions. Biochem. J. 2015, 466, 443–454. [Google Scholar] [CrossRef]
- Bender, T.; Martinou, J.C. The mitochondrial pyruvate carrier in health and disease: To carry or not to carry? Biochim. Biophys. Acta 2016, 1863, 2436–2442. [Google Scholar] [CrossRef]
- Tavoulari, S.; Thangaratnarajah, C.; Mavridou, V.; Harbour, M.E.; Martinou, J.C.; Kunji, E.R. The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Li, C.L.; Wang, M.; Ma, X.Y.; Zhang, W. Nrga1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in arabidopsis. Mol. Plant 2014, 7, 1508–1521. [Google Scholar] [CrossRef]
- Wang, M.; Ma, X.; Shen, J.; Li, C.; Zhang, W. The ongoing story: The mitochondria pyruvate carrier 1 in plant stress response in arabidopsis. Plant Signal. Behav. 2014, 9, 1–4. [Google Scholar] [CrossRef][Green Version]
- Shen, J.L.; Li, C.L.; Wang, M.; He, L.L.; Lin, M.Y.; Chen, D.H.; Zhang, W. Mitochondrial pyruvate carrier 1 mediates abscisic acid-regulated stomatal closure and the drought response by affecting cellular pyruvate content in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 217–229. [Google Scholar] [CrossRef]
- He, L.; Jing, Y.; Shen, J.; Li, X.; Liu, H.; Geng, Z.; Wang, M.; Li, Y.; Chen, D.; Gao, J.; et al. Mitochondrial pyruvate carriers prevent cadmium toxicity by sustaining the TCA cycle and glutathione synthesis. Plant Physiol. 2019, 180, 198–211. [Google Scholar] [CrossRef]
- Shaw, G.C.; Cope, J.J.; Li, L.; Corson, K.; Hersey, C.; Ackermann, G.E.; Gwynn, B.; Lambert, A.J.; Wingert, R.A.; Traver, D.; et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006, 440, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Metzendorf, C.; Wu, W.; Lind, M.I. Overexpression of drosophila mitoferrin in l(2)mbn cells results in dysregulation of Fer1HCH expression. Biochem. J. 2009, 421, 463–471. [Google Scholar] [CrossRef]
- Paradkar, P.N.; Zumbrennen, K.B.; Paw, B.H.; Ward, D.M.; Kaplan, J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 2009, 29, 1007–1016. [Google Scholar] [CrossRef]
- Bashir, K.; Ishimaru, Y.; Nishizawa, N.K. Identification and characterization of the major mitochondrial fe transporter in rice. Plant Signal. Behav. 2011, 6, 1591–1593. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Heazlewood, J.L. Genomic and proteomic analysis of mitochondrial carrier proteins in arabidopsis. Plant Physiol. 2003, 131, 443–453. [Google Scholar] [CrossRef]
- Moore, M.J.; Wofford, J.D.; Dancis, A.; Lindahl, P.A. Recovery of mrs3Δmrs4Δ Saccharomyces cerevisiae cells under iron-sufficient conditions and the role Fe580. Biochemistry 2018, 57, 672–683. [Google Scholar] [CrossRef]
- Brazzolotto, X.; Pierrel, F.; Pelosi, L. Three conserved histidine residues contribute to mitochondrial iron transport through mitoferrins. Biochem. J. 2014, 460, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Christenson, E.T.; Gallegos, A.S.; Banerjee, A. In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J. Biol. Chem. 2018, 293, 3819–3828. [Google Scholar] [CrossRef]
| Dicots | A. thaliana | M. truncatula | G. max | S. lycopersicum | V. vinifera | P. persica | D. carota | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° |
| 1 | 30 | 10 | 46 | 8 | 57 | 8 | 98 | 9 | 23 | 2 | 48 | 12 | 51 | 13 |
| 2 | 20 | 10 | 56 | 4 | 49 | 11 | 56 | 5 | 19 | 2 | 30 | 8 | 44 | 14 |
| 3 | 23 | 9 | 57 | 11 | 46 | 7 | 72 | 7 | 19 | 2 | 27 | 8 | 50 | 4 |
| 4 | 19 | 10 | 44 | 11 | 52 | 10 | 67 | 7 | 24 | 2 | 26 | 2 | 36 | 10 |
| 5 | 27 | 20 | 35 | 9 | 42 | 7 | 67 | 5 | 25 | 3 | 19 | 8 | 42 | 8 |
| 6 | – | – | 49 | 7 | 51 | 9 | 50 | 6 | 22 | 4 | 31 | 12 | 37 | 4 |
| 7 | – | – | 46 | 11 | 45 | 10 | 68 | 1 | 21 | 2 | 22 | 3 | 36 | 14 |
| 8 | – | – | 37 | 21 | 48 | 16 | 66 | 6 | 22 | 2 | 23 | 7 | 32 | 5 |
| 9 | – | – | – | – | 50 | 5 | 73 | 6 | 23 | 4 | – | – | 34 | 2 |
| 10 | – | – | – | – | 52 | 4 | 66 | 4 | 18 | 5 | – | – | – | – |
| 11 | – | – | – | – | 35 | 3 | 57 | 4 | 20 | 1 | – | – | – | – |
| 12 | – | – | – | – | 40 | 2 | 68 | 5 | 23 | 3 | – | – | – | – |
| 13 | – | – | – | – | 46 | 7 | – | – | 24 | 2 | – | – | – | – |
| 14 | – | – | – | – | 49 | 5 | – | – | 30 | 5 | – | – | – | – |
| 15 | – | – | – | – | 52 | 4 | – | – | 20 | 2 | – | – | – | – |
| 16 | – | – | – | – | 38 | 7 | – | – | 22 | 4 | – | – | – | – |
| 17 | – | – | – | – | 42 | 6 | – | – | 17 | 4 | – | – | – | – |
| 18 | – | – | – | – | 58 | 6 | – | – | 29 | 6 | – | – | – | – |
| 19 | – | – | – | – | 51 | 8 | – | – | 24 | 2 | – | – | – | – |
| 20 | – | – | – | – | 48 | 6 | – | – | – | – | – | – | – | – |
| Unknown | – | – | 28 | – | 29 | – | 21 | – | 59 | 3 | 1 | – | 59 | 7 |
| Total | 119 | 59 | 397 | 82 | 978 | 141 | 828 | 65 | 485 | 60 | 227 | 60 | 421 | 81 |
| Monocots | B. distachyon | S. bicolor | Z. mays | O. sativa | H. vulgare | S. italica | M. acuminata | |||||||
| Chr | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° |
| 1 | 75 | 17 | 81 | 15 | 307 | 14 | 43 | 11 | 558 | 5 | 42 | 7 | 28 | 9 |
| 2 | 59 | 17 | 78 | 6 | 244 | 8 | 36 | 7 | 768 | 3 | 49 | 5 | 22 | 6 |
| 3 | 60 | 10 | 74 | 9 | 236 | 7 | 36 | 10 | 700 | 10 | 51 | 7 | 30 | 7 |
| 4 | 49 | 8 | 69 | 9 | 247 | 10 | 36 | 3 | 647 | 9 | 40 | 4 | 30 | 10 |
| 5 | 29 | 4 | 72 | 2 | 224 | 13 | 30 | 8 | 67 | 10 | 47 | 11 | 29 | 6 |
| 6 | – | – | 61 | 5 | 174 | 5 | 31 | 3 | 583 | 5 | 36 | 3 | 35 | 16 |
| 7 | – | – | 66 | 3 | 182 | 4 | 30 | 1 | 657 | 4 | 36 | 5 | 29 | 11 |
| 8 | – | – | 63 | 2 | 181 | 12 | 28 | 3 | – | – | 41 | 3 | 35 | 12 |
| 9 | – | – | 59 | 7 | 160 | 9 | 23 | 6 | – | – | 59 | 18 | 34 | 9 |
| 10 | – | – | 61 | 4 | 151 | 5 | 23 | 2 | – | – | – | – | 34 | 21 |
| 11 | – | – | – | – | – | – | 29 | 5 | – | – | – | – | 26 | 4 |
| 12 | – | – | – | – | – | – | 28 | 2 | – | – | – | – | – | – |
| Unknown | 0 | – | 25 | – | 28 | 3 | 1 | – | 249 | 4 | 4 | – | 140 | – |
| Total | 271 | 56 | 709 | 62 | 2134 | 90 | 374 | 61 | 4229 | 50 | 406 | 63 | 472 | 111 |
| Algae | C. reinhardtii | O. tauri | M. commoda | |||||||||||
| Chr | mbp | MC N° | mbp | MC N° | mbp | MC N° | mbp | MC N° | ||||||
| 1 | 1 | 6 | 1 | 2 | 1 | 5 | 2 | 3 | ||||||
| 2 | 1 | 3 | 1 | 3 | 1 | 3 | 2 | 5 | ||||||
| 3 | 1 | 5 | 1 | 3 | 1 | 5 | 2 | 3 | ||||||
| 4 | 1 | 0 | 0 | 4 | 1 | 0 | 2 | 4 | ||||||
| 5 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 5 | ||||||
| 6 | 1 | 1 | 1 | 9 | 1 | 1 | 1 | 6 | ||||||
| 7 | 1 | 3 | 1 | 2 | 1 | 3 | 1 | 5 | ||||||
| 8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | ||||||
| 9 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | ||||||
| 10 | 1 | 3 | 1 | 3 | 1 | 3 | 1 | 0 | ||||||
| 11 | 1 | 2 | 0 | 1 | 1 | 3 | 1 | 2 | ||||||
| 12 | 1 | 4 | 1 | 2 | 1 | 3 | 1 | 6 | ||||||
| 13 | 1 | 3 | 1 | 0 | 1 | 2 | 1 | 1 | ||||||
| 14 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 3 | ||||||
| 15 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | ||||||
| 16 | 0 | 3 | 1 | 6 | 1 | 2 | 1 | 1 | ||||||
| 17 | 0 | 2 | 1 | 1 | 0 | 3 | 0 | 0 | ||||||
| 18 | 0 | 0 | – | – | 0 | 2 | – | – | ||||||
| 19 | 0 | 0 | – | – | 0 | 0 | – | – | ||||||
| 20 | 1 | 1 | – | – | 0 | 0 | – | – | ||||||
| 21 | 0 | 3 | – | – | – | – | – | – | ||||||
| Unknown | – | – | 10 | 3 | – | – | – | – | ||||||
| Total | 13 | 43 | 21 | 46 | 13 | 39 | 21 | 48 | ||||||
| Subfamilies | Aliases | Main Substrates | Triplets * | References |
|---|---|---|---|---|
| For nucleotides and dinucleotides | ||||
| ADP/ATP | AAC | ADP, ATP | 11 (DNS), 19 (AGT), 23 (KL[G/S]), 84 (TYG), 85 (QRX), 88 (NYV) | [19,80] |
| Coenzyme A/PAP | CoA/PAP | - | 23 (K[V/A]Q), 34 (IVR), 88 ([K/Q]SS) | [65,81] |
| ATP-Mg/Pi | APC | ATP-Mg, ATP-Ca, Pi, AXP | 23 (RQ[Q/A]), 30 (DE[A/T/N]), 84 (EYA), 88 (KDS) | [19,76,82,83,84] |
| Thiamine pyrophosphate | TPC | Thpp, thmp; (d)NDP, (d)NTP | 23 (R[T/S]K), 34 (IT[K/R]), 80 (L[A/T]K), 85 (GAT) | [85,86,87] |
| Pyrimidine nucleotides | PNC | Pyrimidine (deoxy)nucleotides | 19 (G[G/A]K), 27 (CNY), 30 ([D/E]WE), 37 (QQR), 83 ([PEP), 85 (R[I/V][S/T]) | [48,88] |
| FAD/folate | FAD | Folates, FAD | 19 (GGK), 27 (HNY), 30 (DWQ) | [70,89,90] |
| ANT | ANT | ATP, ADP, AMP | 19 (SAK), 30 (DAI), 33 (KAK), 37 (QKR) | [46] |
| NAD+ | NDT/PXN | NAD+, (d)AMP, (d)GMP | 19 (GGK), 27 (CNY), 30 (DWE), 89 (FP[L/F]) | [49,68] |
| GTP/GDP | GGC | GTP, GDP, dgtp, dgdp, ITP, IDP | 22 (EGS), 23 (IEL), 84 (QGK), 85 (RSL), 88 (KLS) | [91] |
| For di-/tri-carboxylates and keto acids | ||||
| Dicarboxylates | DIC | Malate, succinate, phosphate, sulfate, thiosulfate | 26 (TG[C/S]), 27 (H[N/T][S/Q/N]), 33 (K[N/M]K), 88 (RQ[I/L/T]) | [42,92,93] |
| Di-/tri-carboxylates | DTC | Oxoglutarate, citrate | 26 (IGS), 27 (QSL), 33 (KLK), 35 (RRQ), 77 (GTY), 84 (YLH), 88 (RMT), 93 ([K/R]DN) | [50] |
| Citrate/isocitrate | SFC | Citrate, isocitrate, aconitate | 22 (EAG), 84 (KNG), 88 (RNT) | [94,95,96] |
| Citrate | CTP | Citrate, malate, isocitrate, cis-aconitate, PEP | 22 (E[A/S][S/T]), 84 (KN[S/D]), 88 (RRV) | [97,98] |
| 2-oxoglutarate | OGC | 2-Oxoglutarate, malate | 26 (VGS), 27 (QTM), 33 (KLK), 35 (RRR), 77 (GTY), 84 (YVH), 88 (RQT), 93 (TSE) | [43,99] |
| Oxodicarboxylates | ODC | Oxoadipate, oxoglutarate | 22 (EE[A/G]), 77 (PTK), 81 (E[H/N]L) 84 (K[F/W]G), 85 (RNG), 88 (KY[M/L]) | [100,101] |
| Oxaloacetate/sulfate | OAC | Oxaloacetate, sulfate, thiosulfate, a-isopropylmalate | 23 (VAA), 26 (TGM), 30 (E[F/Y]D), 80 (YRR), 84 ([L/M]GH), 88 (RQ[C/S]) | [47,102] |
| For amino acids | ||||
| Glutamate | GC | Glutamate | 22 (GQA), 77 (NTR), 80 (LRV), 84 (EFL), 85 (KSF), 88 (KYA) | [103] |
| Glutamate | BOU | L-Glutamate | - | [104] |
| Aspartate/glutamate | AGC | Aspartate, glutamate, cysteinesulfinate | 22 (GQA), 77 (QCR), 84 (EFQ), 85 (KSF), 88 (KYT) | [45,105] |
| Aspartate/glutamate | UCP1–2 | 23 ([D/E][V/I/S/Q][A/V/T/S]), 88 ([R/K] [D/E][F/M]) | [12,51] | |
| Ornithine | ORC | Ornithine, (lysine, citrulline, arginine, histidine) | 23 ([V/I][A/S]W) but (KSN) in S. cerevisiae, 26 (GL[V/C]) but (ELI) in S. cerevisiae, 84 (EGA), but (QAV) in atbac2 | [106,107,108] |
| Carnitine | CAC | Carnitine, acylcarnitine | 23 (VTW), 85 (FSN) | [44,109] |
| S-adenosylmethionine | SAMC | S-adenosylmethionine, S-adenosylhomocysteine | 19 (G[E/G]G), 23 ([D/E][C/S][A/G]), 26 ([L/F]RT), 80 ([G/A]RW), 85 ([A/S][S/T/D]X), 88 (FQF) | [110,111,112,113] |
| For other substrates | ||||
| Phosphate | PiC, mPT | Phosphate | 19 (CEG), 23 (HDA), 80 (G[R/K]M), 88 (KKQ) | [114,115] |
| Iron | MIT, MRS–4, MFRN–2 | - | 19 (GTG), 22 (E[S/A/H][A/C]), 23 (HDA), 27 ([F/Y][T/N]T) | [12,116] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernie, A.R.; Cavalcanti, J.H.F.; Nunes-Nesi, A. Metabolic Roles of Plant Mitochondrial Carriers. Biomolecules 2020, 10, 1013. https://doi.org/10.3390/biom10071013
Fernie AR, Cavalcanti JHF, Nunes-Nesi A. Metabolic Roles of Plant Mitochondrial Carriers. Biomolecules. 2020; 10(7):1013. https://doi.org/10.3390/biom10071013
Chicago/Turabian StyleFernie, Alisdair R., João Henrique F. Cavalcanti, and Adriano Nunes-Nesi. 2020. "Metabolic Roles of Plant Mitochondrial Carriers" Biomolecules 10, no. 7: 1013. https://doi.org/10.3390/biom10071013
APA StyleFernie, A. R., Cavalcanti, J. H. F., & Nunes-Nesi, A. (2020). Metabolic Roles of Plant Mitochondrial Carriers. Biomolecules, 10(7), 1013. https://doi.org/10.3390/biom10071013






