Adenosine-5′-Phosphosulfate- and Sulfite Reductases Activities of Sulfate-Reducing Bacteria from Various Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures and Cultivation
2.2. Sulfate Determination
2.3. Measurement of Hydrogen Sulfide
2.4. Cell-Free Extracts
2.5. APS Reductase Activity
2.6. Sulfite Reductase Activity
2.7. Kinetic Analysis
2.8. Statistical Analysis
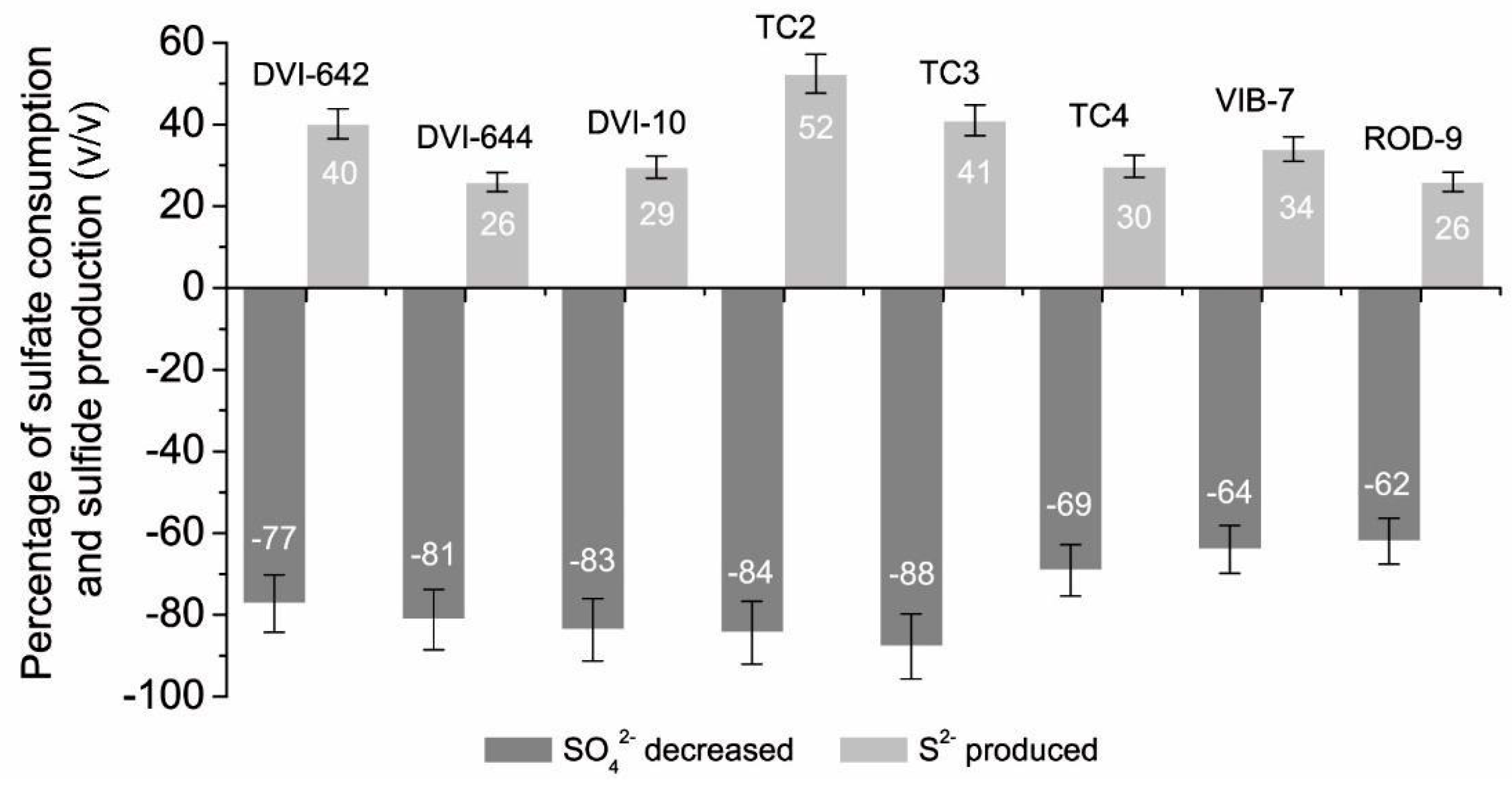
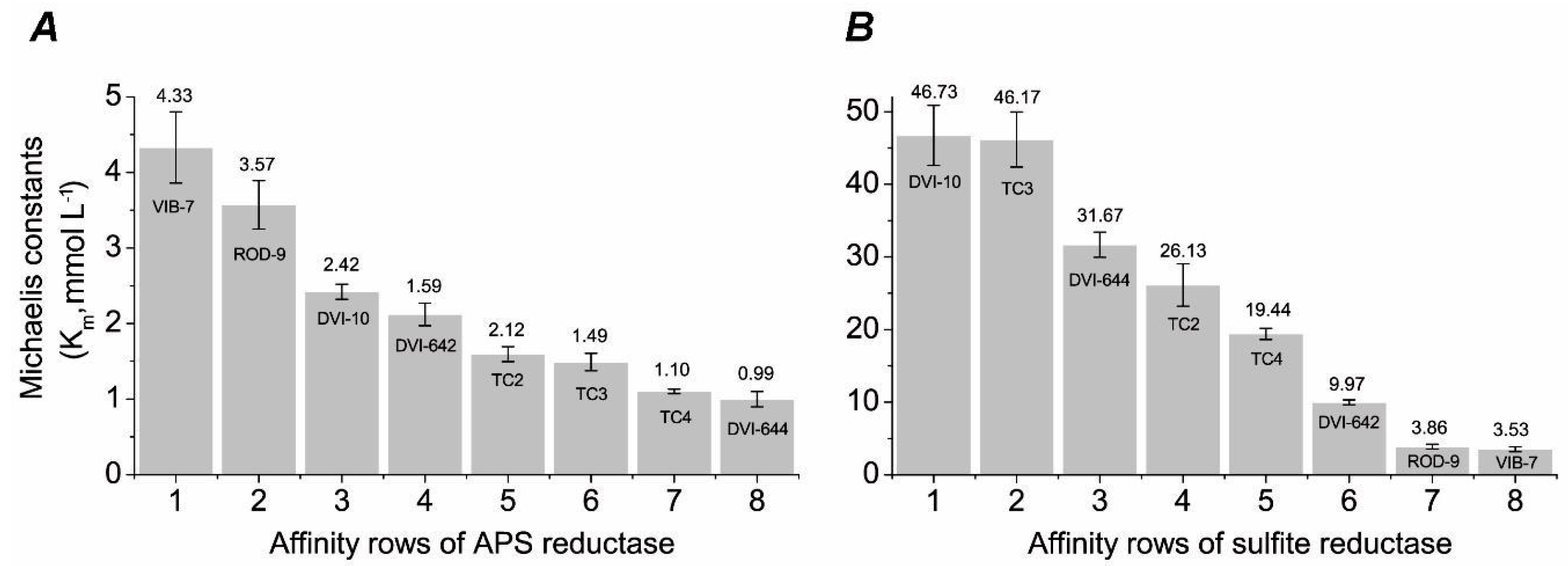
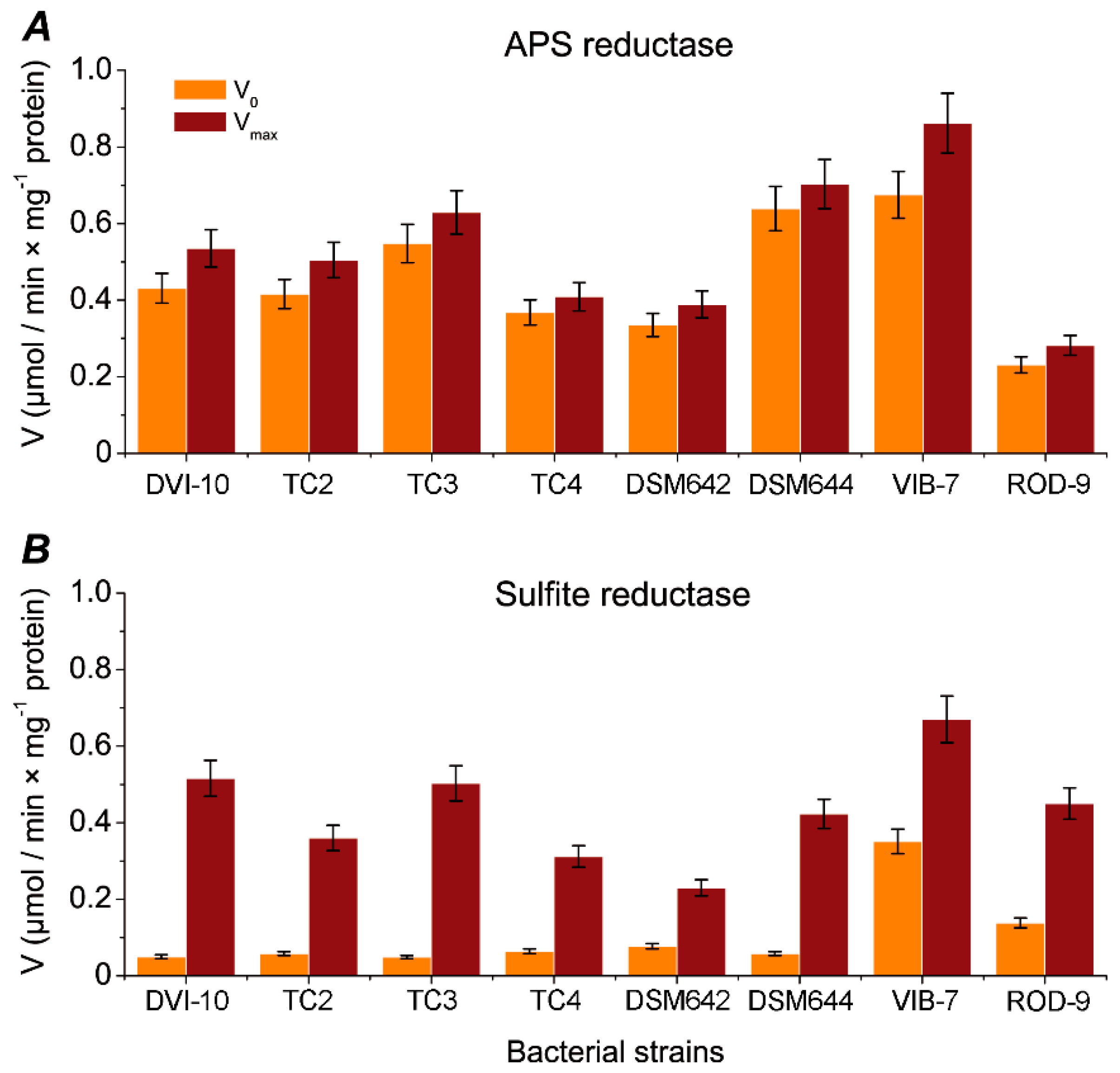
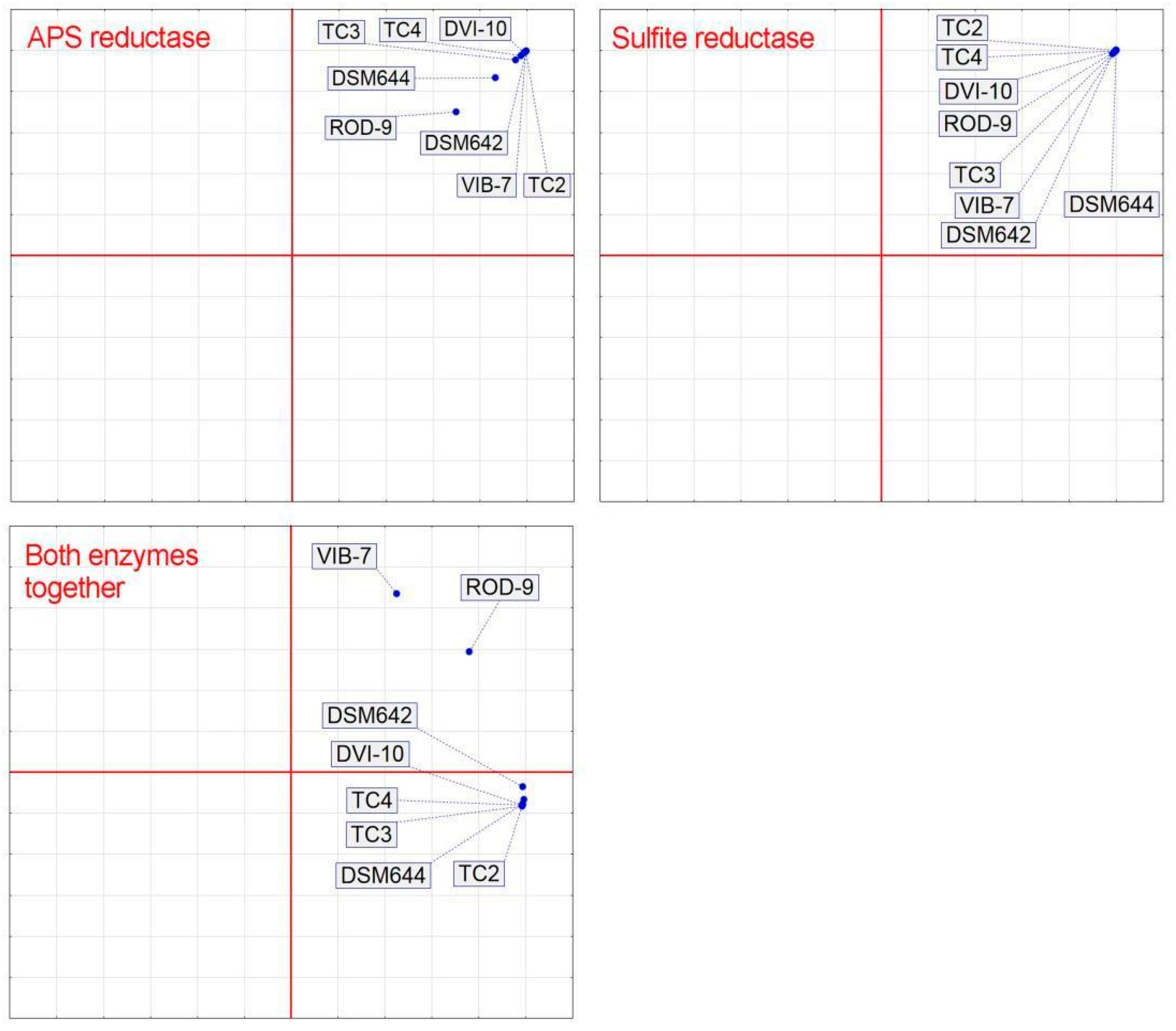
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barton, L.L.; Hamilton, W.A. Sulphate-Reducing Bacteria: Environmental and Engineered Systems; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Pester, M.; Knorr, K.-H.; Friedrich, M.W.; Wagner, M.; Loy, A. Sulfate reducing microorganisms in wetlands—Fameless actors in carbon cycling and climate change. Front. Microbiol. 2012, 3, 72. [Google Scholar] [CrossRef] [PubMed]
- Bowles, M.W.; Mogollón, J.M.; Kasten, S.; Zabel, M.; Hinrichs, K.-U. Global rates of marine sulfate reduction and implications for subsea-floor metabolic activities. Science 2014, 344, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria Desulfovibrio piger Vib-7. Arch. Microbiol. 2019, 201, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen Sulfide as a Toxic Product in the Small–Large Intestine Axis and its Role in IBD Development. J. Clin. Med. 2019, 8, 1054. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Kotrsová, V.; Dordević, D.; Buňková, L.; Vítězová, M.; Amedei, A. Hydrogen Sulfide Effects on the Survival of Lactobacilli with Emphasis on the Development of Inflammatory Bowel Diseases. Biomolecules 2019, 9, 752. [Google Scholar] [CrossRef]
- Iutynska, G.A.; Purish, L.M.; Abdulina, D.R. Corrosive-Relevant Sulfidogenic Microbial Communities of Man-Caused Ecotopes; Lambert Academic Publishing: Saarbrücken, Germany, 2014. [Google Scholar]
- Abdulina, D.R.; Kurmakova, I.N.; Bondar, E.S. Seasonal Dynamics of Bacteria in Corrosive Biofilms Formed on the Surface of Wastewater Treatment Plants. J. Water Chem. Technol. 2019, 41, 44–51. [Google Scholar] [CrossRef]
- Kotrsová, V.; Kushkevych, I. Possible methods for evaluation of hydrogen sulfide toxicity against lactic acid bacteria. Biointerface Res. Appl. Chem. 2019, 9, 4066–4069. [Google Scholar]
- Kushkevych, I.; Leščanová, O.; Dordević, D.; Jančíková, S.; Hošek, J.; Vítězová, M.; Buňková, L.; Drago, L. The Sulfate-Reducing Microbial Communities and Meta-Analysis of Their Occurrence during Diseases of Small–Large Intestine Axis. J. Clin. Med. 2019, 8, 1656. [Google Scholar] [CrossRef]
- Kushkevych, I.; Fafula, R.; Parak, T.; Bartoš, M. Activity of Na+/K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Acta Vet. Brno 2015, 84, 3–12. [Google Scholar] [CrossRef]
- Kushkevych, I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Pol. J. Microbiol. 2015, 64, 107–114. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Kos, J.; Kollár, P.; Jampilek, J. Effect of selected 8-hydroxyquinoline-2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J. Appl. Biomed. 2018, 16, 241–246. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kollar, P.; Suchy, P.; Parak, T.; Pauk, K.; Imramovsky, A. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuro Endocrinol. Lett. 2015, 36, 106–113. [Google Scholar]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Growth and activities of sulphate-reducing bacteria in gut contents of health subjects and patients with ulcerative colitis. Fems. Microbiol. Ecol. 1991, 86, 103–112. [Google Scholar] [CrossRef]
- Coutinho, C.M.L.M.; Coutinho-Silva, R.; Zinkevich, V.; Pearce, C.B.; Ojcius, D.M.; Beech, I. Sulphate-reducing bacteria from ulcerative colitis patients induce apoptosis of gastrointestinal epithelial cells. Microb. Pathog. 2017, 112, 126–134. [Google Scholar] [CrossRef]
- Černý, M.; Vítězová, M.; Vítěz, T.; Bartoš, M.; Kushkevych, I. Variation in the distribution of hydrogen producers from the clostridiales order in biogas reactors depending on different input substrates. Energies 2018, 11, 3270. [Google Scholar] [CrossRef]
- Kushkevych, I.V. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochem. Pol. 2015, 62, 1037–1108. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M.; Kollár, P. Cross-correlation analysis of the Desulfovibrio growth parameters of intestinal species isolated from people with colitis. Biologia 2018, 73, 1137–1143. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Analysis of pH dose-dependent growth of sulfate-reducing bacteria. Open Med. Wars. 2019, 14, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Tomasova, L.; Konopelski, P.; Ufnal, M. Gut bacteria and hydrogen sulfide: The new old players in circulatory system homeostasis. Molecules 2016, 21, 1558. [Google Scholar] [CrossRef] [PubMed]
- Kováč, J.; Vítězová, M.; Kushkevych, I. Metabolic activity of sulfate-reducing bacteria from rodents with colitis. Open Med. Wars. 2018, 13, 344–349. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Fedrová, P.; Vochyanová, Z.; Paráková, L.; Hošek, J. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet. Brno 2017, 86, 405–411. [Google Scholar] [CrossRef]
- Leavitt, W.D.; Bradley, A.S.; Santos, A.A.; Pereira, I.A.C.; Johnston, D.T. Sulfur isotope effects of dissimilatory sulfite reductase. Front. Microbiol. 2015, 6, 1392. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Bartoš, M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017, 12, 82–91. [Google Scholar] [CrossRef]
- Kushkevych, I.; Vítězová, M.; Vítěz, T.; Kováč, J.; Kaucká, P.; Jesionek, W.; Bartoš, M.; Barton, L. A new combination of substrates: Biogas production and diversity of the methanogenic microorganisms. Open Life Sci. 2018, 13, 119–128. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Kollar, P. Analysis of physiological parameters of Desulfovibrio strains from individuals with colitis. Open Life Sci. 2018, 13, 481–488. [Google Scholar] [CrossRef]
- Friedrich, M.W. Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol. 2002, 184, 278–289. [Google Scholar] [CrossRef]
- Fritz, G.; Büchert, T.; Huber, H.; Stetter, K.O.; Kroneck, P.M. Adenylylsulfate reductases from archaea and bacteria are 1:1 alpha beta-heterodimeric iron-sulfur flavoenzymes—high similarity of molecular properties emphasizes their central role in sulfur metabolism. FEBS Lett. 2000, 473, 63–66. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kováč, J.; Vítězová, M.; Vítěz, T.; Bartoš, M. The diversity of sulfate-reducing bacteria in the seven bioreactors. Arch. Microbiol. 2018, 200, 945–950. [Google Scholar] [CrossRef]
- Kováč, J.; Kushkevych, I. New modification of cultivation medium for isolation and growth of intestinal sulfate-reducing bacteria. In Proceedings of the International PhD Students Conference Mendel Net, Brno, Czech Republic, 6–7 November 2019; pp. 702–707. [Google Scholar]
- Kushkevych, I.; Kollar, P.; Ferreira, A.L.; Palma, D.; Duarte, A.; Lopes, M.M.; Bartos, M.; Pauk, K.; Imramovsky, A.; Jampilek, J. Antimicrobial effect of salicylamide derivatives against intestinal sulfate-reducing bacteria. J. Appl. Biomed. 2016, 14, 125–130. [Google Scholar] [CrossRef]
- Dahl, C.; Kredich, N.M.; Deutzmann, R.; Trlfper, H.G. Dissimilatory sulphite reductase from Archaeoglobus fulgidus: Physico-chemical properties of the enzyme and cloning, sequencing and analysis of the reductase genes. Microbiology 1993, 139, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Larsen, Ø.; Lien, T.; Birkeland, N.-K. Dissimilatory sulfite reductase from Archaeoglobus profundus and Desulfotomaculum thermocisternum: Phylogenetic and structural implications from gene sequences. Extremophiles 1999, 3, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Molitor, M.; Dahl, C.; Molitor, I.; Schäfer, U.; Speich, N.; Huber, R.; Deutzmann, R.; Trüper, H.G. A dissimilatory sirohaem-sulfite reductase-type protein from the hyperthermophilic archaeon Pyrobaculum islandicum. Microbiology 1998, 144, 529–541. [Google Scholar] [CrossRef]
- Huang, C.J.; Barret, E.L. Sequence analysis and expression of the Salmonella typhimurium asr operon encoding production of hydrogen sulfide from sulfite. J. Bacteriol. 1991, 173, 1544–1553. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.; Curle, C.; Laishley, E.J. Purification and characterization of an inducible dissimilatory type sulfite reductase from Clostridium pasteurianum. Arch. Microbiol. 1984, 138, 72–78. [Google Scholar] [CrossRef]
- Plugge, C.M.; Zhang, W.; Scholten, J.; Stams, A.J. Metabolic flexibility of sulphate educing bacteria. Front. Microbiol. 2011, 2, 81. [Google Scholar] [CrossRef] [PubMed]
- Abdulina, D.; Kováč, J.; Iutynska, G.; Kushkevych, I. ATP sulfurylase activity of sulfate-reducing bacteria from various ecotopes. 3Biotech 2020, 10, 55. [Google Scholar] [CrossRef]
- Postgate, J.R. The Sulfate-Reducing Bacteria, 2nd ed.; Cambridge University Press: Cambridge, UK, 1984. [Google Scholar]
- Asaulenko, L.H.; Abdulina, D.R.; Purish, L.M. Taxonomic position of certain representatives of sulphate-reducing corrosive microbial community. Mikrobiol. Zhurn. 2010, 72, 3–10. [Google Scholar]
- Purish, L.M.; Asaulenko, L.G.; Abdulina, D.R.; Iutinskaia, G.A. Biodiversity of sulfate-reducing bacteria growing on objects of heating systems. Mikrobiol. Zhurn. 2014, 76, 11–17. [Google Scholar]
- Kolmert, A.; Wikstrom, P.; Hallberg, K.B. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J. Microbiol. Methods 2000, 41, 179–184. [Google Scholar] [CrossRef]
- Sugiyama, M. Reagent Composition for Measuring Hydrogen Sulfide and Method for Measuring Hydrogen. U.S. Patent 6340596 B1 USA, 22 January 2002. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Huisingh, J.; McNeill, J.J.; Matrone, G. Sulfate reduction by a Desulfovibrio species isolated from sheep rumen. Appl. Environ. Microbiol. 1974, 28, 489–497. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Keleti, T. Basic Enzyme Kinetics; Akademiai Kiado: Budapest, Hungary, 1988. [Google Scholar]
- Bailey, N.T.J. Statistical Methods in Biology; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Peck, H.D., Jr.; Deacon, T.E.; Davidson, J.T. Studies on adenosine 5′-phosphosulfate reductase from desulfovibrio desulfuricans and thiobacillus thioparus I. The assay and purification. Biochim. et Biophys. Acta BBA Nucleic Acids Protein Synth. 1965, 96, 429–446. [Google Scholar] [CrossRef]
- Ishimoto, M. Biochemistry of sulfate-reduction. Enzyme system of sulfate-reduction of Desulfovibrio. Seikagaku 1960, 32, 1. [Google Scholar]
- Seki, Y.; Ishimoto, M. Catalytic activity of the chromophore of desulfoviridin, sirohydrochlorin, in sulfite reduction in the presence of iron. J. Biochem. 1979, 86, 273–276. [Google Scholar]
- Seki, Y.; Sogawa, N.; Ishimoto, M. Siroheme as an active catalyst in sulfite reduction. J. Biochem. 1981, 90, 1487–1492. [Google Scholar] [CrossRef]
- Oliveira, T.F.; Vonrhein, C.; Matias, P.M. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J. Biol. Chem. 2008, 283, 34141–34149. [Google Scholar] [CrossRef]
- Canfield, D.E.; Habicht, K.S.; Thamdrup, B.O. The Archean sulfur cycle and the early history of atmospheric oxygen. Science 2000, 288, 658–661. [Google Scholar] [CrossRef]
- Kushkevych, I.; Cejnar, J.; Treml, J.; Dordević, D.; Kollar, P.; Vítězová, M. Recent Advances in Metabolic Pathways of Sulfate Reduction in Intestinal Bacteria. Cells 2020, 9, 698. [Google Scholar] [CrossRef]
- Kushkevych, I.; Kos, J.; Kollar, P.; Kralova, K.; Jampilek, J. Activity of ring-substituted 8-hydroxyquinoline-2-carboxanilides against intestinal sulfate-reducing bacteria Desulfovibrio piger. Med. Chem. Res. 2018, 27, 278–284. [Google Scholar] [CrossRef]
- Jørgensen, B.B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 1982, 296, 643–645. [Google Scholar] [CrossRef]
- Florin, T.H.; Neale, G.; Goretski, S. Sulfate in food and beverages. J. Food Compos. Anal. 1993, 6, 140–151. [Google Scholar] [CrossRef]
- D’Hondt, S.; Jørgensen, B.B.; Miller, D.J. Distributions of microbial activities in deep subseafloor sediments. Science 2004, 306, 2216–2221. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Kakiuchi, R.; Yamaguchi, T.; Takai, K.; Inagaki, F.; Imachi, H. Phylogenetic Diversity of aprA Genes in Subseafloor Sediments on the Northwestern Pacific Margin off Japan. Microbes Environ. 2015, 30, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, М.C.; der Vartanian, D.V.; Peck, H.D. On the nature of the oxidation reduction properties of nitrite reductase from Desulfovibrio desulfuricans. Biochem. Biophys. Res. Commun. 1980, 96, 278–285. [Google Scholar] [CrossRef]
- Peck, H.D.; Legall, J.; Vanbeeumen, J. Biochemistry of dissimilatory sulfate reduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982, 298, 443–466. [Google Scholar]
- Seitz, H.J.; Cypionka, H. Chemolithotrophic growth of Desulfovibrio desulfuricans with hydrogen coupled to ammonification of nitrate or nitrite. Arch. Microbiol. 1986, 146, 63–67. [Google Scholar] [CrossRef]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef]
- Voordouw, G.; Armstrong, S.M.; Reimer, M.F.; Fouts, B.; Telang, A.J.; Shen, Y. Characterization of 16S rRNA genes from oil field microbial communities indicates the presence of a variety of sulfate-reducing, fermentative, and sulfide-oxidizing bacteria. Appl. Environ. Microbiol. 1996, 62, 1623–1629. [Google Scholar] [CrossRef]
- Widdel, F.; Kohring, G.W.; Mayer, F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Arch. Microbiol. 1983, 134, 286–294. [Google Scholar] [CrossRef]
- Klein, M.; Friedrich, M.; Roger, A.J.; Hugenholtz, P.; Fishbain, S.; Abicht, H.; Blackall, L.L.; Stahl, D.A. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 2001, 183, 6028–6035. [Google Scholar] [CrossRef] [PubMed]
- Laue, H.; Friedrich, M.; Ruff, J.; Cook, A.M. Dissimilatory sulfite reductase (desulfoviridin) of the taurine-degrading, non-sulfate-reducing bacterium Bilophila wadsworthia RZATAU contains a fused DsrB-DsrD subunit. J. Bacteriol. 2001, 183, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Loy, A.; Duller, S.; Baranyi, C.; Mussmann, M.; Ott, J.; Sharon, I.; Béjà, O.; Le Paslier, D.; Dahl, C.; Wagner, M. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 2009, 11, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Dordević, D.; Jančíková, S.; Vítězová, M.; Kushkevych, I. Hydrogen sulfide toxicity in the gut environment: Meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes. J. Adv. Res. 2020. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J. Adv. Res. 2020. [Google Scholar] [CrossRef]

| No | Bacterial Strains | Source of Isolation (Country of Origin) | References |
|---|---|---|---|
| 1 | Desulfovibrio desulfuricans DSM642 | Corrosion product (DSMZ collection, Germany) | [40] |
| 2 | Desulfovibrio vulgaris DSM644 | Soil (DSMZ collection, Germany) | |
| 3 | Desulfovibrio sp. 10 (UCM B-11503) | Corrosion products of steel construction of DniproHES (Ukraine) | [41] |
| 4 | Desulfovibrio sp. TC2 | Corrosion products and slime from heat system construction (Kyiv, Ukraine) | [42] |
| 5 | Desulfotomaculum sp. TC3 | ||
| 6 | Desulfomicrobium sp. TC4 | ||
| 7 | Desulfovibrio piger Vib-7 | Human feces from people with colitis (Brno, Czech Republic) | [11] |
| 8 | Desulfomicrobium orale Rod-9 |
| Bacterial Cultures | SO42− at the Beginning (mmol L−1) | SO42− after 24 h Cultivation (mmol L−1) | S2− at the Beginning (mmol L−1) | S2− after 24 h Cultivation (mmol L−1) |
|---|---|---|---|---|
| D. desulfuricans DSM642 | 6.28 ± 0.446 | 1.43 ± 0.092 | 0.72 ± 0.066 | 4.53 ± 0.080 |
| D. vulgaris DSM644 | 7.27 ± 0.379 | 1.37 ± 0.101 | 0.43 ± 0.040 | 4.18 ± 0.248 |
| Desulfovibrio sp. 10 | 5.05 ± 0.121 | 0.83 ± 0.055 | 0.44 ± 0.038 | 3.83 ± 0.265 |
| Desulfovibrio sp. TC2 | 5.56 ± 0.192 | 0.87 ± 0.193 | 1.01 ± 0.092 | 4.86 ± 0.289 |
| Desulfotomaculun sp. TC3 | 6.35 ± 0.142 | 0.78 ± 0.028 | 0.78 ± 0.071 | 4.76 ± 0.298 |
| Desulfomicrobium sp. TC4 | 5.43 ± 0.292 | 1.58 ± 0.156 | 0.43 ± 0.039 | 3.61 ± 0.256 |
| Desulfovibrio piger Vib-7 | 3.50 ± 0.032 | 1.25 ± 0.110 | 0.30 ± 0.027 | 2.21 ± 0.192 |
| Desulfomicrobium orale Rod-9 | 3.53 ± 0.030 | 1.34 ± 0.123 | 0.20 ± 0.018 | 1.97 ± 0.185 |
| Sample | Specific Activity (nkat) | |
|---|---|---|
| APS Reductase | Sulfite Reductase | |
| DSM642 | 2.166 ± 0.020 | 3.833 ± 0.35 |
| DSM644 | 3.900 ± 0.034 | 7.050 ± 0.65 |
| DVI-10 | 2.967 ± 0.016 | 8.600 ± 0.40 |
| DVI-TC2 | 2.800 ± 0.022 | 6.000 ± 0.23 |
| DTM-TC3 | 3.500 ± 0.034 | 8.383 ± 0.37 |
| DMI-TC4 | 2.226 ± 0.019 | 5.200 ± 0.48 |
| VIB-7 | 5.666 ± 0.483 | 0.533 ± 0.026 |
| ROD-9 | 1.833 ±0.200 | 0.466 ± 0.022 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushkevych, I.; Abdulina, D.; Kováč, J.; Dordević, D.; Vítězová, M.; Iutynska, G.; Rittmann, S.K.-M.R. Adenosine-5′-Phosphosulfate- and Sulfite Reductases Activities of Sulfate-Reducing Bacteria from Various Environments. Biomolecules 2020, 10, 921. https://doi.org/10.3390/biom10060921
Kushkevych I, Abdulina D, Kováč J, Dordević D, Vítězová M, Iutynska G, Rittmann SK-MR. Adenosine-5′-Phosphosulfate- and Sulfite Reductases Activities of Sulfate-Reducing Bacteria from Various Environments. Biomolecules. 2020; 10(6):921. https://doi.org/10.3390/biom10060921
Chicago/Turabian StyleKushkevych, Ivan, Daryna Abdulina, Jozef Kováč, Dani Dordević, Monika Vítězová, Galyna Iutynska, and Simon K.-M. R. Rittmann. 2020. "Adenosine-5′-Phosphosulfate- and Sulfite Reductases Activities of Sulfate-Reducing Bacteria from Various Environments" Biomolecules 10, no. 6: 921. https://doi.org/10.3390/biom10060921
APA StyleKushkevych, I., Abdulina, D., Kováč, J., Dordević, D., Vítězová, M., Iutynska, G., & Rittmann, S. K.-M. R. (2020). Adenosine-5′-Phosphosulfate- and Sulfite Reductases Activities of Sulfate-Reducing Bacteria from Various Environments. Biomolecules, 10(6), 921. https://doi.org/10.3390/biom10060921







