AP-TSS: A New Method for the Analysis of RNA Expression from Particular and Challenging Transcription Start Sites
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Extraction and Preparation of RNA
2.3. Production of the Synthetic TERRA
2.4. Reverse Transcription
2.5. Elongation of Specific cDNA
2.6. Digestion of Primers Used for the Reverse Transcription and Template Oligonucleotides by RecJf
2.7. Quantification of Elongated cDNA by qPCR and Data Analysis
3. Results
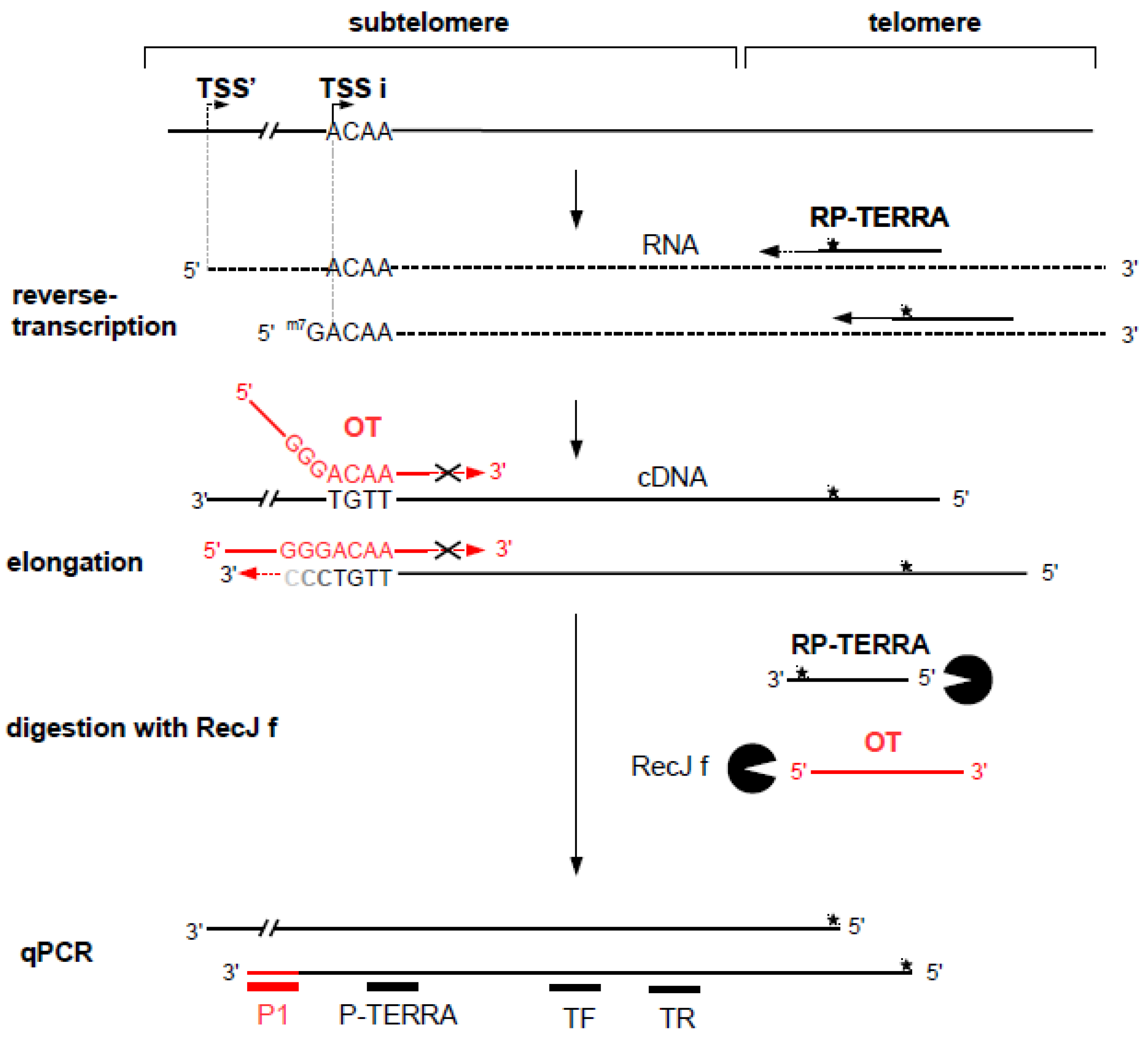
3.1. Principle of the Method of Quantification of Transcript Based on Specific Elongation of the cDNA of Interest (AP-TSS)
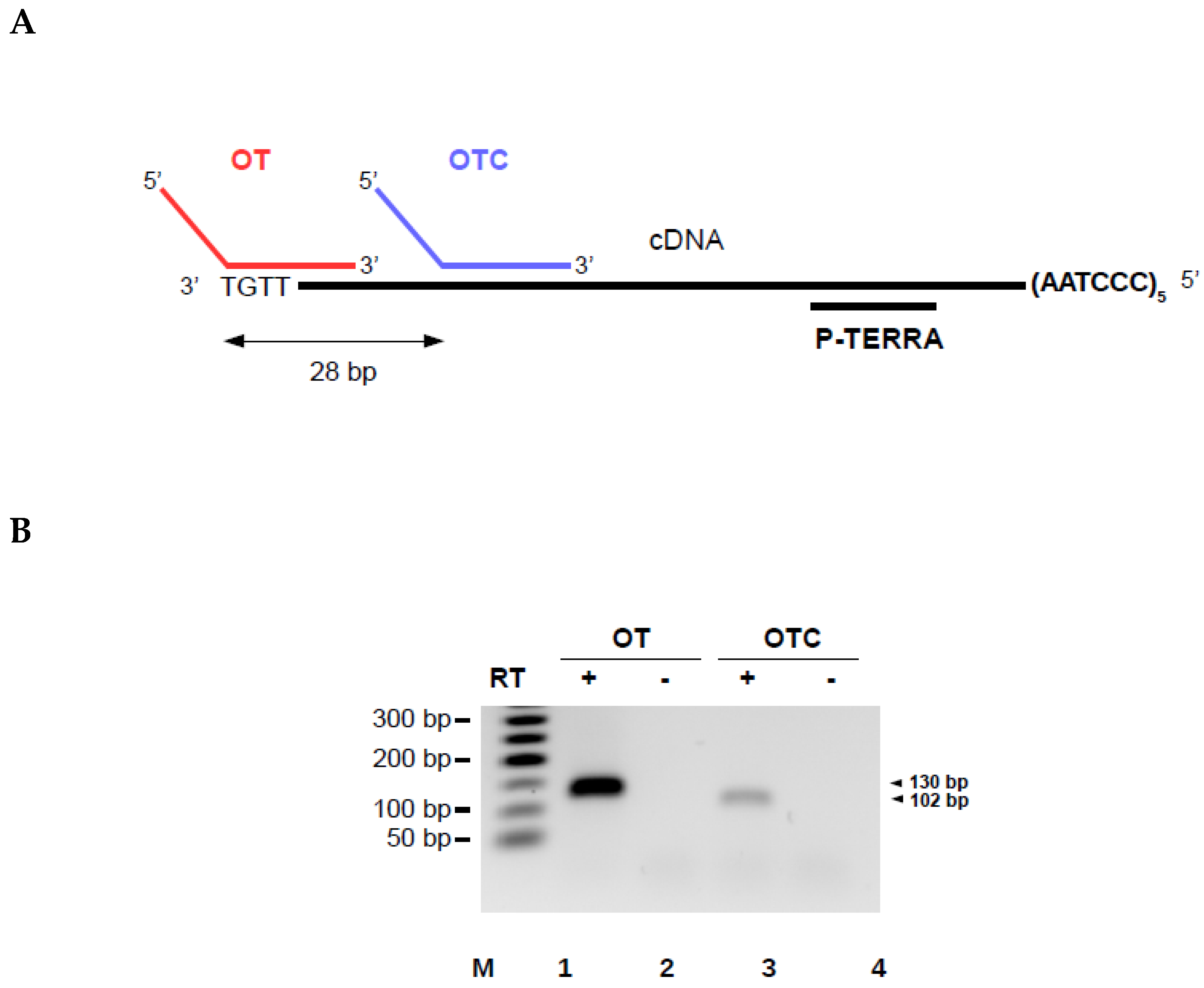
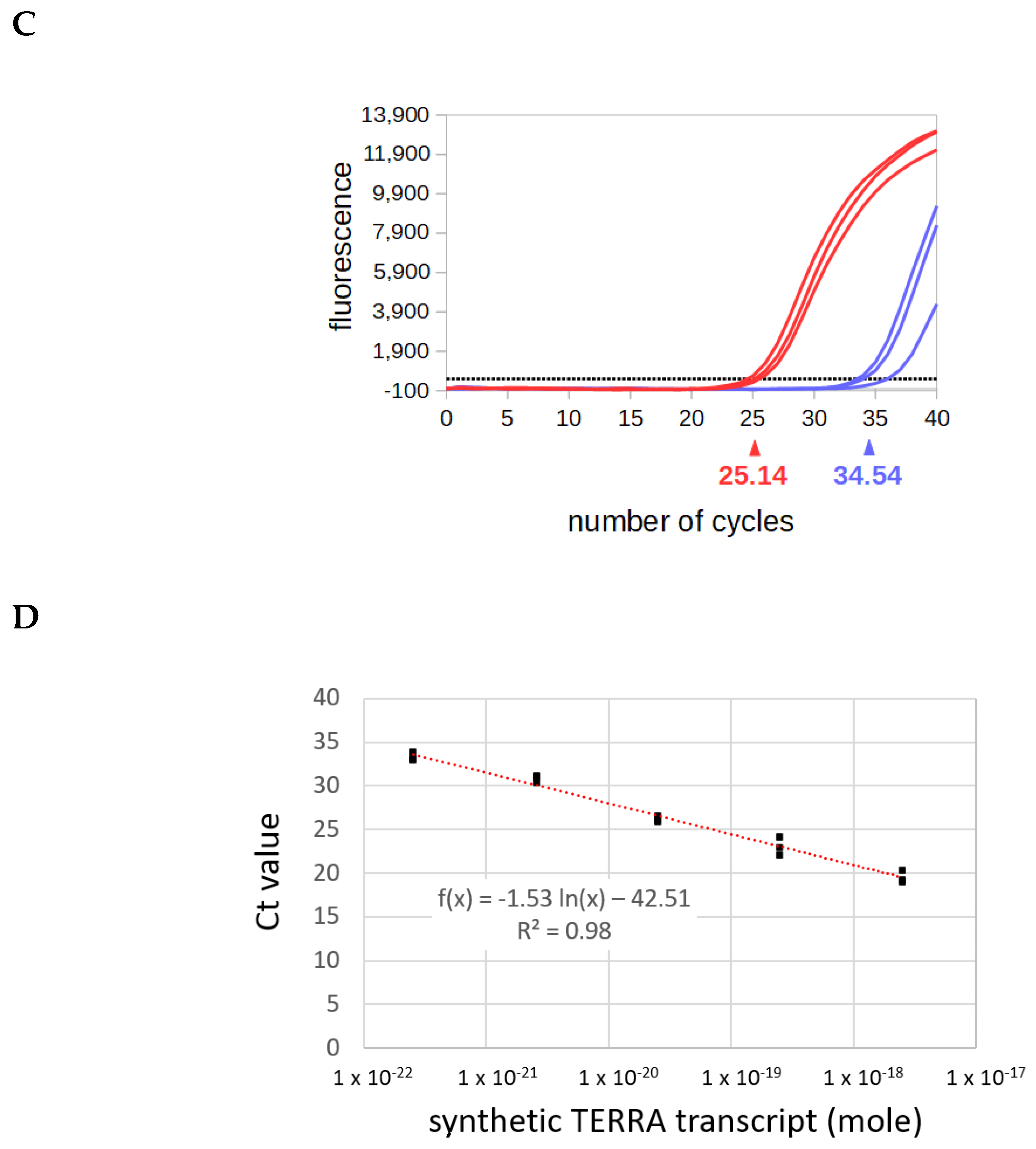
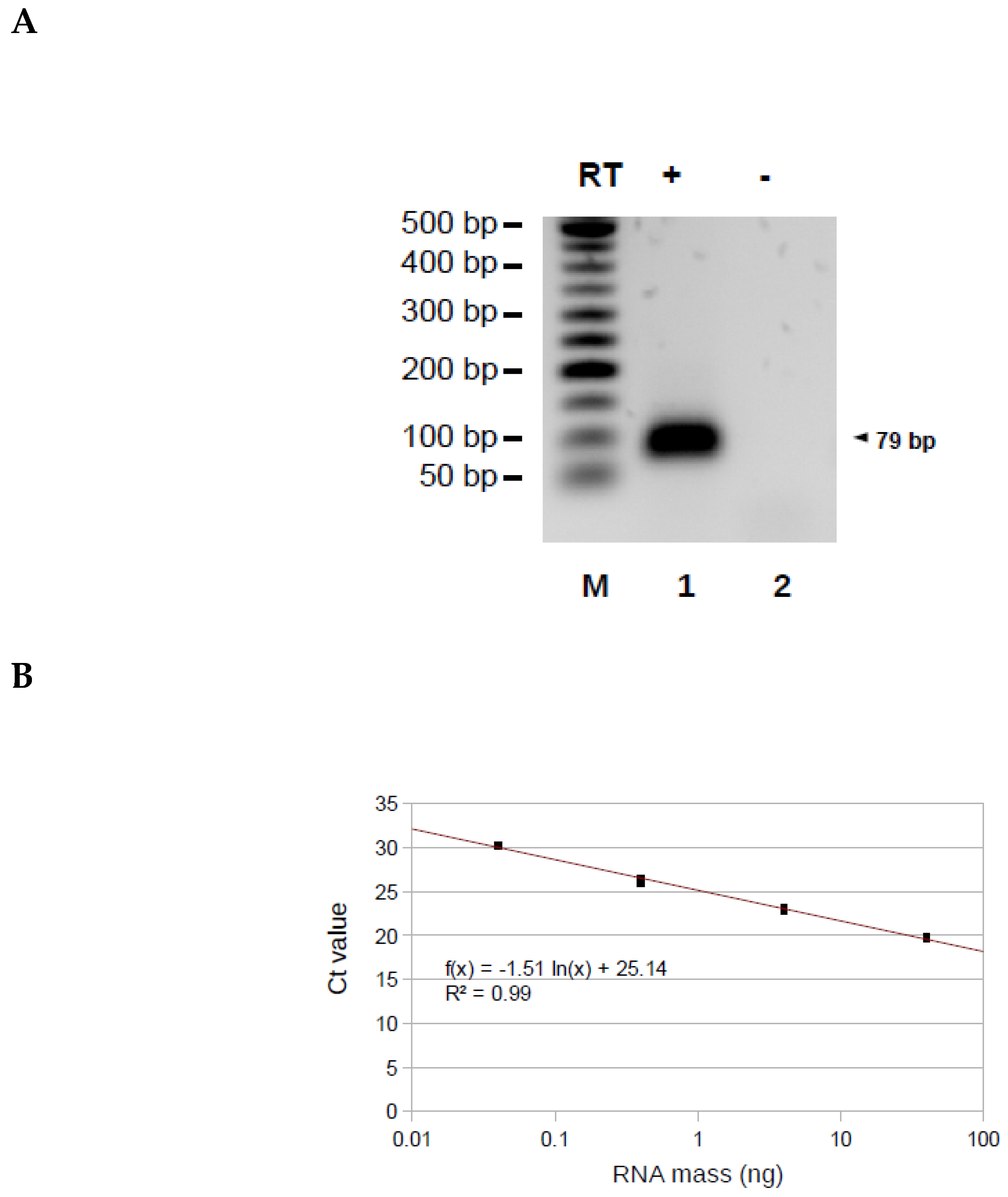
3.2. Validation of the Method on a Synthetic TERRA
3.3. Application of AP-TSS to Compare the Levels of Transcripts from a Particular TERRA TSS Under Different Conditions
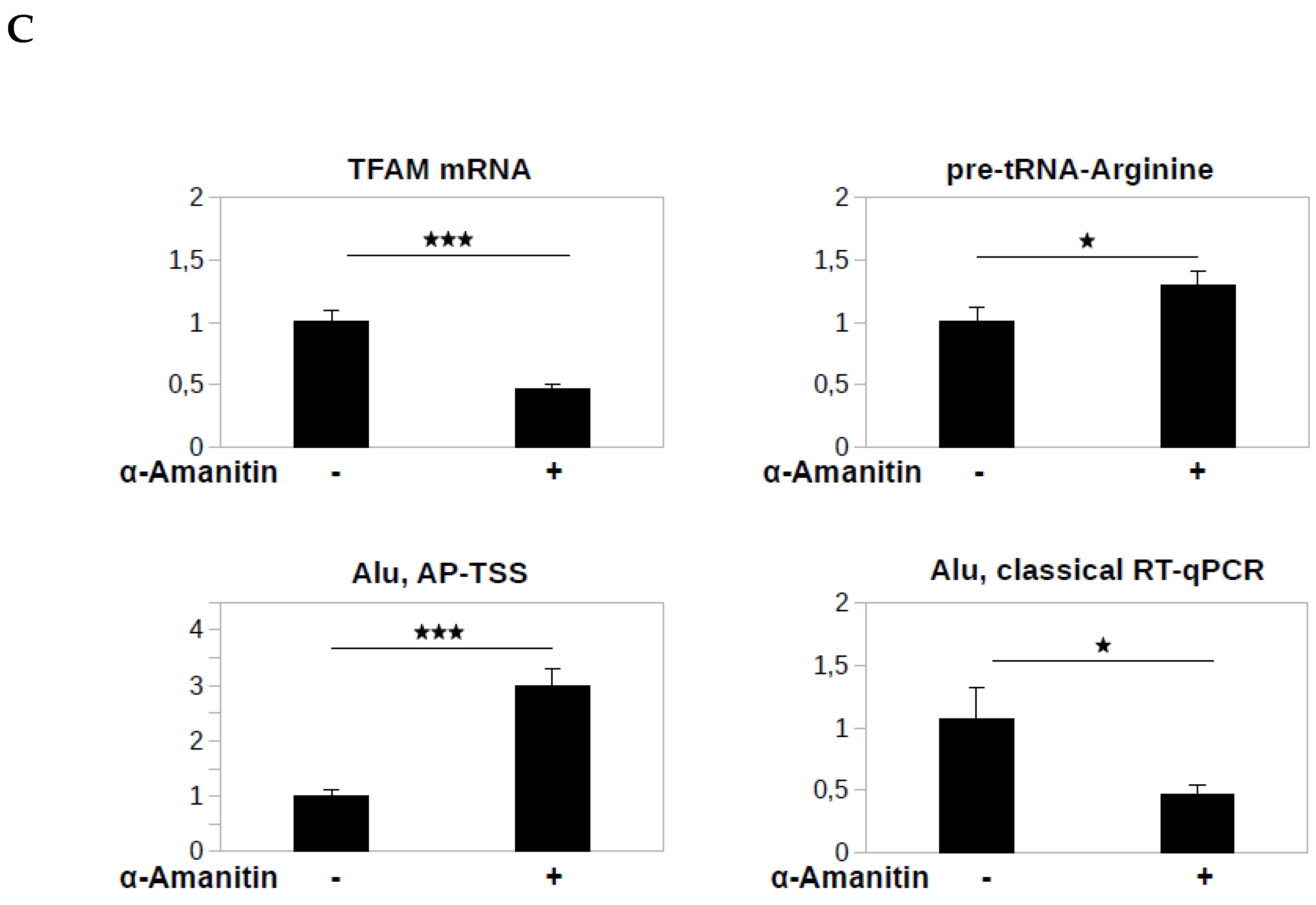
3.4. Application of the Method to the Alu Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-aza-dC | 5-aza-2′-deoxycytidine |
| CAGE | cap analysis of gene expression |
| Ct | cycle threshold |
| DNMT | DNA methyltransferase |
| EtBr | ethidium bromide |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| OT | oligonucleotide template |
| OTC | oligonucleotide template control |
| PAGE | polyacrylamide gel electrophoresis |
| RACE | rapid amplification of cDNA ends |
| RNA-seq | RNA sequencing |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
| SINE | short interspersed nuclear elements |
| TERRA | telomeric repeat-containing RNA |
| TSS | transcription start site |
| TSSi | transcription start site of interest |
| UTR | untranslated region |
References
- Xin, D.; Hu, L.; Kong, X. Alternative promoters influence alternative splicing at the genomic level. PLoS ONE 2008, 3, e2377. [Google Scholar] [CrossRef] [PubMed]
- Davuluri, R.V.; Suzuki, Y.; Sugano, S.; Plass, C.; Huang, T.H.-M. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008, 24, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013, 497, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, K.; Schepeler, T.; Øster, B.; Rasmussen, M.H.; Vang, S.; Wang, K.; Hansen, K.Q.; Lamy, P.; Pedersen, J.S.; Eller, A.; et al. Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis. BMC Genom. 2011, 12, 505. [Google Scholar] [CrossRef] [PubMed]
- Brocks, D.; Schmidt, C.R.; Daskalakis, M.; Jang, H.S.; Shah, N.M.; Li, D.; Li, J.; Zhang, B.; Hou, Y.; Laudato, S.; et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 2017, 49, 1052–1060. [Google Scholar] [CrossRef]
- O’Connell, M.R.; Sarkar, S.; Luthra, G.K.; Okugawa, Y.; Toiyama, Y.; Gajjar, A.H.; Qiu, S.; Goel, A.; Singh, P. Epigenetic changes and alternate promoter usage by human colon cancers for expressing DCLK1-isoforms: Clinical Implications. Sci. Rep. 2015, 5, 14983. [Google Scholar] [CrossRef]
- Garieri, M.; Delaneau, O.; Santoni, F.; Fish, R.J.; Mull, D.; Carninci, P.; Dermitzakis, E.T.; Antonarakis, S.E.; Fort, A. The effect of genetic variation on promoter usage and enhancer activity. Nat. Commun. 2017, 8, 1358. [Google Scholar] [CrossRef]
- Wang, X.; Hou, J.; Quedenau, C.; Chen, W. Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals. Mol. Syst. Biol. 2016, 12, 875. [Google Scholar] [CrossRef]
- Rojas-Duran, M.F.; Gilbert, W.V. Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA 2012, 18, 2299–2305. [Google Scholar] [CrossRef]
- Delloye-Bourgeois, C.; Goldschneider, D.; Paradisi, A.; Therizols, G.; Belin, S.; Hacot, S.; Rosa-Calatrava, M.; Scoazec, J.-Y.; Diaz, J.-J.; Bernet, A.; et al. Nucleolar localization of a netrin-1 isoform enhances tumor cell proliferation. Sci. Signal 2012, 5, ra57. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Frohman, M.A.; Dush, M.K.; Martin, G.R. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 1988, 85, 8998–9002. [Google Scholar] [CrossRef] [PubMed]
- Bower, N.I.; Johnston, I.A. Targeted rapid amplification of cDNA ends (T-RACE)--an improved RACE reaction through degradation of non-target sequences. Nucleic Acids Res. 2010, 38, e194. [Google Scholar] [CrossRef][Green Version]
- Chen, N.; Wang, W.-M.; Wang, H.-L. An efficient full-length cDNA amplification strategy based on bioinformatics technology and multiplexed PCR methods. Sci. Rep. 2016, 6, 19420. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zheng, K.; Chen, H.-C.; Liu, Z.-F. Capping-RACE: A simple, accurate, and sensitive 5′ RACE method for use in prokaryotes. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Kojima, M.; Nishiyori, H.; Nakamura, M.; Fukuda, S.; Tagami, M.; Sasaki, D.; Imamura, K.; Kai, C.; Harbers, M.; et al. CAGE: Cap analysis of gene expression. Nat. Methods 2006, 3, 211–222. [Google Scholar] [CrossRef]
- Montero, J.J.; López de Silanes, I.; Graña, O.; Blasco, M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016, 7, 12534. [Google Scholar] [CrossRef] [PubMed]
- Moravec, M.; Wischnewski, H.; Bah, A.; Hu, Y.; Liu, N.; Lafranchi, L.; King, M.C.; Azzalin, C.M. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016, 17, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Van Beneden, A.; Decottignies, A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 2012, 19, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Farnung, B.O.; Wischnewski, H.; Khoriauli, L.; Vitelli, V.; Chawla, R.; Giulotto, E.; Azzalin, C.M. CpG-island promoters drive transcription of human telomeres. RNA 2009, 15, 2186–2194. [Google Scholar] [CrossRef]
- Porro, A.; Feuerhahn, S.; Delafontaine, J.; Riethman, H.; Rougemont, J.; Lingner, J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014, 5, 5379. [Google Scholar] [CrossRef]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Vavrova-Anderson, J.; Oler, A.J.; Cowling, V.H.; Cairns, B.R.; White, R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015, 6, 6569. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.M.; Liu, W.M.; Schmid, C.W. RNA polymerase III promoter and terminator elements affect AIu RNA expression. Nucleic Acids Res. 1995, 23, 1750–1757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, X.-O.; Gingeras, T.; Weng, Z. Genome-wide analysis of polymerase III–transcribed Alu elements suggests cell-type–specific enhancer function. Genome Res. 2019, 29, 1402–1414. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Goodrich, J.A.; Kugel, J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA 2009, 106, 5569–5574. [Google Scholar] [CrossRef]
- Hu, Q.; Tanasa, B.; Trabucchi, M.; Li, W.; Zhang, J.; Ohgi, K.A.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. DICER- and AGO3-dependent generation of retinoic acid–induced DR2 Alu RNAs regulates human stem cell proliferation. Nat. Struct. Mol. Biol. 2012, 19, 1168–1175. [Google Scholar] [CrossRef]
- Petrie, J.L.; Swan, C.; Ingram, R.M.; Frame, F.M.; Collins, A.T.; Dumay-Odelot, H.; Teichmann, M.; Maitland, N.J.; White, R.J. Effects on prostate cancer cells of targeting RNA polymerase III. Nucleic Acids Res. 2019, 47, 3937–3956. [Google Scholar] [CrossRef]
- Di Ruocco, F.; Basso, V.; Rivoire, M.; Mehlen, P.; Ambati, J.; De Falco, S.; Tarallo, V. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene 2018, 37, 627–637. [Google Scholar] [CrossRef]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef]
- Liu, W.-M.; Chu, W.-M.; Choudary, P.V.; Schmid, C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995, 23, 1758–1765. [Google Scholar] [CrossRef] [PubMed]
- Zajac, P.; Islam, S.; Hochgerner, H.; Lönnerberg, P.; Linnarsson, S. Base preferences in non-templated nucleotide incorporation by MMLV-derived reverse transcriptases. PLoS ONE 2013, 8, e85270. [Google Scholar] [CrossRef] [PubMed]
- Feretzaki, M.; Lingner, J. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods 2017, 114, 39–45. [Google Scholar] [CrossRef]
- Hellmann-Blumberg, U.; Hintz, M.F.; Gatewood, J.M.; Schmid, C.W. Developmental differences in methylation of human Alu repeats. Mol. Cell. Biol. 1993, 13, 4523–4530. [Google Scholar] [CrossRef] [PubMed]
- Paulson, K.E.; Schmid, C.W. Transcriptional inactivity of Alu repeats in HeLa cells. Nucl. Acids Res 1986, 14, 6145–6158. [Google Scholar] [CrossRef] [PubMed]
- Leenen, F.A.D.; Vernocchi, S.; Hunewald, O.E.; Schmitz, S.; Molitor, A.M.; Muller, C.P.; Turner, J.D. Where does transcription start? 5’-RACE adapted to next-generation sequencing. Nucleic Acids Res. 2016, 44, 2628–2645. [Google Scholar] [CrossRef]
- Zhuang, F.; Fuchs, R.T.; Sun, Z.; Zheng, Y.; Robb, G.B. Structural bias in T4 RNA ligase-mediated 3’-adapter ligation. Nucleic Acids Res. 2012, 40, e54. [Google Scholar] [CrossRef]
- Conway, T.; Creecy, J.P.; Maddox, S.M.; Grissom, J.E.; Conkle, T.L.; Shadid, T.M. Unprecedented High-Resolution View of Bacterial Operon Architecture Revealed by RNA Sequencing. mBio 2014, 5, e01442-14. [Google Scholar] [CrossRef]
- Łabaj, P.P.; Leparc, G.G.; Linggi, B.E.; Markillie, L.M.; Wiley, H.S.; Kreil, D.P. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics 2011, 27, i383–i391. [Google Scholar] [CrossRef]
- Zhou, S.; Sternglanz, R.; Neiman, A.M. Developmentally regulated internal transcription initiation during meiosis in budding yeast. PLoS ONE 2017, 12, e0188001. [Google Scholar] [CrossRef]
- Nagai, S.; Davis, R.E.; Mattei, P.J.; Eagen, K.P.; Kornberg, R.D. Chromatin potentiates transcription. Proc. Natl. Acad. Sci. USA 2017, 114, 1536–1541. [Google Scholar] [CrossRef]
- Otsuka, Y.; Kedersha, N.L.; Schoenberg, D.R. Identification of a Cytoplasmic Complex That Adds a Cap onto 5′-Monophosphate RNA. Mol Cell Biol 2009, 29, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Kiss, D.L.; Oman, K.; Bundschuh, R.; Schoenberg, D.R. Uncapped 5’ ends of mRNAs targeted by cytoplasmic capping map to the vicinity of downstream CAGE tags. FEBS Lett. 2015, 589, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Dimont, E.; Ha, T.; Swanson, D.J. FANTOM Consortium; Hide, W.; Goldowitz, D. Relatively frequent switching of transcription start sites during cerebellar development. BMC Genom. 2017, 18, 461. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Issa, J.-P.J.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Bracken, C.P.; Kolle, G.; Szubert, J.M.; Korbie, D.J.; Askarian-Amiri, M.E.; Gardiner, B.B.; Goodall, G.J.; Grimmond, S.M.; et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010, 20, 1639–1650. [Google Scholar] [CrossRef]
- Snider, L.; Asawachaicharn, A.; Tyler, A.E.; Geng, L.N.; Petek, L.M.; Maves, L.; Miller, D.G.; Lemmers, R.J.L.F.; Winokur, S.T.; Tawil, R.; et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: New candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009, 18, 2414–2430. [Google Scholar] [CrossRef]


| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| RP- telomeric repeat-containing RNA (TERRA) | CCCTAACCCTAACCCTAACCCT*A*A*C*C*C*TAA |
| OT | GCACTCGGTGAGTCGTACTACGGGACAACTCGGGGCGCATCAAddC |
| OTC | GCACTCGGTGAGTCGTACTACGGGAAAATGTTTCCCGGTTGCAGCddC |
| P1 | GCACTCGGTGAGTCGTACTACG |
| P-TERRA | CTTTCCCGTTTTCCGCACTG |
| TF | GCAGCCATGAATAATCAAGGT |
| TR | TTCCGCACTGAACCGCTCTAA |
| GAPDH-R | GAAGGTGAAGGTCGGAGTCAAC |
| GAPDH-F | CAGAGTTAAAAGCAGCCCTGGT |
| TERRA-synt-R | CTATAATACGACTCACTATAGACAACTCGGGGCGCATCA |
| TERRA-synt-F | CCCTAACCCTAACCCTAACCCTAACCCTAATTGTGTGCATTAGGAATGCTG |
| RP-Alu | GCGATCTCGGCTC*A*C*T*G*C*AAG |
| OT-Alu | GCACTCGGTGAGTCGTACTACGGGGGCCGGGCGCGGTGGCTCACddC |
| P-Alu | CCGCCTCGGCCTCCCAAAGT |
| Alu-F | ACCATCCCGGCTAAAACGGTGA |
| Alu-R | GCGATCTCGGCTCACTG |
| TFAM-F | GTGGGAGCTTCTCACTCTGG |
| TFAM-R | TAGGGCTTTTTCTCCTGCAA |
| pre-tRNA-Arg-F | GGCTCTGTGGCGCAATGGATA |
| pre-tRNA-Arg-R | TTCGAACCCACAACCTTTGAATTGCTC |
| ARN-28S-F | TGTTAGGACCCGAAAGATGG |
| ARN-28S-R | TCGGAGGGAACCAGCTACTA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Berre, G.; Hossard, V.; Riou, J.-F.; Guieysse-Peugeot, A.-L. AP-TSS: A New Method for the Analysis of RNA Expression from Particular and Challenging Transcription Start Sites. Biomolecules 2020, 10, 827. https://doi.org/10.3390/biom10060827
Le Berre G, Hossard V, Riou J-F, Guieysse-Peugeot A-L. AP-TSS: A New Method for the Analysis of RNA Expression from Particular and Challenging Transcription Start Sites. Biomolecules. 2020; 10(6):827. https://doi.org/10.3390/biom10060827
Chicago/Turabian StyleLe Berre, Gabriel, Virginie Hossard, Jean-Francois Riou, and Anne-Laure Guieysse-Peugeot. 2020. "AP-TSS: A New Method for the Analysis of RNA Expression from Particular and Challenging Transcription Start Sites" Biomolecules 10, no. 6: 827. https://doi.org/10.3390/biom10060827
APA StyleLe Berre, G., Hossard, V., Riou, J.-F., & Guieysse-Peugeot, A.-L. (2020). AP-TSS: A New Method for the Analysis of RNA Expression from Particular and Challenging Transcription Start Sites. Biomolecules, 10(6), 827. https://doi.org/10.3390/biom10060827




