MicroRNA Alterations in a Tg501 Mouse Model of Prion Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Collection
2.3. RNA Extraction
2.4. miRNA Sequencing and Data Analysis
2.5. Quantitative PCR
3. Results
3.1. Small RNA Sequencing
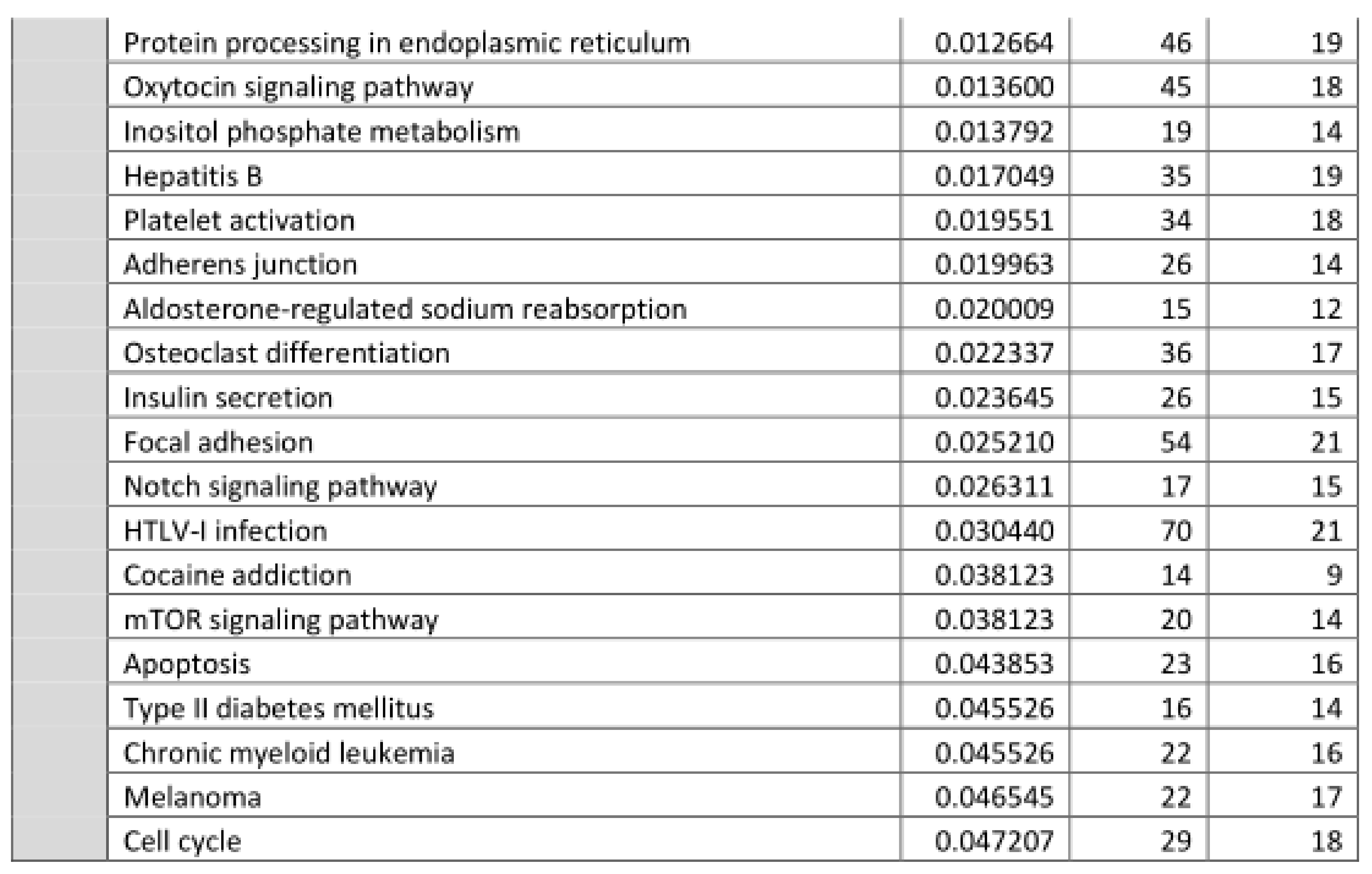
3.2. Functional Annotation of Differentially Expressed miRNAs
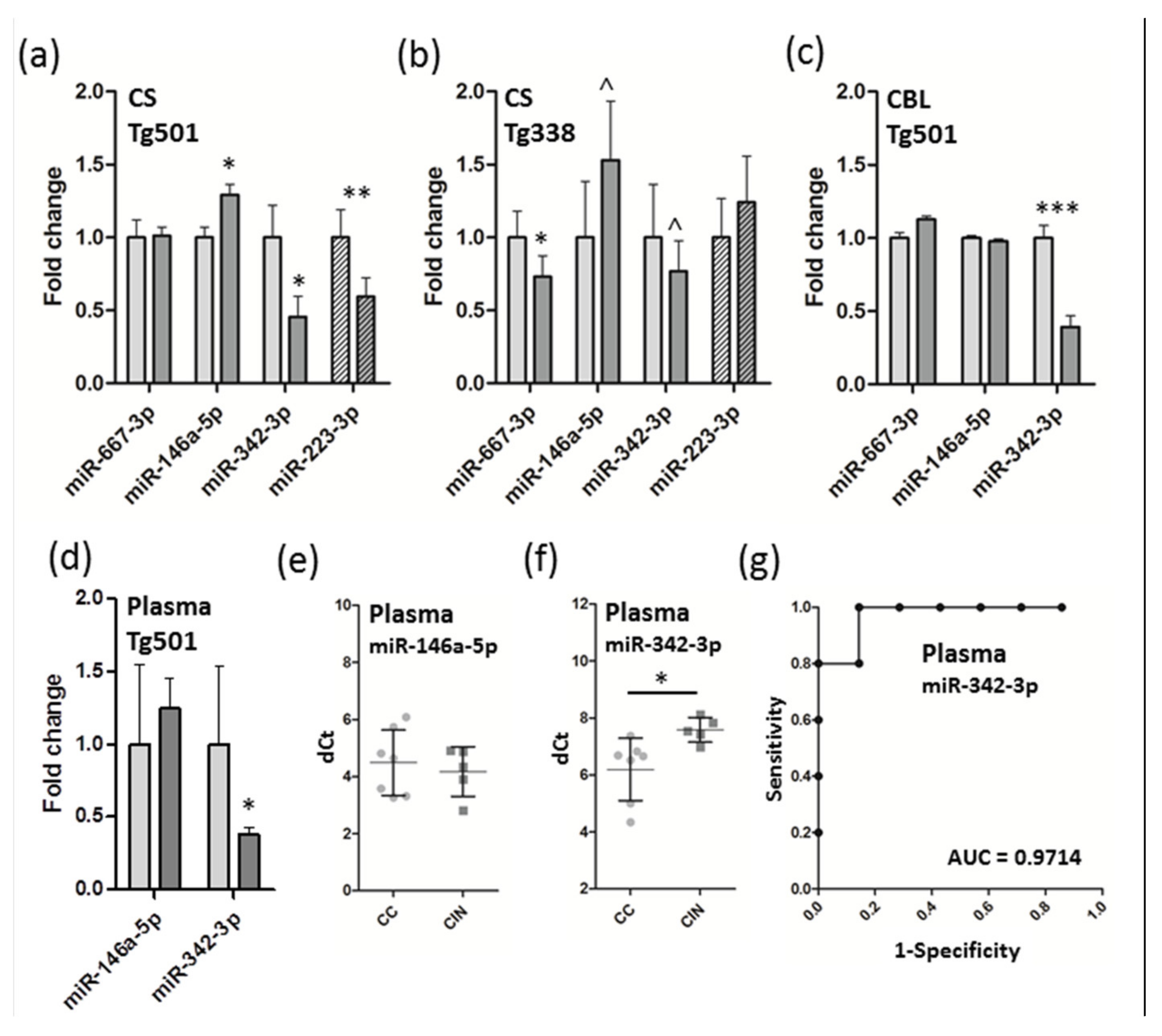
3.3. Verification of miRNA Expression by QRT-PCR and Detection of Candidate Biomarkers from Plasma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
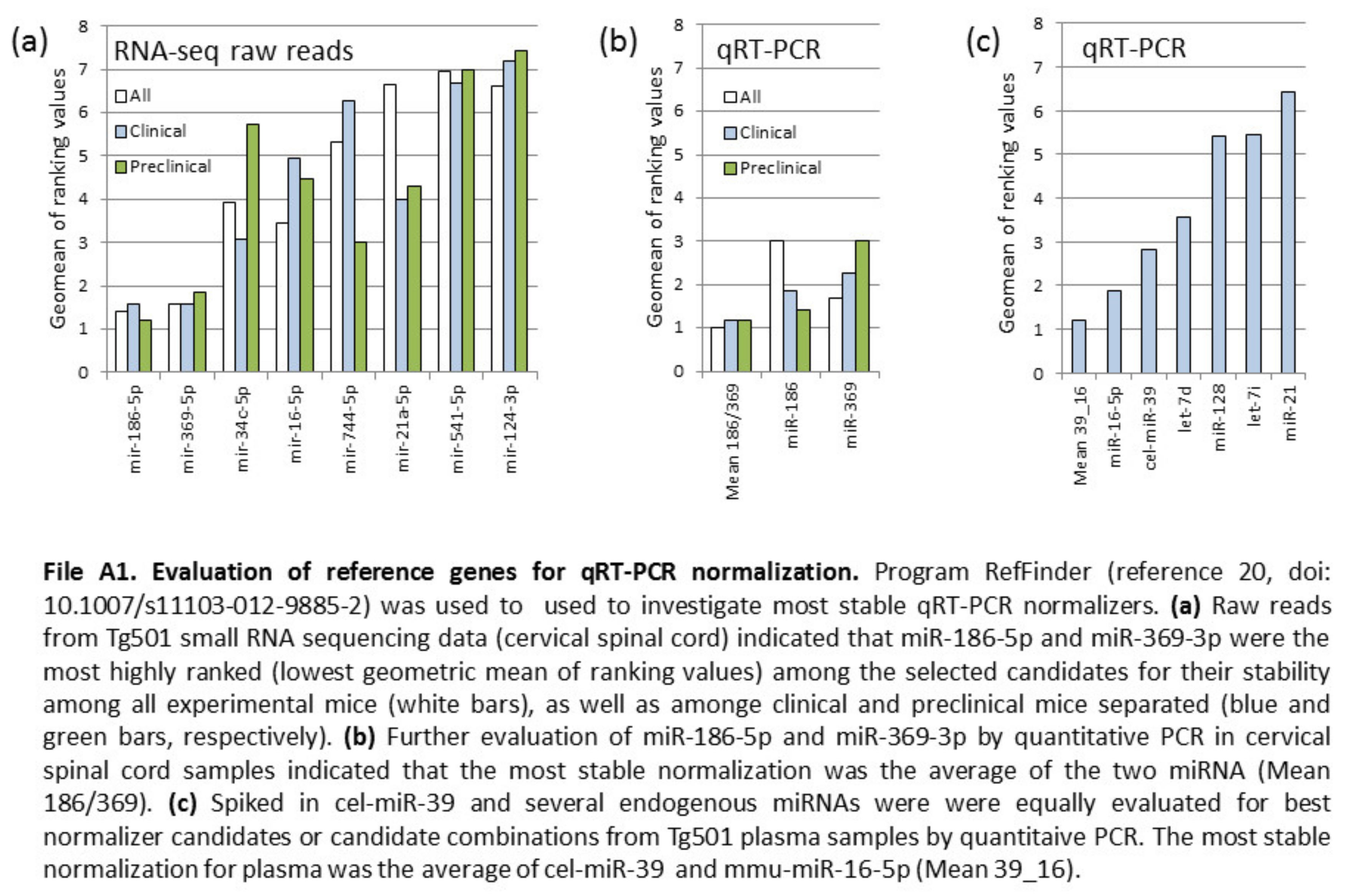
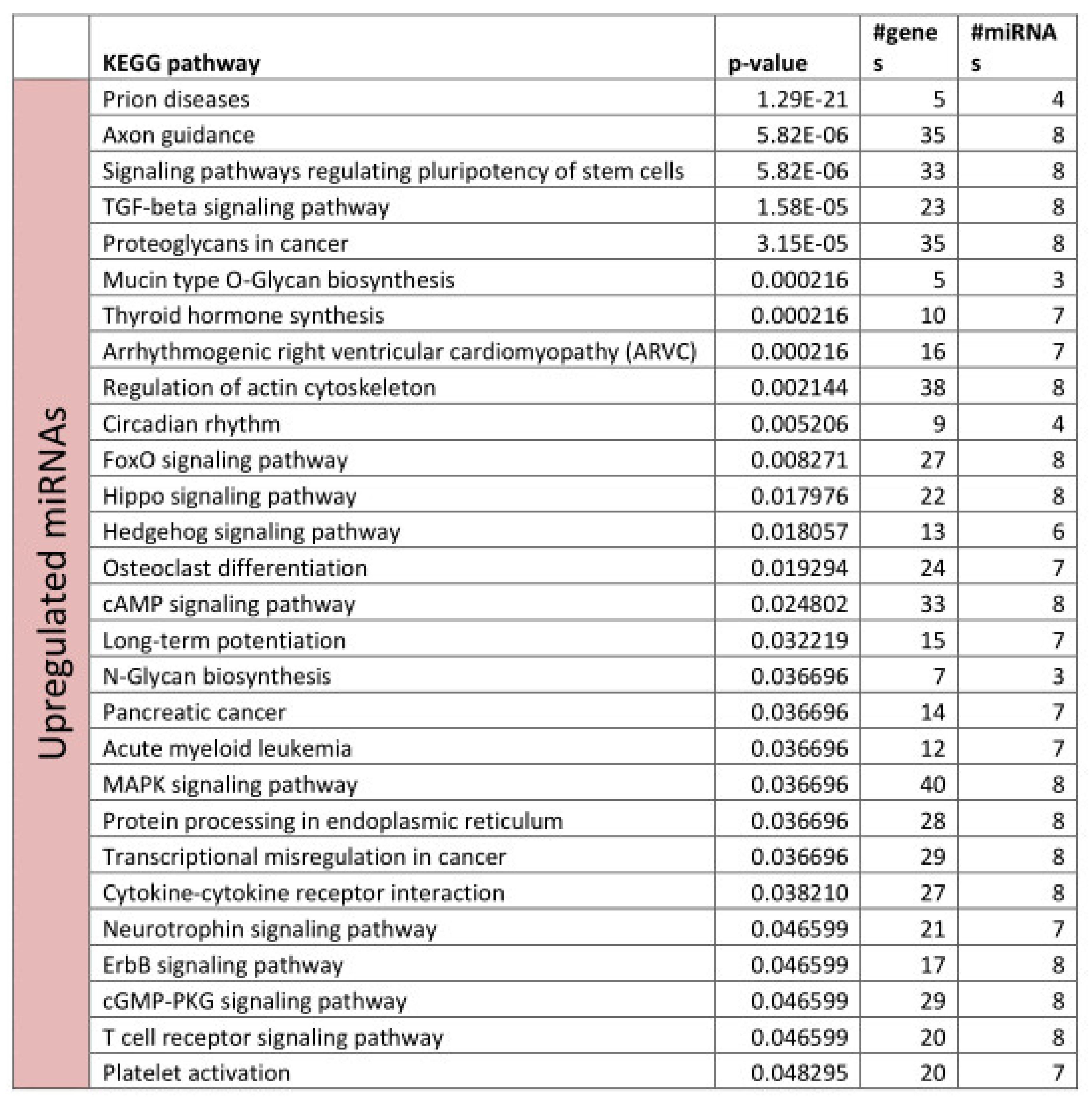
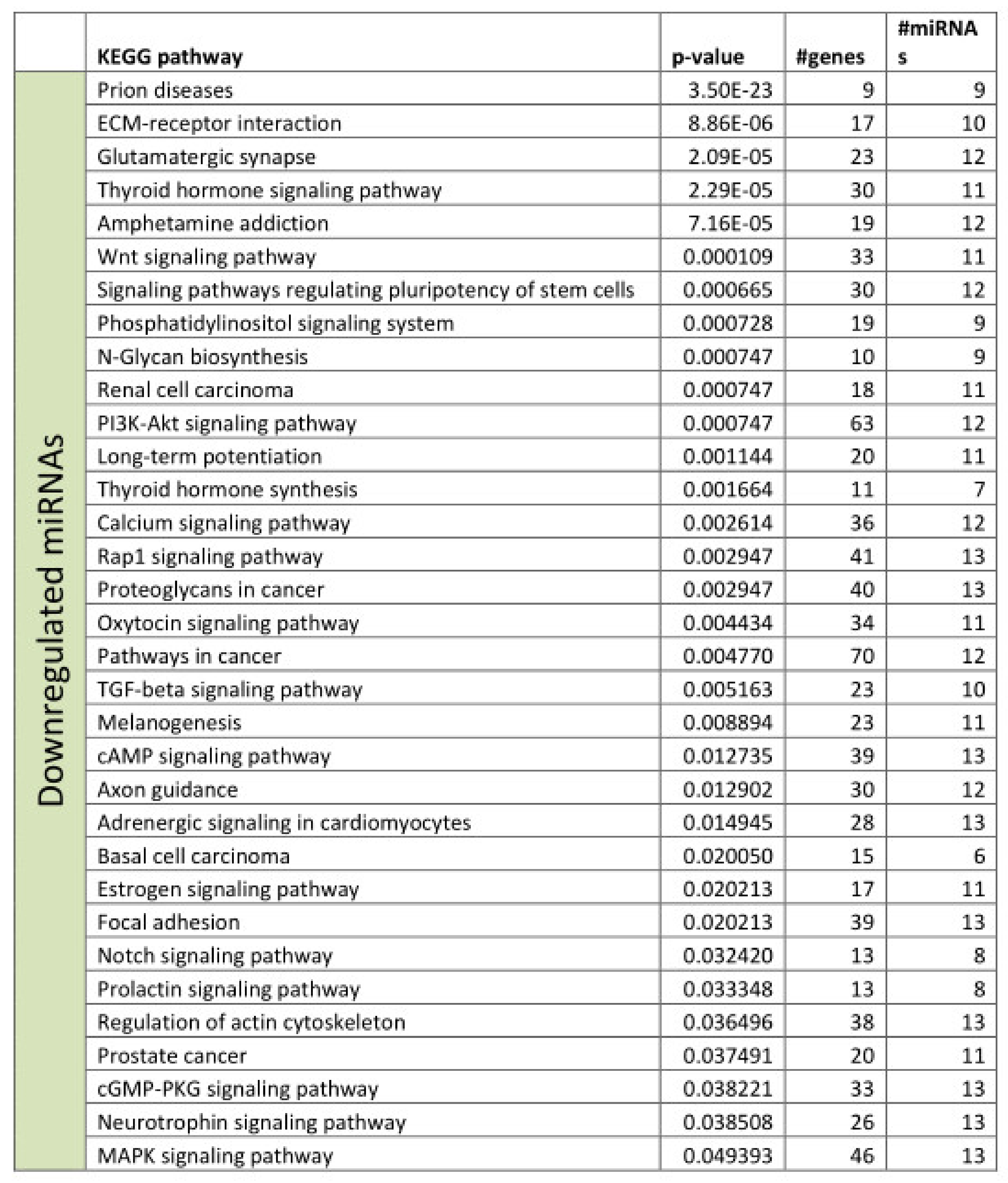
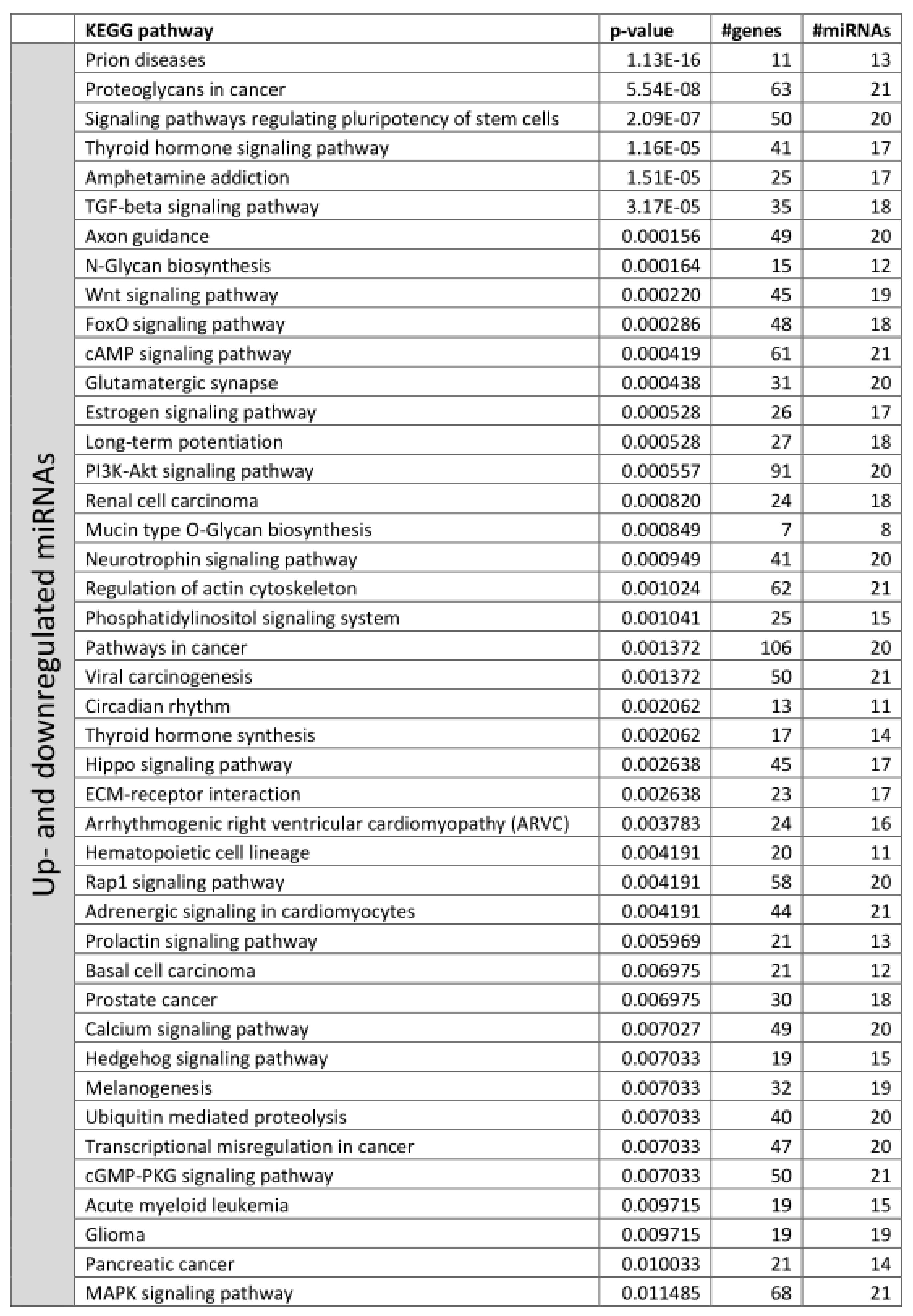
References
- Friedman, R.; Farh, K.K.-H.; Burge, C.B.; Bartel, B. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Kanata, E.; Thüne, K.; Xanthopoulos, K.; Ferrer, I.; Dafou, D.; Zerr, I.; Sklaviadis, T.; Llorens, F. MicroRNA Alterations in the Brain and Body Fluids of Humans and Animal Prion Disease Models: Current Status and Perspectives. Front. Aging Neurosci. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Maciotta, S.; Meregalli, M.; Torrente, Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell. Neurosci. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Llorens, F.; Thüne, K.; Marti, E.; Kanata, E.; Dafou, D.; Diaz-Lucena, D.; Vivancos, A.; Shomroni, O.; Zafar, S.; Schmitz, M.; et al. Regional and subtype-dependent miRNA signatures in sporadic Creutzfeldt-Jakob disease are accompanied by alterations in miRNA silencing machinery and biogenesis. PLOS Pathog. 2018, 14, e1006802. [Google Scholar] [CrossRef] [PubMed]
- Rubio, D.S.; López-Pérez, Ó.; Pablo, Álvaro, D.A.; Bolea, R.; Osta, R.; Badiola, J.J.; Zaragoza, P.; Martín-Burriel, I.; Toivonen, J.M. Increased circulating microRNAs miR-342-3p and miR-21-5p in natural sheep prion disease. J. Gen. Virol. 2017, 98, 305–310. [Google Scholar] [CrossRef]
- Slota, J.A.; Medina, S.J.; Klassen, M.; Gorski, D.; Mesa, C.M.; Robertson, C.; Mitchell, G.; Coulthart, M.B.; Pritzkow, S.; Soto, C.; et al. Identification of circulating microRNA signatures as potential biomarkers in the serum of elk infected with chronic wasting disease. Sci. Rep. 2019, 9, 19705–19715. [Google Scholar] [CrossRef] [PubMed]
- Majer, A.; Medina, S.J.; Niu, Y.; Abrenica, B.; Manguiat, K.J.; Frost, K.L.; Philipson, C.S.; Sorensen, D.L.; Booth, S. Early Mechanisms of Pathobiology Are Revealed by Transcriptional Temporal Dynamics in Hippocampal CA1 Neurons of Prion Infected Mice. PLOS Pathog. 2012, 8, e1003002. [Google Scholar] [CrossRef]
- Aguilar-Calvo, P.; Espinosa, J.C.; Pintado, B.; Gutiérrez-Adán, A.; Alamillo, E.; Miranda, A.; Prieto, I.; Bossers, A.; Andreoletti, O.; Torres, J.M. Role of the goat K222-PrP(C) polymorphic variant in prion infection resistance. J. Virol. 2014, 88, 2670–2676. [Google Scholar] [CrossRef]
- Laude, H.; Vilette, D.; Le Dur, A.; Archer, F.; Soulier, S.; Besnard, N.; Essalmani, R.; Vilotte, J.-L. New in vivo and ex vivo models for the experimental study of sheep scrapie: Development and perspectives. Comptes Rendus Boil. 2002, 325, 49–57. [Google Scholar] [CrossRef]
- Vilotte, J.-L.; Soulier, S.; Essalmani, R.; Stinnakre, M.-G.; Vaiman, D.; Lepourry, L.; Da Silva, J.C.; Besnard, N.; Dawson, M.; Buschmann, A.; et al. Markedly Increased Susceptibility to Natural Sheep Scrapie of Transgenic Mice Expressing Ovine PrP. J. Virol. 2001, 75, 5977–5984. [Google Scholar] [CrossRef]
- López-Pérez, Ó.; Toivonen, J.M.; Otero, A.; Solanas, L.; Zaragoza, P.; Badiola, J.J.; Osta, R.; Bolea, R.; Martín-Burriel, I. Impairment of autophagy in scrapie-infected transgenic mice at the clinical stage. Lab. Investig. 2019, 100, 52–63. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Capece, V.; Vizcaino, J.C.G.; Vidal, R.; Rahman, R.-U.; Centeno, T.P.; Shomroni, O.; Suberviola, I.; Fischer, A.; Bonn, S. Oasis: Online analysis of small RNA deep sequencing data. Bioinform. 2015, 31, 2205–2207. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.-U.; Gautam, A.; Bethune, J.; Sattar, A.; Fiosins, M.; Magruder, D.S.; Capece, V.; Shomroni, O.; Bonn, S. Oasis 2: Improved online analysis of small RNA-seq data. BMC Bioinform. 2018, 19, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Vitsios, D.M.; Enright, A.J. Chimira: Analysis of small RNA sequencing data and microRNA modifications. Bioinform. 2015, 31, 3365–3367. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, 460–466. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, N.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant. Mol. Boil. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Backes, C.; Khaleeq, Q.T.; Meese, E.; Keller, A. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res. 2016, 44, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Glazov, E.A.; McWilliam, S.; Barris, W.C.; Dalrymple, B.P. Origin, Evolution, and Biological Role of miRNA Cluster in DLK-DIO3 Genomic Region in Placental Mammals. Mol. Boil. Evol. 2008, 25, 939–948. [Google Scholar] [CrossRef]
- Winter, J. MicroRNAs of the miR379-410 cluster: New players in embryonic neurogenesis and regulators of neuronal function. Neurogenesis 2015, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, Y.-K.; Lee, K.-P.; Lee, S.-M.; Kang, T.-W.; Kim, H.-J.; Dho, S.H.; Kim, S.-Y.; Kwon, K.-S. Genome-wide profiling of the microRNA-mRNA regulatory network in skeletal muscle with aging. Aging 2014, 6, 524–544. [Google Scholar] [CrossRef] [PubMed]
- Mikovic, J.; Sadler, K.; Butchart, L.; Voisin, S.; Gerlinger-Romero, F.; Della Gatta, P.; Grounds, M.D.; Lamon, S. MicroRNA and Long Non-coding RNA Regulation in Skeletal Muscle From Growth to Old Age Shows Striking Dysregulation of the Callipyge Locus. Front. Genet. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Montag, J.; Hitt, R.; Opitz, L.; Schulz-Schaeffer, W.J.; Hunsmann, G.; Motzkus, D. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Molecular Neurodegeneration 2009, 4, 36–43. [Google Scholar] [CrossRef]
- Saba, R.; Goodman, C.D.; Huzarewich, R.L.C.H.; Robertson, C.A.; Booth, S. A miRNA Signature of Prion Induced Neurodegeneration. PLoS ONE 2008, 3, 1–11. [Google Scholar] [CrossRef]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef]
- Wang, L.-L.; Min, L.; Guo, Q.-D.; Zhang, J.-X.; Jiang, H.-L.; Shao, S.; Xing, J.-G.; Yin, L.-L.; Liu, J.-H.; Liu, R.; et al. Profiling microRNA from Brain by Microarray in a Transgenic Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Fu, Y.; Hu, X.; Zheng, C.; Sun, G.; Xu, J.; Liu, R.; Luo, S.; Cao, P. Intrahippocampal miR-342-3p inhibition reduces beta-amyloid plaques and ameliorates learning and memory in Alzheimer’s disease. Metab Brain Dis. 2019, 34, 1355–1363. [Google Scholar] [CrossRef]
- Tan, L.; Yu, J.; Tan, M.-S.; Liu, Q.-Y.; Wang, H.-F.; Zhang, W.; Jiang, T.; Tan, L. Genome-Wide Serum microRNA Expression Profiling Identifies Serum Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 40, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Doecke, J.D.; Sharples, R.; Villemagne, V.L.; Fowler, C.J.; Rembach, A.; Martins, R.N.; Rowe, C.C.; Macaulay, S.L.; Masters, C.L.; et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 2014, 20, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Nuzziello, N.; Introna, A.; Consiglio, A.; Licciulli, F.; D’Errico, E.; Scarafino, A.; Distaso, E.; Simone, I.L. Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2018, 11, 1–16. [Google Scholar] [CrossRef]
- Boese, A.S.; Saba, R.; Campbell, K.; Majer, A.; Medina, S.; Burton, L.; Booth, T.F.; Chong, P.; Westmacott, G.; Dutta, S.M.; et al. MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol. Cell. Neurosci. 2016, 71, 13–24. [Google Scholar] [CrossRef]
- Gao, C.; Wei, J.; Zhang, B.-Y.; Shi, Q.; Chen, C.; Wang, J.; Shi, Q.; Dong, X.-P. MiRNA expression profiles in the brains of mice infected with scrapie agents 139A, ME7 and S15. Emerg. Microbes Infect. 2016, 5, 115–125. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Dua, P.; Pogue, A.I.; Eicken, C.; Hill, J.M. Upregulation of micro RNA-146a (miRNA-146a), a marker for inflammatory neurodegeneration, in sporadic Creutzfeldt-Jakob disease (sCJD) and Gerstmann-Straussler-Scheinker (GSS) syndrome. J. Toxicol. Environ. Heal. Part. A 2011, 74, 1460–1468. [Google Scholar] [CrossRef]
- Saba, R.; Gushue, S.; Huzarewich, R.L.C.H.; Manguiat, K.; Medina, S.; Robertson, C.; Booth, S. MicroRNA 146a (miR-146a) Is Over-Expressed during Prion Disease and Modulates the Innate Immune Response and the Microglial Activation State. PLoS ONE 2012, 7, 1–16. [Google Scholar] [CrossRef]
- Sierksma, A.; Lu, A.; Salta, E.; Eynden, E.V.; Callaerts-Vegh, Z.; D’hooge, R.; Blum, D.; Buée, L.; Fiers, M.; De Strooper, B. Deregulation of neuronal miRNAs induced by amyloid-beta or TAU pathology. Mol. Neurodegener. 2018, 13, 54–69. [Google Scholar] [CrossRef]
- Lukiw, W.J.; Alexandrov, P.N. Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer’s disease (AD) brain. Mol. Neurobiol. 2012, 46, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Butovsky, O.; Jedrychowski, M.P.; Cialic, R.; Krasemann, S.; Murugaiyan, G.; Fanek, Z.; Greco, D.J.; Wu, P.M.; Doykan, C.E.; Kiner, O.; et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 2014, 77, 75–99. [Google Scholar] [CrossRef] [PubMed]
- Maffioletti, E.; Milanesi, E.; Ansari, A.; Zanetti, O.; Galluzzi, S.; Geroldi, C.; Gennarelli, M.; Bocchio-Chiavetto, L. miR-146a Plasma Levels Are Not Altered in Alzheimer’s Disease but Correlate With Age and Illness Severity. Front. Aging Neurosci 2020, 11, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Denk, J.; Boelmans, K.; Siegismund, C.S.; Lassner, D.; Arlt, S.; Jahn, H. MicroRNA Profiling of CSF Reveals Potential Biomarkers to Detect Alzheimer’s Disease. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Li, J.; Huang, L.; Chen, X.; Li, D.; Wang, T.; Hu, C.; Xu, J.; Zhang, C.; Zen, K.; et al. Serum MicroRNA Profiles Serve as Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Dis. Markers 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Sanchez-Juan, P.; Bishop, M.T.; Kovacs, G.G.; Calero, M.; Aulchenko, Y.S.; Ladogana, A.; Boyd, A.; Lewis, V.; Ponto, C.; Calero, O.; et al. A Genome Wide Association Study Links Glutamate Receptor Pathway to Sporadic Creutzfeldt-Jakob Disease Risk. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef]
- Juźwik, C.A.; Drake, S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 10–16. [Google Scholar] [CrossRef]
- Jia, L.-H.; Liu, Y.-N. Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 2016, 34, 233–237. [Google Scholar] [CrossRef]
- Lusardi, T.A.; Phillips, J.I.; Wiedrick, J.T.; Harrington, C.A.; Lind, B.; Lapidus, J.A.; Quinn, J.F.; Saugstad, J.A. MicroRNAs in Human Cerebrospinal Fluid as Biomarkers for Alzheimer’s Disease. J. Alzheimer’s Dis. 2016, 55, 1223–1233. [Google Scholar] [CrossRef]
- Bauernfeind, F.; Rieger, A.; Schildberg, F.A.; Knolle, P.A.; Schmid-Burgk, J.L.; Hornung, V. NLRP3 Inflammasome Activity Is Negatively Controlled by miR-223. J. Immunol. 2012, 189, 4175–4181. [Google Scholar] [CrossRef]
- Duan, Y.; Kelley, N.; He, Y. Role of the NLRP3 inflammasome in neurodegenerative diseases and therapeutic implications. Neural Regen. Res. 2020, 15, 1249–1250. [Google Scholar] [CrossRef]
- Cheng, C.; Li, W.; Zhang, Z.; Yoshimura, S.; Hao, Q.; Zhang, C.; Wang, Z. MicroRNA-144 Is Regulated by Activator Protein-1 (AP-1) and Decreases Expression of Alzheimer Disease-related A Disintegrin and Metalloprotease 10 (ADAM10)*. J. Boil. Chem. 2013, 288, 13748–13761. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Russelakis-Carneiro, M.; Maundrell, K.; Castilla, J.; Soto, C. Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J. 2003, 22, 5435–5445. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Schwarz, A.; Neidhold, S.; Burwinkel, M.; Riemer, C.; Simon, D.; Kopf, M.; Otto, M.; Baier, M. Role of Interleukin-1 in Prion Disease-Associated Astrocyte Activation. Am. J. Pathol. 2004, 165, 671–678. [Google Scholar] [CrossRef]
- Taylor, D.R.; Hooper, N.M. The prion protein and lipid rafts (Review). Mol. Membr. Boil. 2006, 23, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G.; Gasque, P.; Ströbel, T.; Lindeck-Pozza, E.; Strohschneider, M.; Ironside, J.W.; Budka, H.; Guentchev, M. Complement activation in human prion disease. Neurobiol. Dis. 2004, 15, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Mays, C.E.; Armijo, E.; Morales, R.; Kramm, C.; Flores, A.; Tiwari, A.; Bian, J.; Telling, G.C.; Pandita, T.K.; Hunt, C.R.; et al. Prion disease is accelerated in mice lacking stress-induced heat shock protein 70 (HSP70). J. Boil. Chem. 2019, 294, 13619–13628. [Google Scholar] [CrossRef]
- Llorens, F.; Ansoleaga, B.; Garcia-Esparcia, P.; Zafar, S.; Grau-Rivera, O.; López-González, I.; Blanco, R.; Carmona, M.; Yagüe, J.; Nos, C.; et al. PrP mRNA and protein expression in brain and PrP(c) in CSF in Creutzfeldt-Jakob disease MM1 and VV2. Prion. 2013, 7, 383–393. [Google Scholar] [CrossRef]
- Caughey, B.; Raymond, G.J. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 1993, 67, 643–650. [Google Scholar] [CrossRef]
- Lehmann, S. Sulfated Glycans Stimulate Endocytosis of the Cellular Isoform of the Prion Protein, PrP^C., in Cultured Cells. J. Boil. Chem. 1995, 270, 30221–30229. [Google Scholar] [CrossRef]
- Filali, H.; Martín-Burriel, I.; Harders, F.; Varona, L.; Lyahyai, J.; Zaragoza, P.; Pumarola, M.; Badiola, J.J.; Bossers, A.; Bolea, R. Gene Expression Profiling and Association with Prion-Related Lesions in the Medulla Oblongata of Symptomatic Natural Scrapie Animals. PLoS ONE 2011, 6, 1–12. [Google Scholar] [CrossRef]
- López-Pérez, Ó; Bernal-Martín, M.; Hernaiz, A.; Llorens, F.; Betancor, M.; Otero, A.; Toivonen, J.M.; Zaragoza, P.; Zerr, I.; Badiola, J.J.; et al. BAMBI and CHGA in Prion Diseases: Neuropathological Assessment and Potential Role as Disease Biomarkers. Biomol. 2020, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.M.; Basu, U.; Khaniya, B.; Taniguchi, M.; Williams, J.L.; Moore, S.; Guan, L.L. Gene Expression in the Medulla Following Oral Infection of Cattle with Bovine Spongiform Encephalopathy. J. Toxicol. Environ. Heal. Part. A 2011, 74, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Marbiah, M.M.; Harvey, A.; West, B.T.; Louzolo, A.; Banerjee, P.; Alden, J.; Grigoriadis, A.; Hummerich, H.; Kan, H.-M.; Cai, Y.; et al. Identification of a gene regulatory network associated with prion replication. EMBO J. 2014, 33, 1527–1547. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1–47. [Google Scholar] [CrossRef]
- Liu, S.; Hossinger, A.; Göbbels, S.; Vorberg, I. Prions on the run: How extracellular vesicles serve as delivery vehicles for self-templating protein aggregates. Prion 2017, 11, 98–112. [Google Scholar] [CrossRef]
- Beraldo, F.H.; Arantes, C.P.; Santos, T.G.; Machado, C.F.; Roffe, M.; Hajj, G.N.; Lee, K.S.; Magalhaes, A.C.; Caetano, F.A.; Mancini, G.L.; et al. Metabotropic glutamate receptors transduce signals for neurite outgrowth after binding of the prion protein to laminin gamma1 chain. FASEB J. 2011, 25, 265–279. [Google Scholar] [CrossRef]
- Um, J.W.; Kaufman, A.C.; Kostylev, M.; Heiss, J.K.; Stagi, M.; Takahashi, H.; Kerrisk, M.E.; Vortmeyer, A.; Wisniewski, T.; Koleske, A.J.; et al. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron. 2013, 79, 887–902. [Google Scholar] [CrossRef]
- Lee, W.S.; Bae, Y.C.; Suk, K.; Lee, W.-H. Axon Guidance Molecules Guiding Neuroinflammation. Exp. Neurobiol. 2019, 28, 311–319. [Google Scholar] [CrossRef]
- Kim, T.-K.; Lee, I.; Cho, J.-H.; Canine, B.; Keller, A.; Price, N.D.; Hwang, D.; Carlson, G.A.; Hood, L. Core transcriptional regulatory circuits in prion diseases. Mol. Brain 2020, 13, 1–14. [Google Scholar] [CrossRef]
- Tian, C.; Liu, D.; Chen, C.; Xu, Y.; Gong, H.-S.; Chen, C.; Shi, Q.; Zhang, B.-Y.; Han, J.; Dong, X. Global transcriptional profiling of the postmortem brain of a patient with G114V genetic Creutzfeldt-Jakob disease. Int. J. Mol. Med. 2013, 31, 676–688. [Google Scholar] [CrossRef]
- Lopez-Bendito, G.; Flames, N.; Ma, L.; Fouquet, C.; Di Meglio, T.; Chédotal, A.; Tessier-Lavigne, M.; Marin, O. Robo1 and Robo2 Cooperate to Control the Guidance of Major Axonal Tracts in the Mammalian Forebrain. J. Neurosci. 2007, 27, 3395–3407. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Regulation/Stage | Log2 FC 1 | p-value | padj 2 | Hi-conf 3 |
|---|---|---|---|---|---|
| mmu-miR-667-3p | down/clinical | −0.57 | 2.38 × 10−10 | 1.45 × 10−7 | x |
| mmu-miR-146a-5p | up/clinical | 1.15 | 7.46 × 10−10 | 2.28 × 10−7 | x |
| mmu-miR-877-5p | down/clinical | −0.62 | 2.22 × 10−9 | 4.52 × 10−7 | |
| mmu-miR-10a-5p | up/clinical | 0.88 | 1.67 × 10−8 | 2.55 × 10−6 | x |
| mmu-miR-142a-5p | up/clinical | 0.54 | 4.81 × 10−7 | 5.87 × 10−5 | |
| mmu-miR-3085-3p | down/clinical | −0.71 | 1.40 × 10−6 | 1.24 × 10−4 | x |
| mmu-miR-331-3p | down/clinical | −0.66 | 1.42 × 10−6 | 1.24 × 10−4 | |
| mmu-miR-7046-3p | down/clinical | −0.77 | 2.72 × 10−6 | 2.07 × 10−4 | x |
| mmu-miR-1839-5p | up/clinical | 0.33 | 2.06 × 10−5 | 1.40 × 10−3 | x |
| mmu-miR-326-3p | down/clinical | −0.41 | 2.69 × 10−5 | 1.64 × 10−3 | x |
| mmu-miR-434-3p | down/clinical | −0.41 | 7.06 × 10−5 | 3.92 × 10−3 | x |
| mmu-miR-149-5p | down/clinical | −0.41 | 1.03 × 10−5 | 5.24 × 10−3 | x |
| mmu-miR-7b-5p | up/clinical | 0.81 | 1.26 × 10−4 | 5.78 × 10−3 | x |
| mmu-miR-764-5p | down/clinical | −0.69 | 1.32 × 10−4 | 5.78 × 10−3 | x |
| mmu-miR-146b-5p | up/clinical | 0.81 | 1.85 × 10−4 | 7.56 × 10−3 | x |
| mmu-miR-3475-3p | down/clinical | −0.26 | 2.07 × 10−4 | 7.90 × 10−3 | |
| mmu-miR-758-3p | down/clinical | −0.31 | 2.79 × 10−4 | 1.00 × 10−2 | x |
| mmu-miR-7a-5p | up/clinical | 0.76 | 4.45 × 10−4 | 1.44 × 10−2 | x |
| mmu-miR-384-5p | up/clinical | 0.43 | 4.48 × 10−4 | 1.44 × 10−2 | x |
| mmu-miR-410-3p | down/clinical | −0.18 | 7.15 × 10−4 | 2.18 × 10−2 | x |
| mmu-miR-342-3p | down/clinical | −1.02 | 1.00 × 10−3 | 2.91 × 10−2 | x |
| mmu-miR-872-3p | down/clinical | −0.35 | 1.11 × 10−3 | 3.07 × 10−2 | x |
| mmu-miR-342-5p | down/clinical | −0.97 | 1.65 × 10−3 | 4.38 × 10−2 | x |
| mmu-miR-223-3p | down/preclinical | −0.70 | 1.01 × 10−4 | 3.46 × 10−2 | x |
| mmu-miR-151-3p | up/preclinical | 0.12 | 1.40 × 10−4 | 3.46 × 10−2 | x |
| mmu-miR-144-5p | down/preclinical | −0.70 | 1.70 × 10−4 | 3.46 × 10−2 | x |
| Rank | KEGG pathway | padj | #genes | #miRNAs |
|---|---|---|---|---|
| 1 | Prion diseases | 1.3 × 10−21 | 5 | 4 |
| 2 | Axon guidance | 5.8 × 10−6 | 35 | 8 |
| 3 | Signaling regulating pluripotency of stem cells | 5.8 × 10−6 | 33 | 8 |
| 4 | TGF-beta signaling pathway | 1.6 × 10−5 | 23 | 8 |
| 5 | Proteoglycans in cancer | 3.2 × 10−5 | 35 | 8 |
| 6 | Mucin type O-Glycan biosynthesis | 2.2 × 10−4 | 5 | 3 |
| 7 | Thyroid hormone synthesis | 2.2 × 10−4 | 10 | 7 |
| 8 | Arrhythmogenic right ventricular cardiomyopathy | 2.2 × 10−4 | 16 | 7 |
| Rank | KEGG pathway | padj | #genes | #miRNAs |
|---|---|---|---|---|
| 1 | Prion diseases | 3.5 × 10−23 | 9 | 9 |
| 2 | ECM-receptor interaction | 8.9 × 10−6 | 17 | 10 |
| 3 | Glutamatergic synapse | 2.1 × 10−5 | 23 | 12 |
| 4 | Thyroid hormone signaling pathway | 2.3 × 10−5 | 30 | 11 |
| 5 | Amphetamine addiction | 7.2 × 10−5 | 19 | 12 |
| 6 | Wnt signaling pathway | 1.1 × 10−4 | 33 | 11 |
| 7 | Signaling regulating pluripotency of stem cells | 6.7 × 10−4 | 30 | 12 |
| 8 | Phosphatidylinositol signaling system | 7.3 × 10−4 | 19 | 9 |
| 9 | N-Glycan biosynthesis | 7.5 × 10−4 | 10 | 9 |
| 10 | Renal cell carcinoma | 7.5 × 10−4 | 18 | 11 |
| 11 | PI3K-Akt signaling pathway | 7.5 × 10−4 | 63 | 12 |
| 12 | Long-term potentiation | 1.0 × 10−3 | 20 | 11 |
| Stage | miRNA | Tg5011 | Pub2 | Disease; Model(s) | Tissues or Fractions3 | Refs. |
|---|---|---|---|---|---|---|
| Clinical Tg501 | miR-667-3p | ↓ | ↓ | Prion (RML)—infected mice | FB synaptoneurosomes | [35] |
| miR-146a-5p | ↑ | ↑↓ | sCJD; prion(RML/22A/ sCJD-MM1)—infected mice, prion(M1000)-infected neuronal cells | B, FC, CBL, FB synaptoneurosomes, neuronal cells, and exosomes | [4,7,27,28,35,38] | |
| miR-877-5p | ↓ | ↓ | sCJD; prion(sCJD-MM1)—infected mice | FC, CBL | [4] | |
| miR-142a-5p | ↑ | ↑ | Prion(RML)—infected mice | FB synaptoneurosomes | [35] | |
| miR-331-3p | ↓ | ↓ | sCJD; prion(sCJD-MM1)—infected mice | FC, C | [4] | |
| miR-149-5p | ↓ | ↓ | Prion(RML)—infected mice | FB, SN | [35] | |
| miR-7b-5p | ↑ | ↑ | Prion(RML)—infected mice | SN | [7,35] | |
| miR-146b-5p | ↑ | ↓ | Prion(RML)—infected mice | HC synaptoneurosomes | [35] | |
| miR-7a-5p | ↑ | ↑ | Prion(RML)-infected mice | FB synaptoneurosomes | [35] | |
| miR-342-3p | ↓ | ↑↓ | sCJD, sheep scrapie; prion(22A/sCJD-MM1)—infected mice, prion(BSE)—infected macaques, prion(M1000)—infected neuronal cells | FC, plasma; B,C, CBL, BP, neuronal cell-derived exosomes | [4,5,26,28,45] | |
| miR-342-5p | ↓ | ↓ | Prion (RML)—infected mice | HC synaptoneurosomes | [35] | |
| Preclin. Tg501 | miR-223-3p | ↓ | ↑↓ | Asymptomatic CWD elk; scrapie-infected hamster | Serum | [6] |
| miR-151-3p | ↑ | ↑ | Asymptomatic CWD elk | Serum | [6] | |
| miR-144-5p | ↓ | ↑ | Asymptomatic CWD elk | Serum | [6] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toivonen, J.M.; Sanz-Rubio, D.; López-Pérez, Ó.; Marín-Moreno, A.; Bolea, R.; Osta, R.; Badiola, J.J.; Zaragoza, P.; Espinosa, J.-C.; Torres, J.-M.; et al. MicroRNA Alterations in a Tg501 Mouse Model of Prion Disease. Biomolecules 2020, 10, 908. https://doi.org/10.3390/biom10060908
Toivonen JM, Sanz-Rubio D, López-Pérez Ó, Marín-Moreno A, Bolea R, Osta R, Badiola JJ, Zaragoza P, Espinosa J-C, Torres J-M, et al. MicroRNA Alterations in a Tg501 Mouse Model of Prion Disease. Biomolecules. 2020; 10(6):908. https://doi.org/10.3390/biom10060908
Chicago/Turabian StyleToivonen, Janne M., David Sanz-Rubio, Óscar López-Pérez, Alba Marín-Moreno, Rosa Bolea, Rosario Osta, Juan J. Badiola, Pilar Zaragoza, Juan-Carlos Espinosa, Juan-Maria Torres, and et al. 2020. "MicroRNA Alterations in a Tg501 Mouse Model of Prion Disease" Biomolecules 10, no. 6: 908. https://doi.org/10.3390/biom10060908
APA StyleToivonen, J. M., Sanz-Rubio, D., López-Pérez, Ó., Marín-Moreno, A., Bolea, R., Osta, R., Badiola, J. J., Zaragoza, P., Espinosa, J.-C., Torres, J.-M., & Martín-Burriel, I. (2020). MicroRNA Alterations in a Tg501 Mouse Model of Prion Disease. Biomolecules, 10(6), 908. https://doi.org/10.3390/biom10060908






