Biological Manganese Removal by Novel Halotolerant Bacteria Isolated from River Water
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Enrichment and Isolation
2.2. Manganese Removal by Five Isolated Bacteria
2.3. Manganese Removal by Five Isolated Bacteria
2.4. Electron Microscopic Analysis and Characterization of Mn Precipitates
2.5. Analytical Methods
2.6. Phylogenetic Study of Isolated Strains
3. Results and Discussion
3.1. Isolation of Mn Removal Bacteria
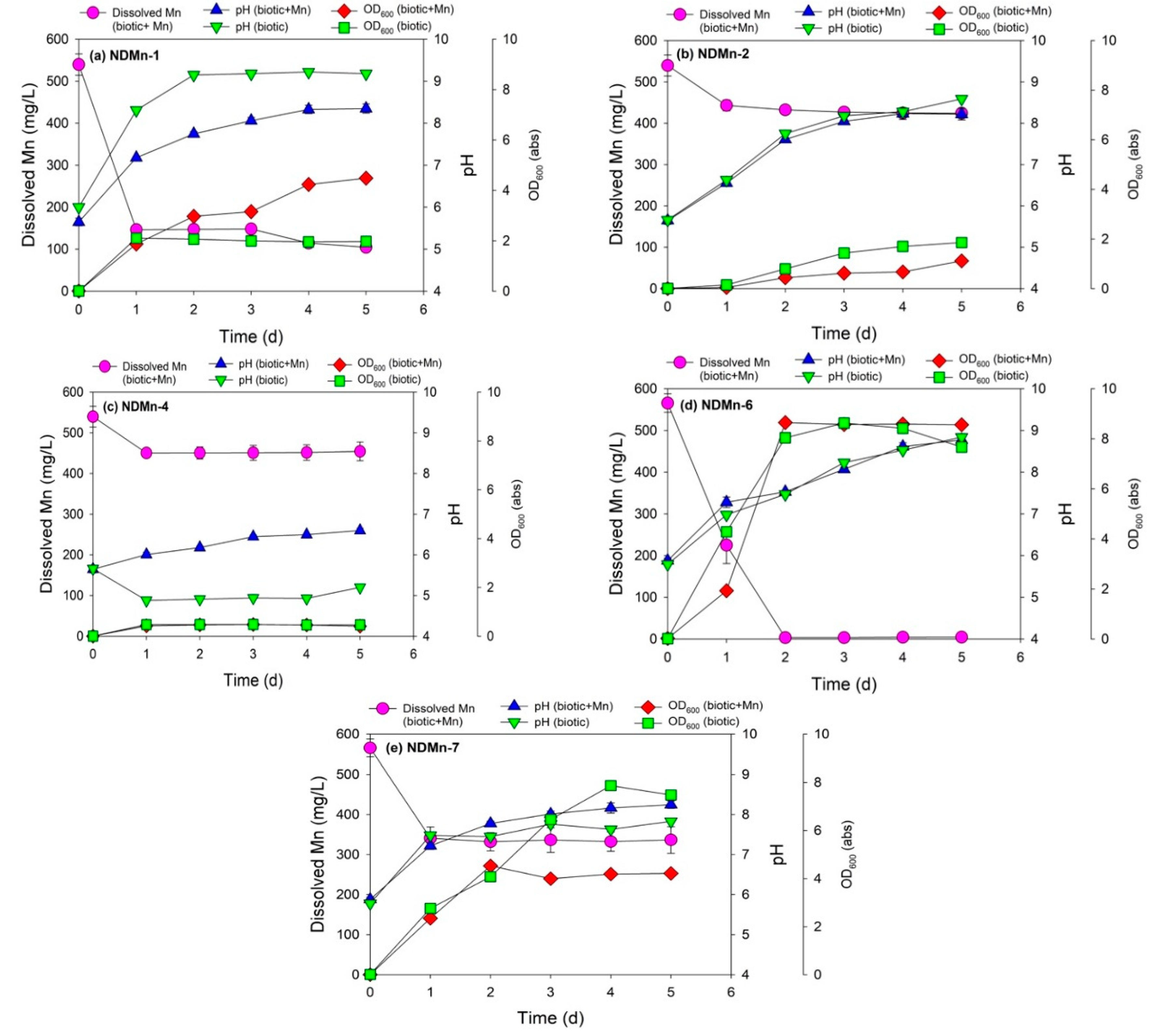
3.2. Cell Growth and Mn Removal by Five Isolated Bacterial Strains
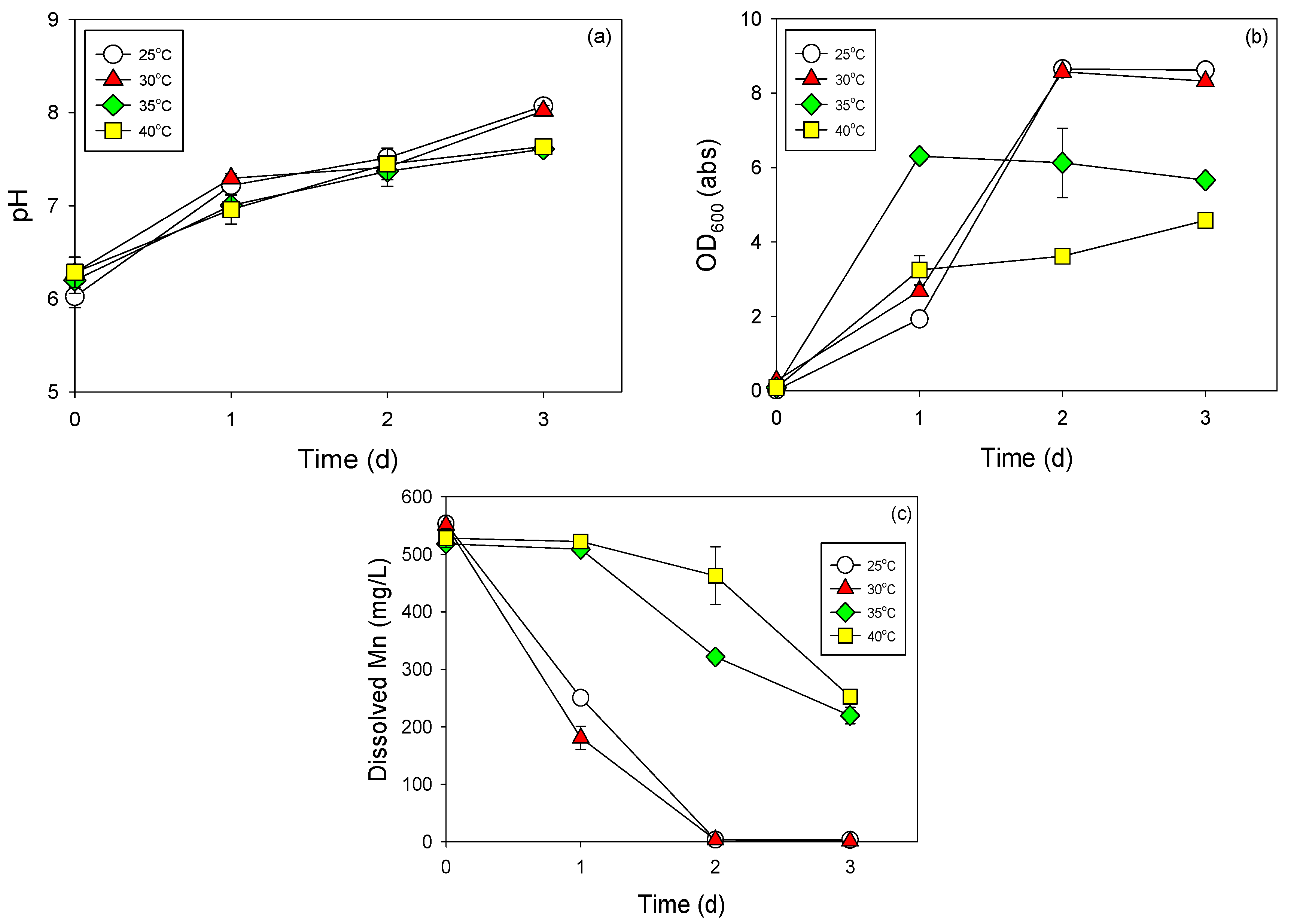
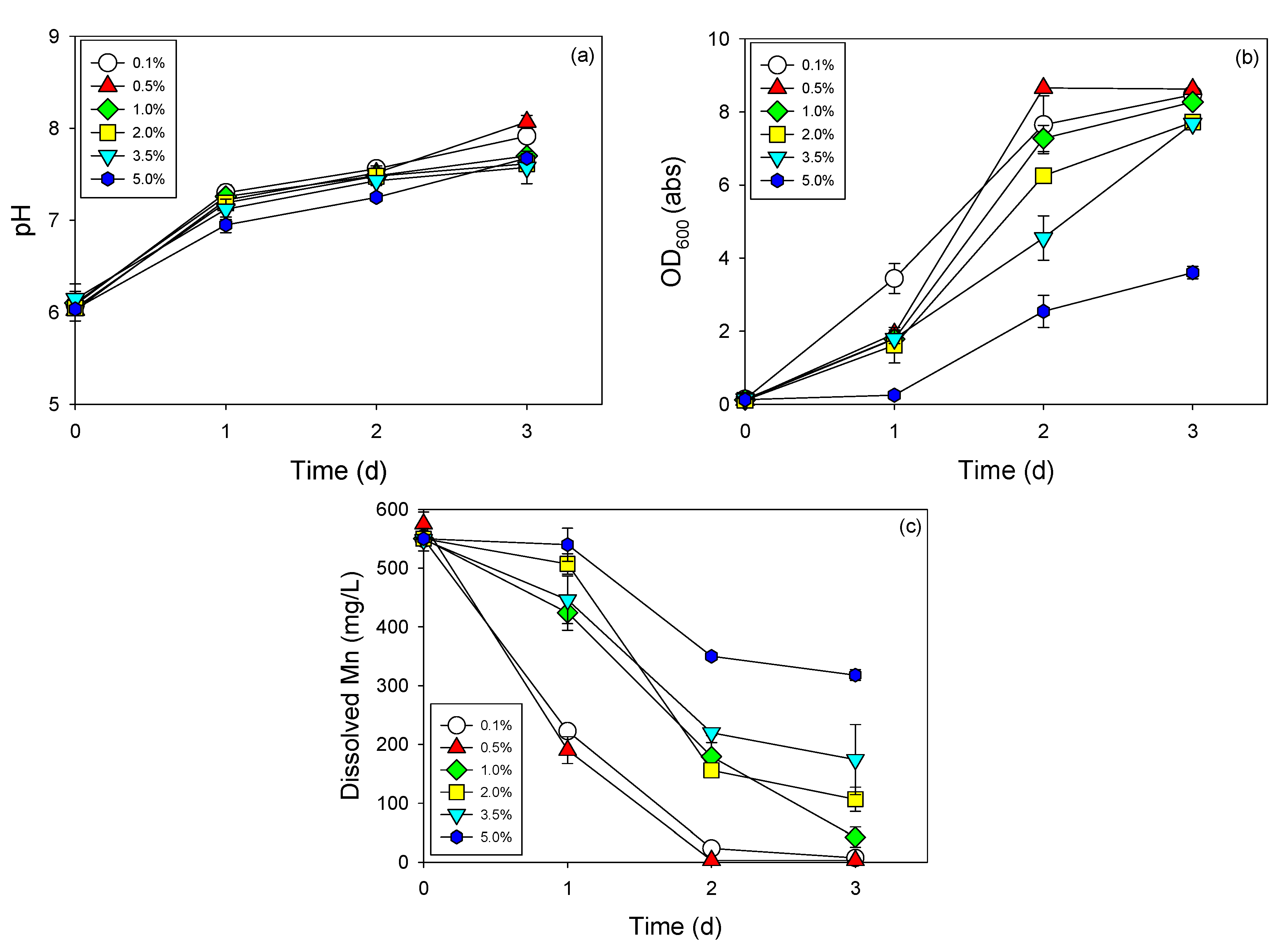
3.3. Cell Growth and Mn Removal by Strain NDMn-6 at Various Temperatures and Salinities
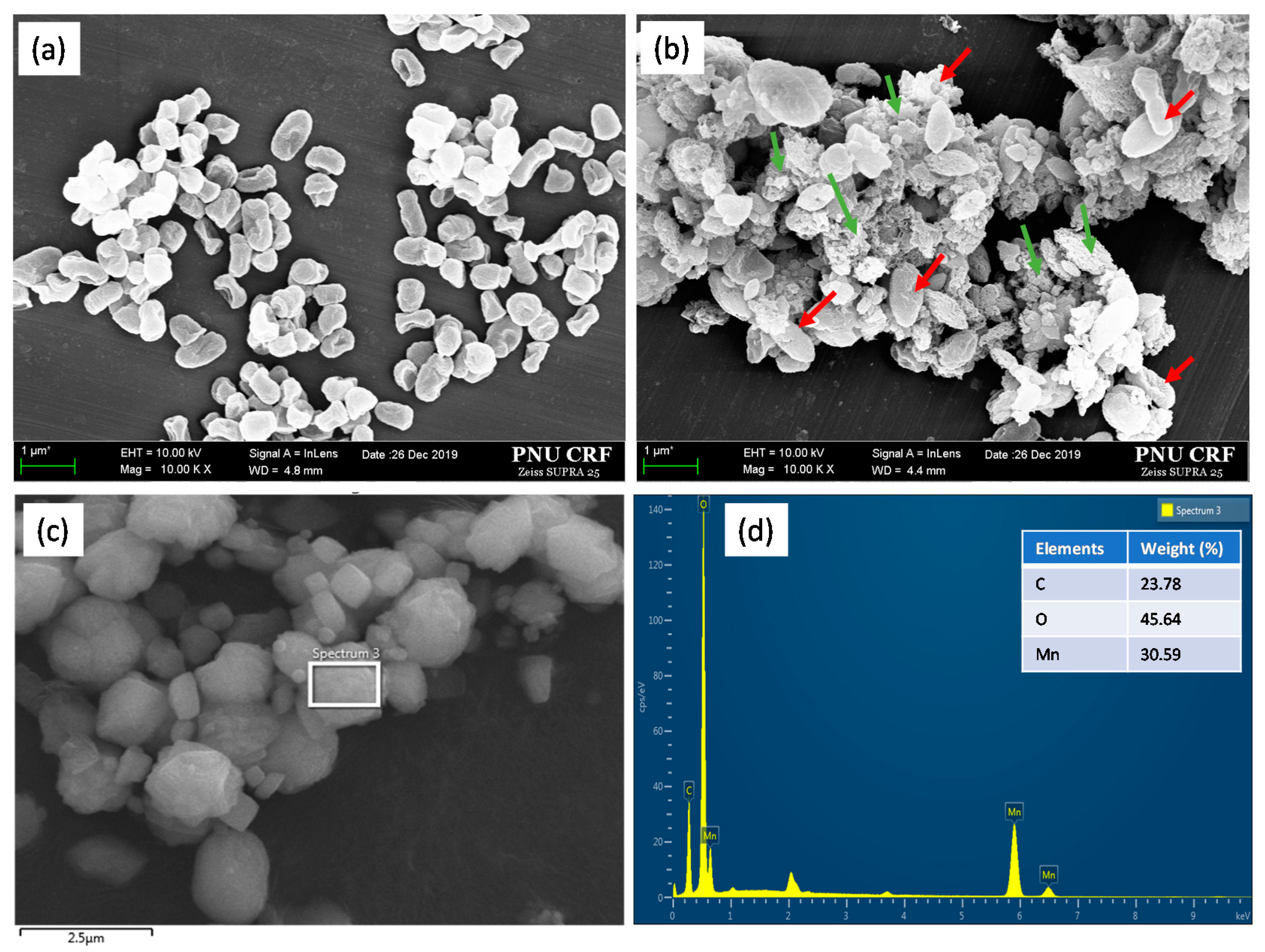
3.4. Electron Microscopic Analysis and Characteristics of Mn Precipitates Produced by Strain NDMn-6
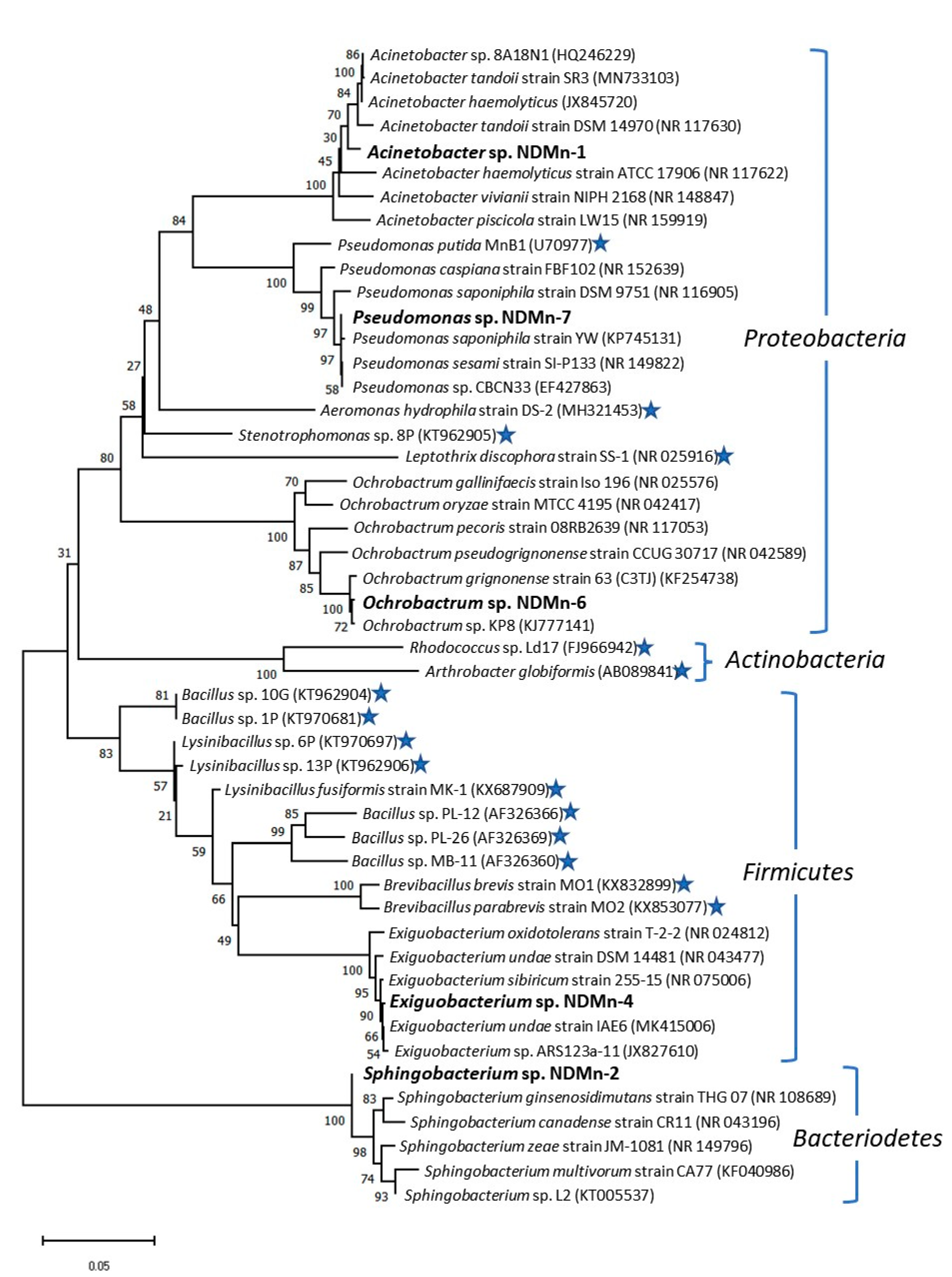
3.5. Phylogenetic Analysis of Isolated Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ehrlich, H.L.; Newman, D.K. Geomicrobiology, 5th ed.; Ehrlich, H.L., Newman, D.K., Eds.; CRC Press: New York, NY, USA, 2009. [Google Scholar]
- Post, J.E. Manganese oxide minerals: Crystal structures and economic and environmental significance. Proc. Natl. Acad. Sci. USA 1999, 96, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Bowen, H.J.M. Environmental Chemistry of the Elements; Academic Press: London, UK, 1979. [Google Scholar]
- Erikson, K.M.; Aschner, M. Manganese: Its Role in Disease and Health. In Metal Ions in Life Sciences; De Gruyter: Berlin, Germany, 2019; Volume 19. [Google Scholar]
- O’neal, S.L.; Zheng, W. Manganese Toxicity Upon Overexposure: A Decade in Review. Curr. Environ. Health Rep. 2015, 2, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Crossgrove, J.; Zheng, W. Manganese toxicity upon overexposure. NMR Biomed. 2004, 17, 544–553. [Google Scholar] [CrossRef]
- Millaleo, R.; Reyes-Díaz, M.; Ivanov, A.G.; Mora, M.L.; Alberdi, M. Manganese as essential and toxic element for plants: Transport, accumulation and resistance mechanisms. J. Soil Sci. Plant Nutr. 2010, 10, 476–494. [Google Scholar] [CrossRef]
- Walter, E.; Alsaffar, S.; Livingstone, C.; Ashley, S.L. Manganese toxicity in critical care: Case report, literature review and recommendations for practice. J. Intensive Care Soc. 2016, 17, 252–257. [Google Scholar] [CrossRef]
- WHO. Manganese in Drinking-water Background Document for Development of WHO Guidelines for Drinking-Water Quality; WHO: Geneva, Switzerland, 2011.
- Das, A.P.; Sukla, L.B.; Pradhan, N.; Nayak, S. Manganese biomining: A review. Bioresour. Technol. 2011, 102, 7381–7387. [Google Scholar] [CrossRef]
- Tobiason, J.E.; Bazilio, A.; Goodwill, J.; Mai, X.; Nguyen, C. Manganese Removal from Drinking Water Sources. Curr. Pollut. Rep. 2016, 2, 168–177. [Google Scholar] [CrossRef]
- Beukes, L.S.; Schmidt, S. Isolation and characterization of a manganese-oxidizing bacterium from a biofiltration system for the treatment of borehole water in KwaZulu-Natal (South Africa). Eng. Life Sci. 2012, 12, 544–552. [Google Scholar] [CrossRef]
- Barboza, N.R.; Amorim, S.S.; Santos, P.A.; Reis, F.D.; Cordeiro, M.M.; Guerra-Sá, R.; Leão, V.A. Indirect Manganese Removal by Stenotrophomonas sp. and Lysinibacillus sp. Isolated from Brazilian Mine Water. BioMed Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef]
- Hosseinkhani, B.; Emtiazi, G. Synthesis and characterization of a novel extracellular biogenic manganese oxide (bixbyite-like Mn2O3) nanoparticle by isolated Acinetobacter sp. Curr. Microbiol. 2011, 63, 300–305. [Google Scholar] [CrossRef]
- Burger, M.S.; Krentz, C.A.; Mercer, S.S.; Gagnon, G.A. Manganese removal and occurrence of manganese oxidizing bacteria in full-scale biofilters. J. Water Supply Res. Technol. AQUA 2008, 57, 351–359. [Google Scholar] [CrossRef]
- Tang, W.; Gong, J.; Wu, L.; Li, Y.; Zhang, M.; Zeng, X. DGGE diversity of manganese mine samples and isolation of a Lysinibacillus sp. efficient in removal of high Mn (II) concentrations. Chemosphere 2016, 165, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Y.; Qin, Z.; Luo, P.; Ma, Z.; Tan, M.; Kang, H.; Huang, Z. A novel manganese oxidizing bacterium-Aeromonas hydrophila strain DS02: Mn(II) oxidization and biogenic Mn oxides generation. J. Hazard. Mater. 2019, 367, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Akob, D.M.; Bohu, T.; Beyer, A.; Schäffner, F.; Händel, M.; Johnson, C.A.; Merten, D.; Büchel, G.; Totsche, K.U.; Küsel, K. Identification of Mn(II)-oxidizing bacteria from a low-pH contaminated former uranium mine. Appl. Environ. Microbiol. 2014, 80, 5086–5097. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, X.; Liu, B.; Xie, G.; Xing, D. Characterization of manganese oxidation by Brevibacillus at different ecological conditions. Chemosphere 2018, 205, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.K.; Choi, W.; Ha, Y.; Gu, Y.; Lee, C.; Park, J.; Jang, G.; Shin, C.; Cho, S. Microbial tellurite reduction and production of elemental tellurium nanoparticles by novel bacteria isolated from wastewater. J. Ind. Eng. Chem. 2019, 78, 246–256. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Nguyen, T.H.; Ha, M.; Kang, H.Y. Kinetics of microbial selenite reduction by novel bacteria isolated from activated sludge. J. Environ. Manag. 2019, 236, 746–754. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Tran, H.T.; Park, Y.; Yu, J.; Lee, T. Microbial arsenite oxidation with oxygen, nitrate, or an electrode as the sole electron acceptor. J. Ind. Microbiol. Biotechnol. 2017, 44, 857–868. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecule. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Lund, P.; Tramonti, A.; De Biase, D. Coping with low pH: Molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 2014, 38, 1091–1125. [Google Scholar] [CrossRef]
- Freitas, R.M.; Perilli, T.A.G.; Ladeira, A.C.Q. Oxidative Precipitation of Manganese from Acid Mine Drainage by Potassium Permanganate. J. Chem. 2013, 2013, 287257. [Google Scholar] [CrossRef]
- Cameron, F.J.; Jones, M.V.; Edwards, C. Effects of salinity on bacterial iron oxidation. Curr. Microbiol. 1984, 10, 353–356. [Google Scholar] [CrossRef]
- Caspi, R.; Tebo, B.M.; Haygood, M.G. c-type cytochromes and manganese oxidation in Pseudomonas putida MnB1. Appl. Environ. Microbiol. 1998, 64, 3549–3555. [Google Scholar] [CrossRef]
- Siering, P.L.; Ghiorse, W.C. Phylogeny of the Sphaerotilus-Leptothrix group inferred from morphological comparisons, genomic fingerprinting, and 16S ribosomal DNA sequence analyses. Int. J. Syst. Bacteriol. 1996, 46, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.A.; Tebo, B.M. Enzymatic manganese(II) oxidation by metabolically dormant spores of diverse Bacillus species. Appl. Environ. Microbiol. 2002, 68, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Salaün, S.; Kervarec, N.; Potin, P.; Haras, D.; Piotto, M.; La Barre, S. Whole-cell spectroscopy is a convenient tool to assist molecular identification of cultivatable marine bacteria and to investigate their adaptive metabolism. Talanta 2010, 80, 1758–1770. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Mochida, K.; Asahara, M.; Suzuki, M.; Kasai, H.; Yokota, A. Alicyclobacillus pomorum sp. nov., a novel thermo-acidophilic, endospore-forming bacterium that does not possess ω-alicyclic fatty acids, and emended description of the genus Alicyclobacillus. Int. J. Syst. Evol. Microbiol. 2003, 53, 1537–1544. [Google Scholar] [CrossRef]

| Environmental Factors | Temperature | Salinity |
|---|---|---|
| Various temperature experiment | 20 °C | 0.5% |
| 25 °C | ||
| 30 °C | ||
| 37 °C | ||
| 40 °C | ||
| Various salinity experiment | 25 °C | 0.1% |
| 0.5% | ||
| 1.0% | ||
| 3.5% | ||
| 5.0% |
| Bacterial Strains | Culture Media | Final Mn Concentration (mg/L) | Final pH (with Mn) | Final pH (without Mn) | Final OD600 (with Mn, abs) | Final OD600 (without Mn, abs) |
|---|---|---|---|---|---|---|
| NDMn-1 | Basal salt | 104.48 ± 5.13 | 8.35 ± 0.11 | 9.18 ± 0.02 | 4.49 ± 0.07 | 1.97 ± 0.01 |
| NDMn-2 | Basal salt | 424.43 ± 6.68 | 8.22 ± 0.14 | 8.59 ± 0.05 | 1.12 ± 0.01 | 1.86 ± 0.03 |
| NDMn-4 | Basal salt | 454.35 ± 18.80 | 6.60 ± 0.06 | 5.20 ± 0.11 | 0.41 ± 0.00 | 0.47 ± 0.01 |
| NDMn-6 | Nutrient broth | 4.52 ± 0.78 | 8.77 ± 0.06 | 8.84 ± 0.09 | 8.56 ± 0.02 | 7.67 ± 0.02 |
| NDMn-7 | Nutrient broth | 336.71 ± 27.12 | 8.25 ± 0.09 | 7.83 ± 0.05 | 4.22 ± 0.01 | 7.48 ± 0.01 |
| Bacterial Strains | Mn Removal Efficiency (%) | Mn Removal Rate (mg/L/d) | Reference |
|---|---|---|---|
| Lysinibacillus sp. MK-1 | 94.7 | 7.4 | [16] |
| Brevibacillus brevis MO1 | 83.6 | 4.6 | [19] |
| Brevibacillus parabrevis MO2 | 94.0 | 5.2 | |
| Acinetobacter sp. | 90.0 | 7.1 | [14] |
| Aeromonas hydrophila strain DS02 | 89.6 | 82.1 | [17] |
| Bacillus sp. 1G | 58.5 | 4.0 | [13] |
| Bacillus sp. 10G | 63.0 | 4.4 | |
| Stenotrophomonas sp. 7P | 70.9 | 5.0 | |
| Stenotrophomonas sp. 8P | 66.4 | 4.6 | |
| Lysinibacillus sp. 6P | 65.6 | 4.0 | |
| Lysinibacillus sp. 6P | 82.7 | 5.3 | |
| Acinetobacter sp. NDMn-1 | 80.6 | 147.7 | This study |
| Ochrobactrum sp. NDMn-6 | 99.1 | 181.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.K.; Ha, M.-G.; Kang, H.Y.; Nguyen, D.D. Biological Manganese Removal by Novel Halotolerant Bacteria Isolated from River Water. Biomolecules 2020, 10, 941. https://doi.org/10.3390/biom10060941
Nguyen VK, Ha M-G, Kang HY, Nguyen DD. Biological Manganese Removal by Novel Halotolerant Bacteria Isolated from River Water. Biomolecules. 2020; 10(6):941. https://doi.org/10.3390/biom10060941
Chicago/Turabian StyleNguyen, Van Khanh, Myung-Gyu Ha, Ho Young Kang, and Dinh Duc Nguyen. 2020. "Biological Manganese Removal by Novel Halotolerant Bacteria Isolated from River Water" Biomolecules 10, no. 6: 941. https://doi.org/10.3390/biom10060941
APA StyleNguyen, V. K., Ha, M.-G., Kang, H. Y., & Nguyen, D. D. (2020). Biological Manganese Removal by Novel Halotolerant Bacteria Isolated from River Water. Biomolecules, 10(6), 941. https://doi.org/10.3390/biom10060941





