Concise Polygenic Models for Cancer-Specific Identification of Drug-Sensitive Tumors from Their Multi-Omics Profiles
Abstract
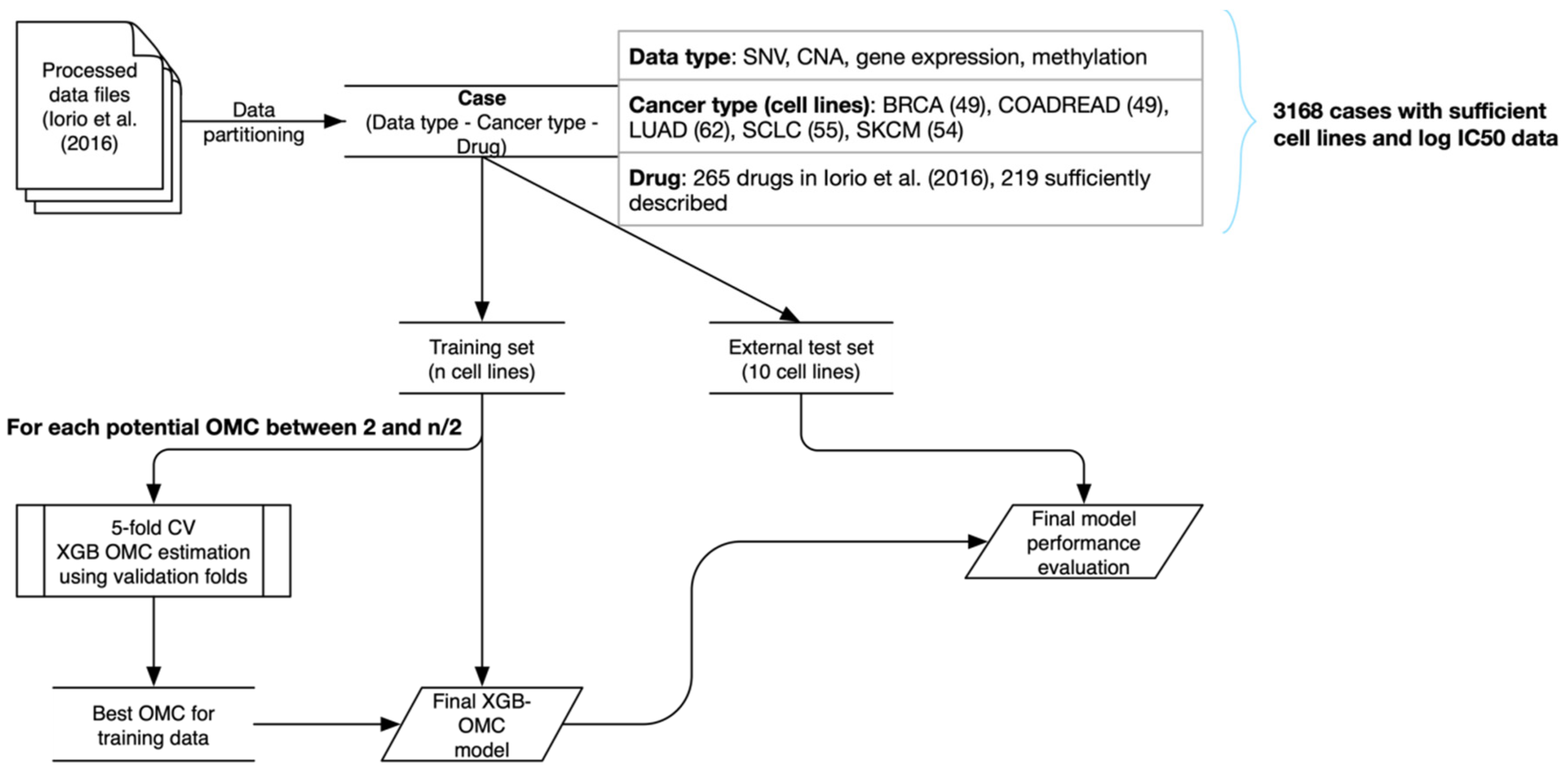
1. Background
2. Data Description
3. Results and Discussion
3.1. Building Predictive Cancer-Specific Models
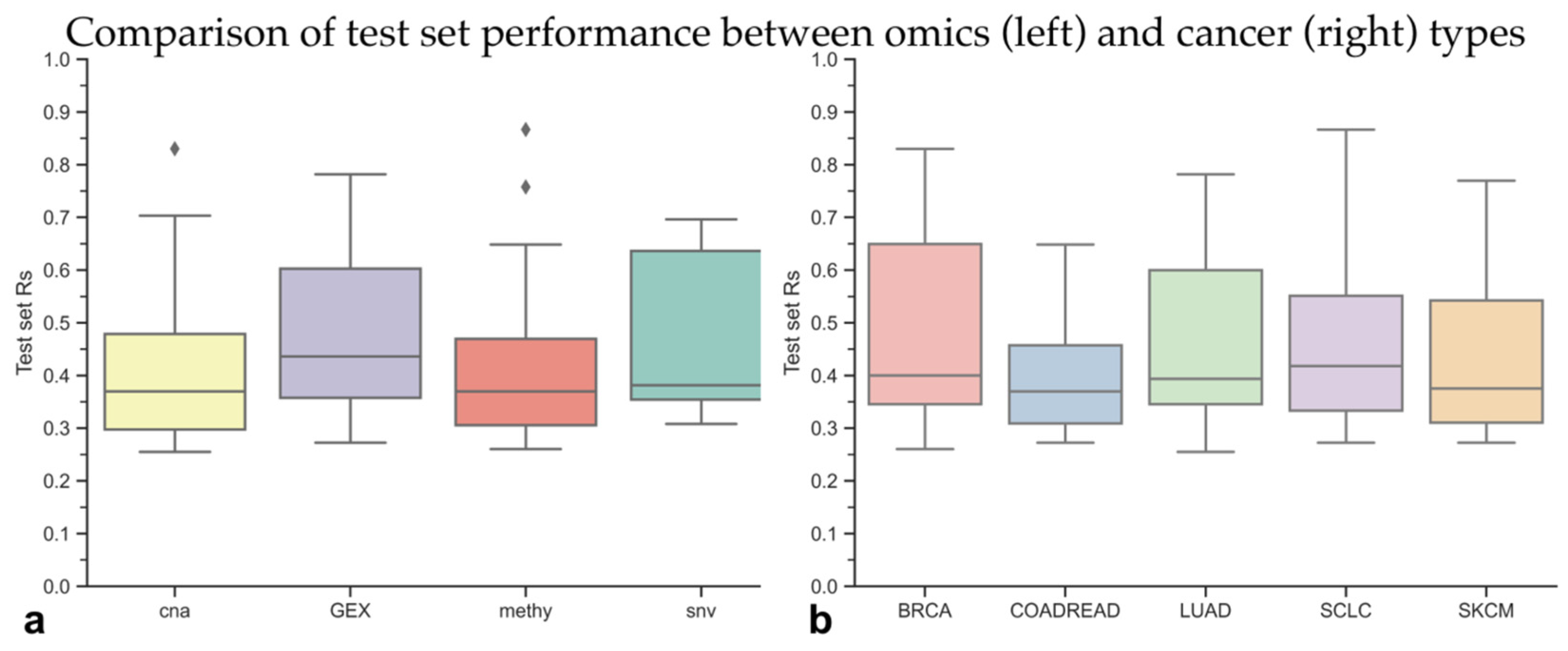
3.2. Identifying the Most Predictive Molecular Profile for Each Drug and Cancer Type
3.3. The Best XGB-OMC Model per Cancer Type as Case Studies
4. Methods
4.1. GDSC
4.2. Cell Line Information (Metadata)
4.3. Drug Response (Continuous Variable)
4.4. Single-Nucleotide Variants (SNV, Binary Feature)
4.5. Copy Number Alterations (CNA, Binary Feature)
4.6. Transcriptome (GEX, Continuous Feature)
4.7. Methylome (Methy, Continuous Feature)
4.8. Generation of Non-Overlapping Training and Test Sets
4.9. Optimal Model Complexity Estimation
4.10. Model Selection by Stratified Five-Fold Cross-Validation on the Training Set
4.11. Pre-Ranking Feature Input
4.12. Obtaining the OMC for a Given Drug Fold
4.13. XGBoost (XGB)
4.14. Final Model Performance Estimation
4.15. Pathway Enrichment Analysis
4.16. Statistical Tests
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCLE | Cancer Cell Line Encyclopedia |
| CTRP | Cancer Therapeutics Response Portal |
| GDSC | Genomics of Drug Sensitivity in Cancer |
| GO BP | Gene Ontology – Biological Process |
| OMC | Optimal Model Complexity |
| PDX | Patient-Derived Xenografts |
| RF | Random Forest |
| XGB | Extreme Gradient Boosting |
References
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.G.; Seashore-Ludlow, B.; Cheah, J.H.; Adams, D.J.; Price, E.V.; Gill, S.; Javaid, S.; Coletti, M.E.; Jones, V.L.; Bodycombe, N.E.; et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016, 12, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Gonçalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Ali, M.; Aittokallio, T. Machine learning and feature selection for drug response prediction in precision oncology applications. Biophys. Rev. 2019, 11, 31–39. [Google Scholar] [CrossRef]
- Azuaje, F. Computational models for predicting drug responses in cancer research. Brief. Bioinform. 2017, bbw065. [Google Scholar] [CrossRef]
- De Niz, C.; Rahman, R.; Zhao, X.; Pal, R. Algorithms for Drug Sensitivity Prediction. Algorithms 2016, 9, 77. [Google Scholar] [CrossRef]
- Chen, B.; Butte, A.J. Leveraging big data to transform target selection and drug discovery. Clin. Pharmacol. Ther. 2016, 99, 285–297. [Google Scholar] [CrossRef]
- Costello, J.C.; Heiser, L.M.; Georgii, E.; Gönen, M.; Menden, M.P.; Wang, N.J.; Bansal, M.; Ammad-Ud-Din, M.; Hintsanen, P.; Khan, S.A.; et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol. 2014, 32, 1202–1212. [Google Scholar] [CrossRef]
- Ammad-ud-din, M.; Georgii, E.; Gönen, M.; Laitinen, T.; Kallioniemi, O.; Wennerberg, K.; Poso, A.; Kaski, S. Integrative and personalized QSAR analysis in cancer by kernelized Bayesian matrix factorization. J. Chem. Inf. Model. 2014, 54, 2347–2359. [Google Scholar] [CrossRef] [PubMed]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Naulaerts, S.; Dang, C.; Ballester, P.J. Precision and recall oncology: Combining multiple gene mutations for improved identification of drug-sensitive tumours. Oncotarget 2017, 8, 97025–97040. [Google Scholar] [CrossRef] [PubMed]
- Menden, M.P.; Iorio, F.; Garnett, M.; McDermott, U.; Benes, C.H.; Ballester, P.J.; Saez-Rodriguez, J. Machine Learning Prediction of Cancer Cell Sensitivity to Drugs Based on Genomic and Chemical Properties. PLoS ONE 2013, 8, e61318. [Google Scholar] [CrossRef] [PubMed]
- Riddick, G.; Song, H.; Ahn, S.; Walling, J.; Borges-Rivera, D.; Zhang, W.; Fine, H.A. Predicting in vitro drug sensitivity using Random Forests. Bioinformatics 2011, 27, 220–224. [Google Scholar] [CrossRef]
- Ballester, P.; Nguyen, L.; Dang, C.C. Systematic assessment of multi-gene predictors of pan-cancer cell line sensitivity to drugs exploiting gene expression data. F1000Research 2016, 5, 2927. [Google Scholar]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Model selection and overfitting. Nat. Methods 2016, 13, 703–704. [Google Scholar] [CrossRef]
- Li, H.; Peng, J.; Leung, Y.; Leung, K.S.; Wong, M.H.; Lu, G.; Ballester, P.J.P. The Impact of Protein Structure and Sequence Similarity on the Accuracy of Machine-Learning Scoring Functions for Binding Affinity Prediction. Biomolecules 2018, 8, 12. [Google Scholar] [CrossRef]
- Sidorov, P.; Naulaerts, S.; Ariey-Bonnet, J.; Pasquier, E.; Ballester, P.J. Predicting Synergism of Cancer Drug Combinations Using NCI-ALMANAC Data. Front. Chem. 2019, 7, 509. [Google Scholar] [CrossRef]
- Li, H.; Peng, J.; Sidorov, P.; Leung, Y.; Leung, K.S.; Wong, M.H.; Lu, G.; Ballester, P.J. Classical scoring functions for docking are unable to exploit large volumes of structural and interaction data. Bioinformatics 2019, 35, 3989–3995. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Shen, A.; Ding, J.; Geng, M. Molecularly targeted cancer therapy: Some lessons from the past decade. Trends Pharmacol. Sci. 2014, 35, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining—KDD’16; ACM Press: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Sheridan, R.P.; Wang, W.M.; Liaw, A.; Ma, J.; Gifford, E.M. Extreme Gradient Boosting as a Method for Quantitative Structure-Activity Relationships. J. Chem. Inf. Model. 2016, 56, 2353–2360. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Heidemeyer, M.; Ban, F.; Cherkasov, A.; Ester, M. SimBoost: A read-across approach for predicting drug-target binding affinities using gradient boosting machines. J. Cheminform. 2017, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Simm, J.; Zakeri, P.; Moreau, Y.; Saez-Rodriguez, J. Linking drug target and pathway activation for effective precision therapy using multi-task learning. Sci. Rep. 2018, 8, 8322. [Google Scholar] [CrossRef]
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; GilLopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Haury, A.C.; Gestraud, P.; Vert, J.P. The Influence of Feature Selection Methods on Accuracy, Stability and Interpretability of Molecular Signatures. PLoS ONE 2011, 6, e28210. [Google Scholar] [CrossRef]
- Rücker, C.; Rücker, G.; Meringer, M. Y-randomization and its variants in QSPR/QSAR. J. Chem. Inf. Model. 2007, 47, 2345–2357. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef]
- Poirier, J.T.; Gardner, E.E.; Connis, N.; Moreira, A.L.; De Stanchina, E.; Hann, C.L.; Rudin, C.M. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015, 34, 5869–5878. [Google Scholar] [CrossRef]
- Codignola, A.; Tarroni, P.; Clementi, F.; Pollo, A.; Lovallo, M.; Carbone, E.; Sher, E. Calcium channel subtypes controlling serotonin release from human small cell lung carcinoma cell lines. J. Biol. Chem. 1993, 268, 26240–26247. [Google Scholar] [PubMed]
- Taniwaki, M.; Daigo, Y.; Ishikawa, N.; Takano, A.; Tsunoda, T.; Yasui, W.; Inai, K.; Kohno, N.; Nakamura, Y. Gene expression profiles of small-cell lung cancers: Molecular signatures of lung cancer. Int. J. Oncol. 2006, 29, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Hubaux, R.; Thu, K.L.; Coe, B.P.; Macaulay, C.; Lam, S.; Lam, W.L. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J. Thorac. Oncol. 2013, 8, 1102–1106. [Google Scholar] [CrossRef]
- Bilir, B.; Kucuk, O.; Moreno, C.S. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J. Transl. Med. 2013, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.Y.; Chen, L.H.; Lin, Y.F.; Lai, L.C.; Chuang, E.Y.; Tsai, M.H.; Shen, C.Y.; Chen, L.H.; Lin, Y.F.; Lai, L.C.; et al. Mitomycin C treatment induces resistance and enhanced migration via phosphorylated Akt in aggressive lung cancer cells. Oncotarget 2016, 7, 79995–80007. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Santamaria, R.; Xu, W.; Cols, M.; Chen, K.; Puga, I.; Shan, M.; Xiong, H.; Bussel, J.B.; Chiu, A.; et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol. 2010, 11, 836–845. [Google Scholar] [CrossRef]
- Kang, Y.H.; Lee, K.A.; Ryu, C.J.; Lee, H.G.; Lim, J.S.; Park, S.N.; Paik, S.G.; Yoon, D.Y. Mitomycin C induces apoptosis via Fas/FasL dependent pathway and suppression of IL-18 in cervical carcinoma cells. Cancer Lett. 2006, 237, 33–44. [Google Scholar] [CrossRef]
- Siegel, D.; Yan, C.; Ross, D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem. Pharmacol. 2012, 83, 1033–1040. [Google Scholar] [CrossRef]
- Shkreta, L.; Chabot, B. The RNA splicing response to DNA damage. Biomolecules 2015, 5, 2935–2977. [Google Scholar] [CrossRef]
- Al-Saffar, N.M.S.; Jackson, L.E.; Raynaud, F.I.; Clarke, P.A.; De Molina, A.R.; Lacal, J.C.; Workman, P.; Leach, M.O. The phosphoinositide 3-kinase inhibitor PI-103 downregulates choline kinase α leading to phosphocholine and total choline decrease detected by magnetic resonance spectroscopy. Cancer Res. 2010, 70, 5507–5517. [Google Scholar] [CrossRef]
- Prevo, R.; Deutsch, E.; Sampson, O.; Diplexcito, J.; Cenge, K.; Harper, J.; O’Neill, P.; McKenna, W.G.; Patel, S.; Bernhard, E.J. Class I PI3 kinase inhibition by the pyridinylfuranopyrimidine inhibitor PI-103 enhances tumor radiosensitivity. Cancer Res. 2008, 68, 5915–5923. [Google Scholar] [CrossRef]
- Raynaud, F.I.; Eccles, S.; Clarke, P.A.; Hayes, A.; Nutley, B.; Alix, S.; Henley, A.; Di-Stefano, F.; Ahmad, Z.; Guillard, S.; et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007, 67, 5840–5850. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef]
- Kojima, K.; Shimanuki, M.; Shikami, M.; Samudio, I.J.; Ruvolo, V.; Corn, P.; Hanaoka, N.; Konopleva, M.; Andreeff, M.; Nakakuma, H. The dual PI3 kinase/mTOR inhibitor PI-103 prevents p53 induction by Mdm2 inhibition but enhances p53-mediated mitochondrial apoptosis in p53 wild-type AML. Leukemia 2008, 22, 1728–1736. [Google Scholar] [CrossRef]
- Keisner, S.V.; Shah, S.R. Pazopanib: The newest tyrosine kinase inhibitor for the treatment of advanced or metastatic renal cell carcinoma. Drugs 2011, 71, 443–454. [Google Scholar] [CrossRef]
- Fruehauf, J.P.; Alger, B.; Parmakhtiar, B.; Jakowatz, J.G.; Bettis, C.; Chuang, T.; Ein-Gal, S.Y. A phase II single arm study of pazopanib and paclitaxel as first-line treatment for unresectable stage III and stage IV melanoma: Interim analysis. ASCO Meet. Abstr. 2012, 30, 8524. [Google Scholar] [CrossRef]
- Kumar, R.; Knick, V.B.; Rudolph, S.K.; Johnson, J.H.; Crosby, R.M.; Crouthamel, M.C.; Hopper, T.M.; Miller, C.G.; Harrington, L.E.; Onori, J.A.; et al. Pharmacokinetic-pharmacodynamic correlation from mouse to human with pazopanib, a multikinase angiogenesis inhibitor with potent antitumor and antiangiogenic activity. Mol. Cancer Ther. 2007, 6, 2012–2021. [Google Scholar] [CrossRef]
- Preussner, J.; Bayer, J.; Kuenne, C.; Looso, M. ADMIRE: Analysis and visualization of differential methylation in genomic regions using the Infinium HumanMethylation450 Assay. Epigene. Chromatin 2015, 8, 51. [Google Scholar] [CrossRef][Green Version]
- Smedley, D.; Haider, S.; Ballester, B.; Holland, R.; London, D.; Thorisson, G.; Kasprzyk, A. BioMart--biological queries made easy. BMC Genom. 2009, 10, 22. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 1 June 2016).
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013, 41, 77–83. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Expansion of the Gene Ontology Consortium Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, 331–338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2016, 14, 3–13. [Google Scholar] [CrossRef]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef] [PubMed]
- Vincent, K.M.; Findlay, S.D.; Postovit, L.M. Assessing breast cancer cell lines as tumour models by comparison of mRNA expression profiles. Breast Cancer Res. 2015, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Zhang, Z.; Wang, F.; Gruener, R.F.; Nath, A.; Morrison, G.; Bhutra, S.; Grossman, R.L.; Huang, R.S. Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res. 2017, 27, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Naulaerts, S.; Bomane, A.; Bruna, A.; Ghislat, G.; Ballester, P. Machine learning models to predict in vivo drug response via optimal dimensionality reduction of tumour molecular profiles. bioRxiv 2018. [Google Scholar]
- Bomane, A.; Gonçalves, A.; Ballester, P. Paclitaxel response can be predicted with interpretable multi-variate classifiers exploiting DNA-methylation and miRNA data. Front. Genet. 2019, 10, 1041. [Google Scholar] [CrossRef]
- Ding, Z.; Zu, S.; Gu, J. Evaluating the molecule-based prediction of clinical drug responses in cancer. Bioinformatics 2016, 32, 2891–2895. [Google Scholar] [CrossRef]
- Jang, I.S.; Neto, E.C.; Guinney, J.; Friend, S.H.; Margolin, A.A. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac. Symp. Biocomput. 2014, 63–74. [Google Scholar]
- Papillon-Cavanagh, S.; De Jay, N.; Hachem, N.; Olsen, C.; Bontempi, G.; Aerts, H.J.W.L.; Quackenbush, J.; Haibe-Kains, B. Comparison and validation of genomic predictors for anticancer drug sensitivity. J. Am. Med. Inform. Assoc. 2013, 20, 597–602. [Google Scholar] [CrossRef]
- Kurilov, R.; Haibe-Kains, B.; Brors, B. Assessment of modelling strategies for drug response prediction in cell lines and xenografts. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Vakiani, E.; Solit, D.B. KRAS and BRAF: Drug targets and predictive biomarkers. J. Pathol. 2011, 223, 219–229. [Google Scholar] [CrossRef]
- Ciombor, K.K.; Bekaii-Saab, T. Selumetinib for the treatment of cancer. Expert Opin. Investig. Drugs 2015, 24, 111–123. [Google Scholar] [CrossRef]
- Gross, A.M.; Wolters, P.; Baldwin, A.; Dombi, E.; Fisher, M.J.; Weiss, B.D.; Kim, A.; Blakeley, J.O.; Whitcomb, P.; Holmblad, M.; et al. Phase II study of the MEK 1/2 inhibitor selumetinib (AZD6244, ARRY-142886) in children with neurofibromatosis type 1 (NF1) and inoperable plexiform neurofibromas (PN). J. Clin. Oncol. 2018, 36, 10503. [Google Scholar] [CrossRef]
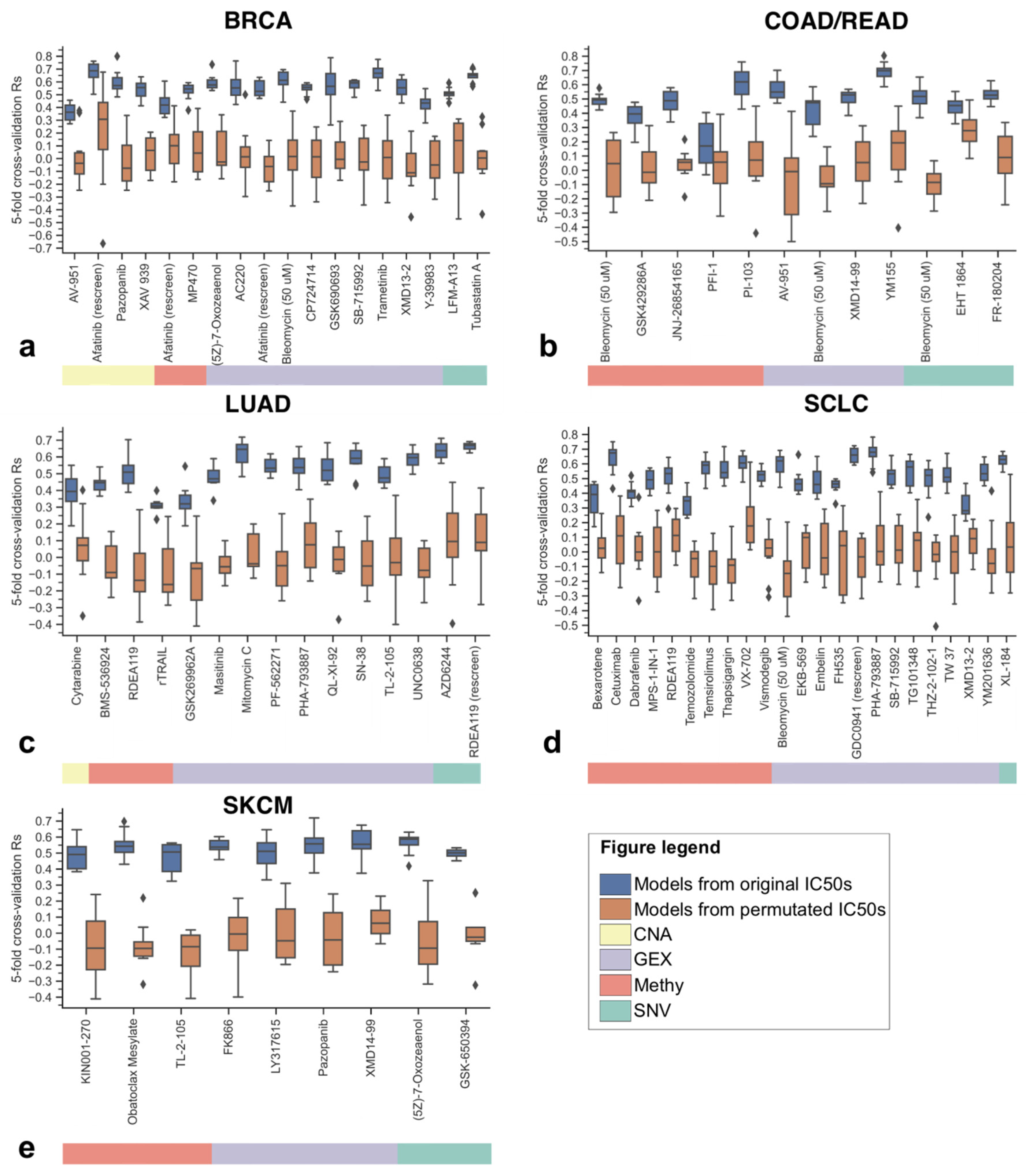
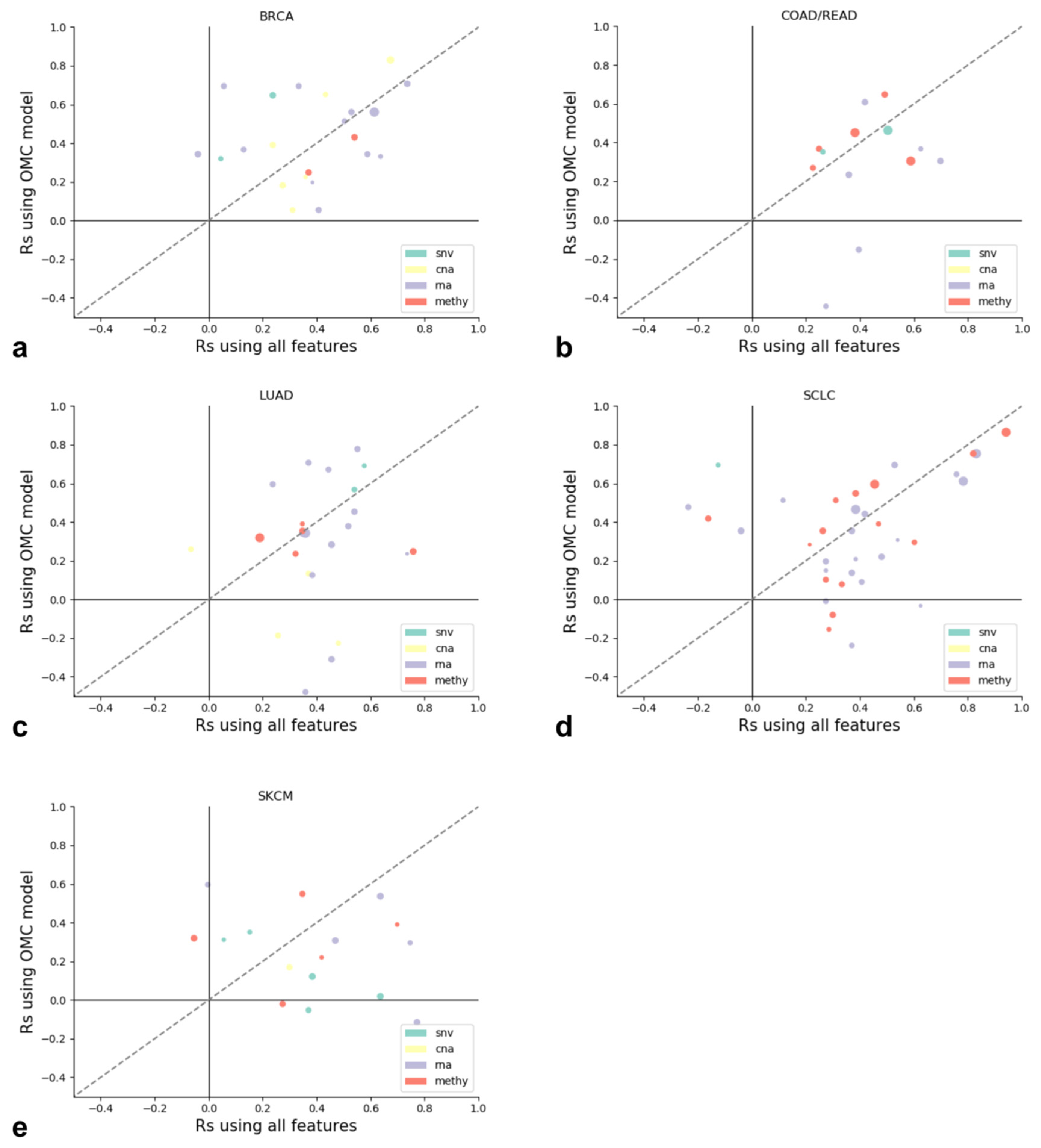
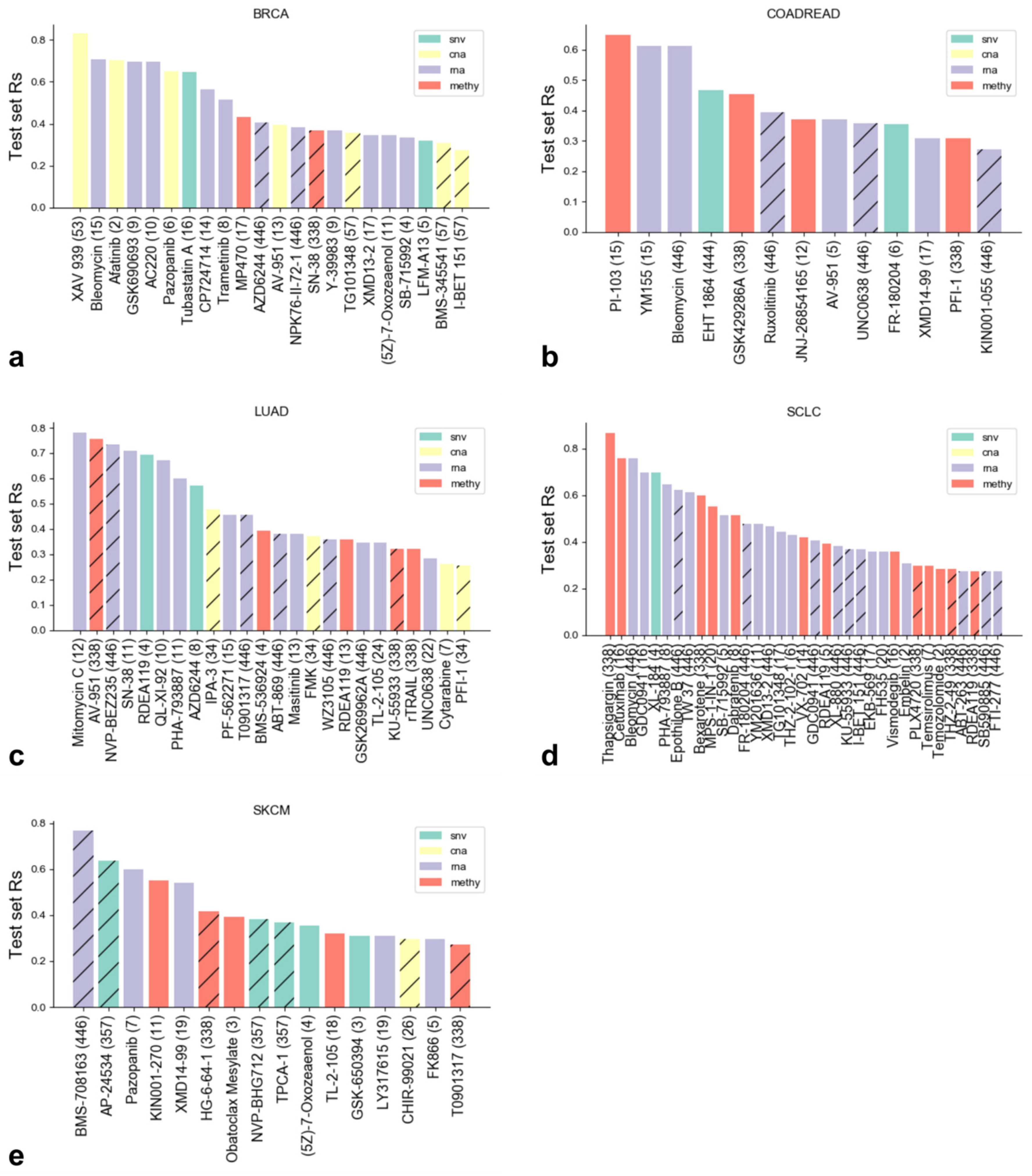
| Profile | Cancer Type | Drug Name | OMC | Test Rs (XGB-OMC) | Test Rs (XGB-all) | Test R2 (XGB-OMC) | Test R2 (XGB-all) |
|---|---|---|---|---|---|---|---|
| Methy | SCLC | Thapsigargin | 338 (all) | 0.867 | 0.939 | 0.535 | 0.609 |
| CNA | BRCA | XAV 939 | 53 (all) | 0.830 | 0.673 | 0.347 | 0.382 |
| GEX | LUAD | Mitomycin C | 12 | 0.782 | 0.552 | 0.465 | 0.280 |
| Methy | COAD/READ | PI-103 | 15 | 0.648 | 0.491 | 0.395 | 0.296 |
| GEX | SKCM | Pazopanib | 7 | 0.600 | −0.006 | 0.321 | −0.231 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naulaerts, S.; Menden, M.P.; Ballester, P.J. Concise Polygenic Models for Cancer-Specific Identification of Drug-Sensitive Tumors from Their Multi-Omics Profiles. Biomolecules 2020, 10, 963. https://doi.org/10.3390/biom10060963
Naulaerts S, Menden MP, Ballester PJ. Concise Polygenic Models for Cancer-Specific Identification of Drug-Sensitive Tumors from Their Multi-Omics Profiles. Biomolecules. 2020; 10(6):963. https://doi.org/10.3390/biom10060963
Chicago/Turabian StyleNaulaerts, Stefan, Michael P. Menden, and Pedro J. Ballester. 2020. "Concise Polygenic Models for Cancer-Specific Identification of Drug-Sensitive Tumors from Their Multi-Omics Profiles" Biomolecules 10, no. 6: 963. https://doi.org/10.3390/biom10060963
APA StyleNaulaerts, S., Menden, M. P., & Ballester, P. J. (2020). Concise Polygenic Models for Cancer-Specific Identification of Drug-Sensitive Tumors from Their Multi-Omics Profiles. Biomolecules, 10(6), 963. https://doi.org/10.3390/biom10060963






