Heparin Blocks the Inhibition of Tissue Kallikrein 1 by Kallistatin through Electrostatic Repulsion
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cloning, Mutagenesis, Expression, and Purification of Recombinant Proteins
2.3. Crystallization, Data Collection, and Structure Determination
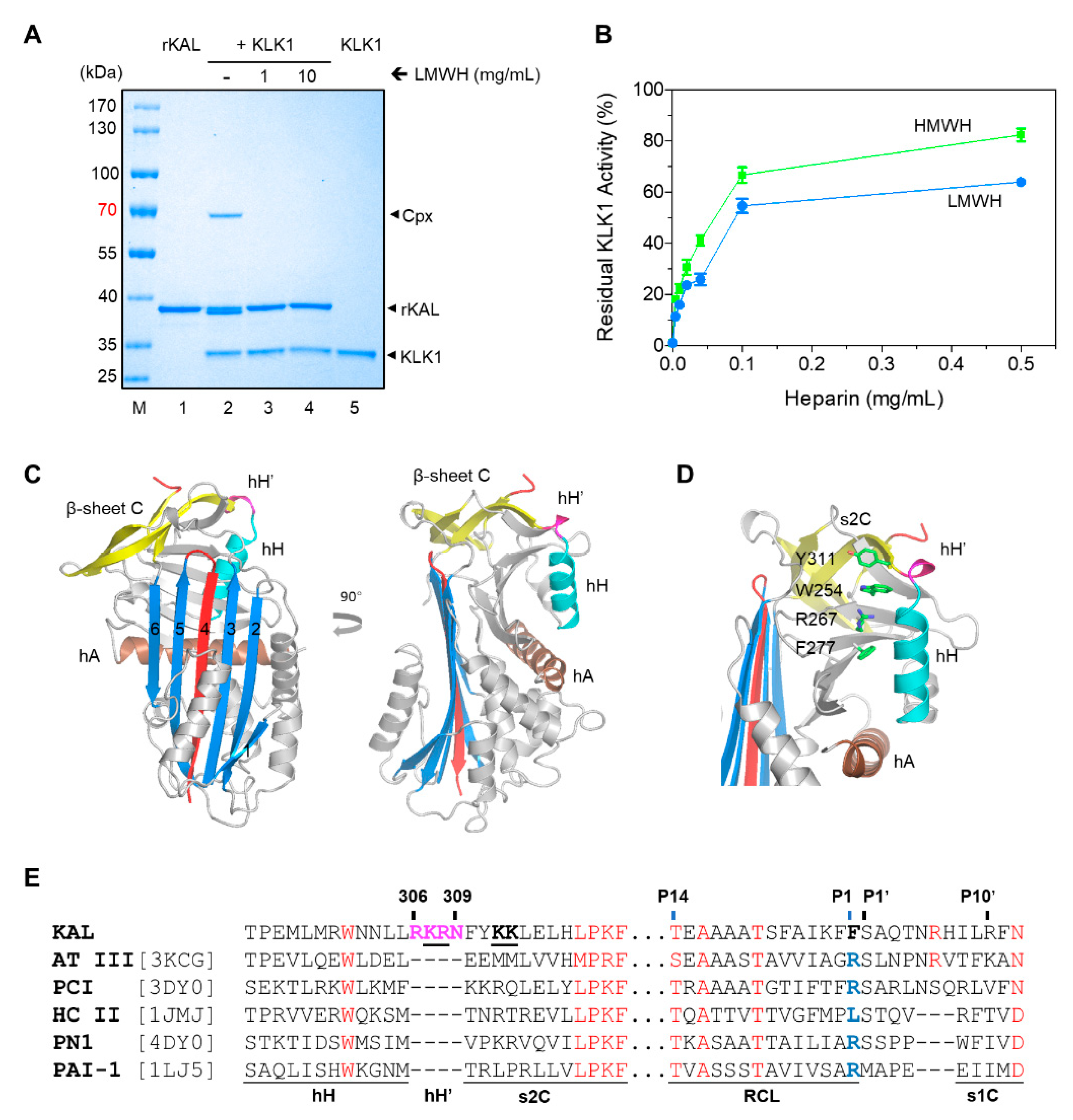
2.4. Stoichiometries of Inhibition and Rates of Inhibition
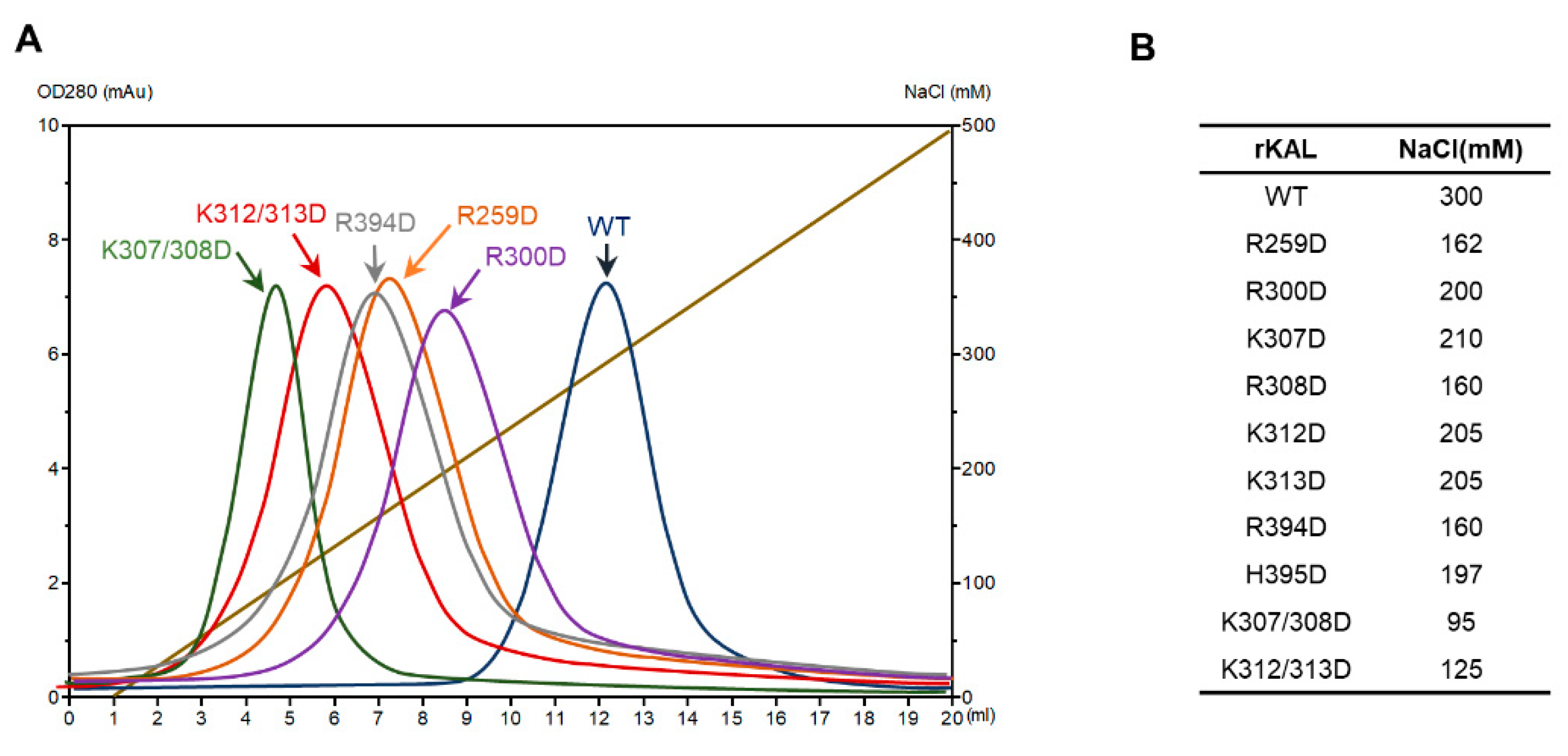
2.5. Heparin Affinity Chromatography of Kallistatin Variants
2.6. Model of the Kallistatin-KLK1-Heparin Ternary Complex
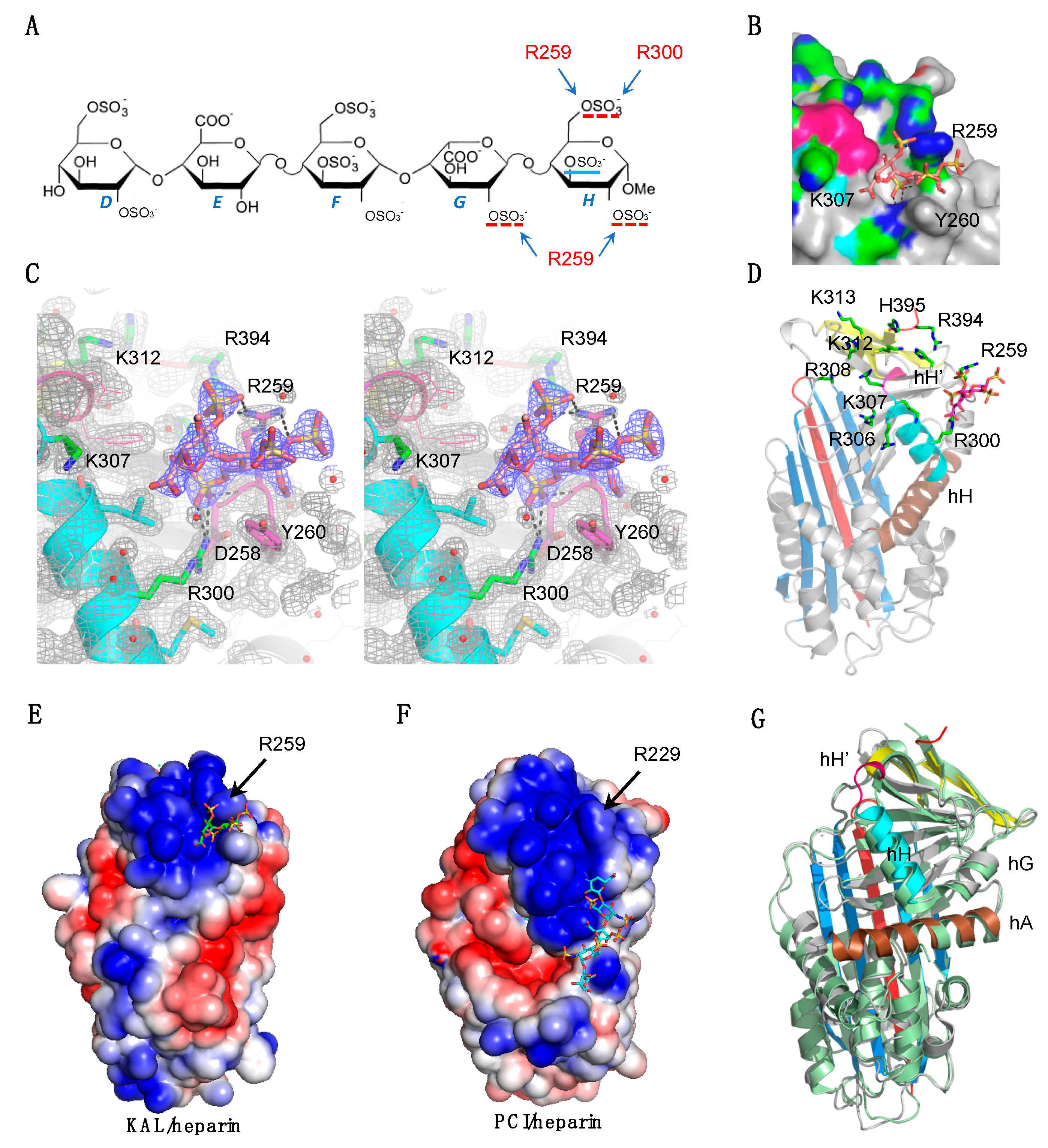
3. Results
3.1. The Overall Structures of Kallistatin and Its Complex with Heparin
3.2. Heparin Binding Site of Kallistatin
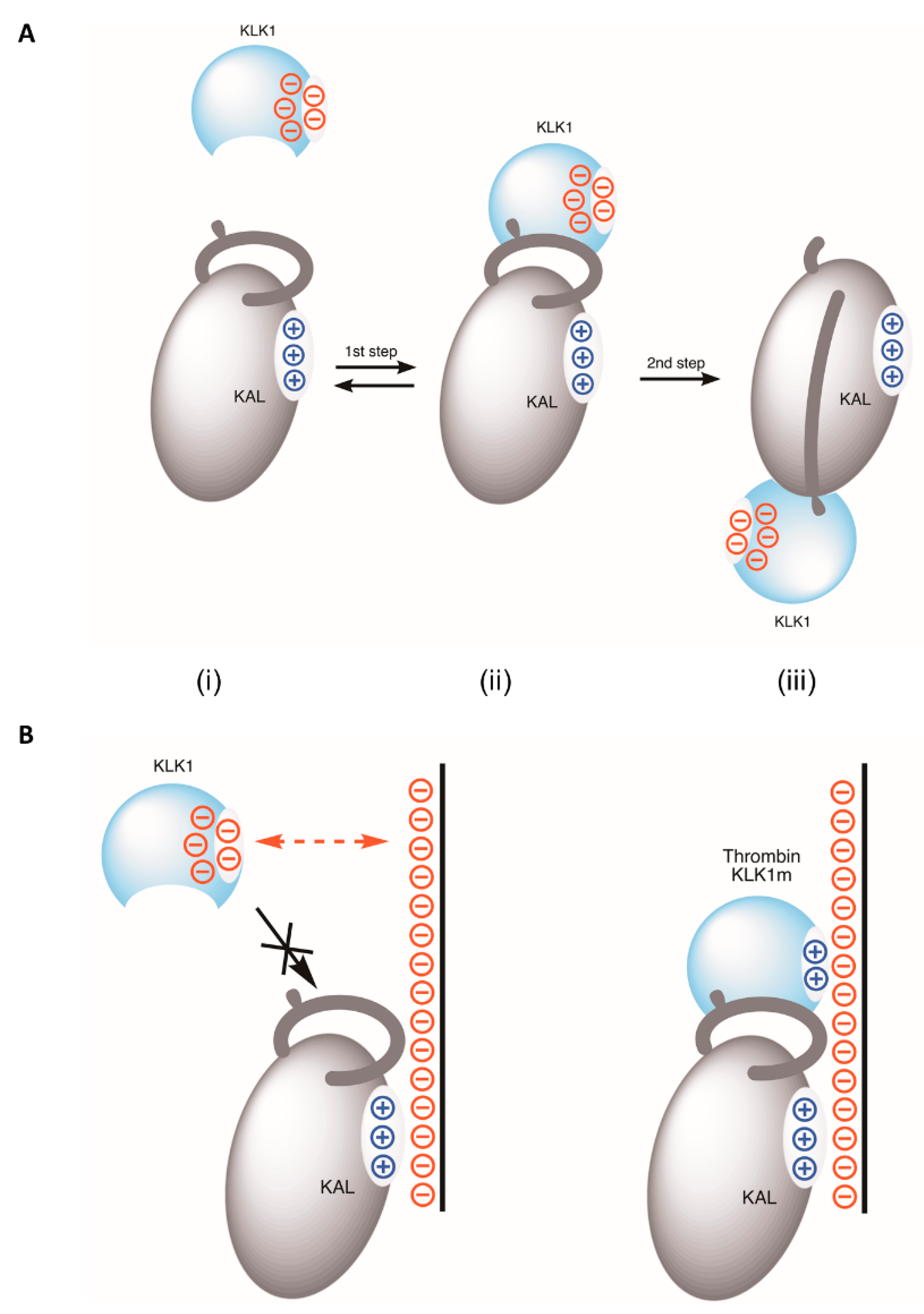
3.3. The Mechanism of Heparin in Blocking KLK1/Kallistatin Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chao, J.; Li, P.; Chao, L. Kallistatin suppresses cancer development by multi-factorial actions. Crit. Rev. Oncol. Hematol. 2017, 113, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Chai, K.X.; Chen, L.M.; Xiong, W.; Chao, S.; Woodleymiller, C.; Wang, L.X.; Lu, H.S.; Chao, L. Tissue kallikrein-binding protein is a serpin. I. Purification, characterization, and distribution in normotensive and spontaneously hypertensive rats. J. Biol. Chem. 1990, 265, 16394–16401. [Google Scholar] [PubMed]
- Chai, K.X.; Chen, L.M.; Chao, J.; Chao, L. Kallistatin: A novel human serine proteinase inhibitor. Molecular cloning, tissue distribution, and expression in Escherichia coli. J. Biol. Chem. 1993, 268, 24498–24505. [Google Scholar] [PubMed]
- Chao, J.; Li, P.; Chao, L. Kallistatin: Double-edged role in angiogenesis, apoptosis and oxidative stress. Biol. Chem. 2017, 398, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.; Carrell, R.W. Implications of the three-dimensional structure of alpha 1-antitrypsin for structure and function of serpins. Biochemistry 1989, 28, 8951–8966. [Google Scholar] [CrossRef]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef]
- Huntington, J.A.; Read, R.J.; Carrell, R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 2000, 407, 923. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the size of active sites in proteases I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Gettins, P.G.W. Serpin Structure, Mechanism, and Function. Chem. Rev. 2003, 34, 4751–4804. [Google Scholar] [CrossRef]
- Luo, L.Y.; Jiang, W. Inhibition profiles of human tissue kallikreins by serine protease inhibitors. Biol. Chem. 2006, 387, 813–816. [Google Scholar] [CrossRef]
- Gettins, P.G.; Olson, S.T. Exosite determinants of serpin specificity. J. Biol. Chem. 2009, 284, 20441–20445. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.C.; Chao, L.; Chao, J. Roles of the P1, P2, and P3 residues in determining inhibitory specificity of kallistatin toward human tissue kallikrein. J. Biol. Chem. 2000, 275, 38457–38466. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.C.; Chao, L.; Chao, J. Reactive-site specificity of human kallistatin toward tissue kallikrein probed by site-directed mutagenesis. Biochim. Biophys. Acta 2000, 1479, 237–246. [Google Scholar] [CrossRef]
- Raspi, G. Kallikrein and kallikrein-like proteinases: Purification and determination by chromatographic and electrophoretic methods. J. Chromatogr. B Biomed. Appl. 1996, 684, 265–287. [Google Scholar] [CrossRef]
- Diamandis, E.P.; Yousef, G.M.; Luo, L.Y.; Magklara, A.; Obiezu, C.V. The new human kallikrein gene family: Implications in carcinogenesis. Trends Endocrin. Metab. Tem. 2000, 11, 54–60. [Google Scholar] [CrossRef]
- Yousef, G.M.; Diamandis, E.P. Human tissue kallikreins: A new enzymatic cascade pathway? Biol. Chem. 2002, 383, 1045. [Google Scholar] [CrossRef]
- Akiyama, K.; Nakamura, T.; Iwanaga, S.; Hara, M. The chymotrypsin-like activity of human prostate-specific antigen, gamma-seminoprotein. FEBS Lett. 1987, 225, 168. [Google Scholar] [CrossRef]
- Alexander-Curtis, M.; Pauls, R.; Chao, J.; Volpi, J.J.; Bath, P.M.; Verdoorn, T.A. Human tissue kallikrein in the treatment of acute ischemic stroke. Ther. Adv. Neurol. Disord. 2019, 12, 1756286418821918. [Google Scholar] [CrossRef]
- Johnson, D.J.; Li, W.; Adams, T.E.; Huntington, J.A. Antithrombin–S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation. EMBO J. 2006, 25, 2029–2037. [Google Scholar] [CrossRef]
- Storoni, L.C.; McCoy, A.J.; Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 432–438. [Google Scholar] [CrossRef]
- Dementiev, A.; Petitou, M.; Herbert, J.M.; Gettins, P.G. The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity. Nat. Struct. Mol. Biol. 2004, 11, 863. [Google Scholar] [CrossRef]
- Jin, L.; Abrahams, J.P.; Skinner, R.; Petitou, M.; Pike, R.N.; Carrell, R.W. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. USA 1997, 94, 14683–14688. [Google Scholar] [CrossRef] [PubMed]
- Ersdalbadju, E.; Lu, A.; Zuo, Y.; Picard, V.; Bock, S.C. Identification of the antithrombin III heparin binding site. J. Biol. Chem. 1997, 272, 19393. [Google Scholar] [CrossRef] [PubMed]
- Baglin, T.P.; Carrell, R.W.; Church, F.C.; Esmon, C.T.; Huntington, J.A. Crystal Structures of Native and Thrombin-Complexed Heparin Cofactor II Reveal a Multistep Allosteric Mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 11079–11084. [Google Scholar] [CrossRef]
- Tollefsen, D.M. Heparin cofactor II. Oxyg. Transport. Tissue XXXIII 1997, 425, 35–44. [Google Scholar]
- Ragg, H.; Ulshöfer, T.; Gerewitz, J. Glycosaminoglycan-mediated leuserpin-2/thrombin interaction. Structure-function relationships. J. Biol. Chem. 1990, 265, 22386. [Google Scholar] [PubMed]
- Li, W.; Huntington, J.A. Crystal structures of protease nexin-1 in complex with heparin and thrombin suggest a 2-step recognition mechanism. Blood 2012, 120, 459–467. [Google Scholar] [CrossRef]
- Stone, S.R.; Brown-Luedi, M.L.; Rovelli, G.; Guidolin, A.; Mcglynn, E.; Monard, D. Localization of the heparin-binding site of glia-derived nexin/protease nexin-1 by site-directed mutagenesis. Biochemistry 1994, 33, 7731. [Google Scholar] [CrossRef]
- Keijer, J.; Linders, M.; Wegman, J.J.; Ehrlich, H.J.; Mertens, K.; Pannekoek, H. On the target specificity of plasminogen activator inhibitor 1: The role of heparin, vitronectin, and the reactive site. Blood 1991, 78, 1254–1261. [Google Scholar] [CrossRef]
- Yang, L.; Ding, Q.; Huang, X.; Olson, S.T.; Rezaie, A.R. Characterization of the heparin-binding site of the protein z-dependent protease inhibitor. Biochemistry 2012, 51, 4078–4085. [Google Scholar] [CrossRef]
- Huang, X.; Rezaie, A.R.; Broze, G.J.; Olson, S.T. Heparin Is a Major Activator of the Anticoagulant Serpin, Protein Z-dependent Protease Inhibitor. J. Biol. Chem. 2011, 286, 8740–8751. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Cai, H.; Wu, J.; Wei, Z.; Zhang, F.; Huang, X.; Ma, L.; Feng, L.; Zhang, R.; Wang, Y.; et al. Heparin Binds Lamprey Angiotensinogen and Promotes Thrombin Inhibition through a Template Mechanism. J. Biol. Chem. 2016, 291, 24900–24911. [Google Scholar] [CrossRef] [PubMed]
- Rein, C.M.; Desai, U.R.; Church, F.C. Serpin-glycosaminoglycan interactions. Methods Enzymol. 2011, 501, 105. [Google Scholar] [PubMed]
- Huntington, J.A. Thrombin inhibition by the serpins. J. Thromb Haemost 2013, 11 (Suppl. 1), 254–264. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huntington, J.A. The heparin binding site of protein C inhibitor is protease-dependent. J. Biol. Chem. 2008, 283, 36039–36045. [Google Scholar] [CrossRef]
- Li, W.; Adams, T.E.; Nangalia, J.; Esmon, C.T.; Huntington, J.A. Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-Å structure of the heparin-bridged complex. Proc. Natl. Acad. Sci. USA 2008, 105, 4661. [Google Scholar] [CrossRef]
- Chen, V.C.; Chao, L.; Pimenta, D.C.; Bledsoe, G.; Juliano, L.; Chao, J. Identification of a major heparin-binding site in kallistatin. J. Biol. Chem. 2001, 276, 1276–1284. [Google Scholar] [CrossRef]
- Lin, F.; Zhou, A.; Wei, Z. Crystallization and crystallographic studies of kallistatin. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 1135–1138. [Google Scholar] [CrossRef]
- Zakabunin, A.I.; Mishukova, O.V.; Khrapov, E.A.; Sergievichev, D.S.; Boyarskikh, U.A.; Sverdlov, E.D.; Filipenko, M.L. Cloning and expression of the gene of the human chymotrypsin-like kallikrein-7 protease in Escherichia coli and isolation of the recombinant protein. Mol. Gen. Microbiol. Virol. 2007, 22, 64–67. [Google Scholar] [CrossRef]
- Takayama, T.K.; McMullen, B.A.; Nelson, P.S.; Matsumura, M.; Fujikawa, K. Characterization of hK4 (Prostase), a Prostate-Specific Serine Protease:? Activation of the Precursor of Prostate Specific Antigen (pro-PSA) and Single-Chain Urokinase-Type Plasminogen Activator and Degradation of Prostatic Acid Phosphatase\\r. Biochemistry 2001, 40, 15341–15348. [Google Scholar] [CrossRef]
- Battye, T.G.G.; Luke, K.; Owen, J.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A new graphical interface for diffraction-image processing withMOSFLM. Acta Crystallogr. 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; Mccoy, A. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011, 67, 235–242. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Storoni, L.C.; Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Ritchie, A.W.; Webb, L.J. Optimizing Electrostatic Field Calculations with the Adaptive Poisson–Boltzmann Solver to Predict Electric Fields at Protein–Protein Interfaces II: Explicit Near-Probe and Hydrogen-Bonding Water Molecules. J. Phys. Chem. B 2014, 118, 7692–7702. [Google Scholar] [CrossRef]
- Zhou, G.X.; Chao, L.; Chao, J. Kallistatin: A novel human tissue kallikrein inhibitor. Purification, characterization, and reactive center sequence. J. Biol. Chem. 1992, 267, 25873–25880. [Google Scholar]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Ellisdon, A.M.; Zhang, Q.; Henstridge, M.A.; Johnson, T.K.; Warr, C.G.; Law, R.H.; Whisstock, J.C. High resolution structure of cleaved Serpin 42 Da from Drosophila melanogaster. BMC Struct. Biol. 2014, 14, 14. [Google Scholar] [CrossRef]
- Zhou, A.; Wei, Z.; Read, R.J.; Carrell, R.W. Structural mechanism for the carriage and release of thyroxine in the blood. Proc. Natl. Acad. Sci. USA 2006, 103, 13321–13326. [Google Scholar] [CrossRef] [PubMed]
- Ecke, S.; Geiger, M.; Binder, B.R. Heparin binding of protein-C inhibitor--analysis of the effect of heparin on the interaction of protein-C inhibitor with tissue kallikrein. Eur. J. Biochem. 1997, 248, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Ulbricht, D.; Tindall, C.A.; Oertwig, K.; Hanke, S.; Sträter, N.; Heiker, J.T. Kallikrein-related peptidase 14 is the second KLK protease targeted by the serpin vaspin. Biol. Chem. 2018, 399, 1079–1084. [Google Scholar] [CrossRef]
- Ulbricht, D.; Oertwig, K.; Arnsburg, K.; Saalbach, A.; Pippel, J.; Sträter, N.; Heiker, J.T. Basic Residues of beta-Sheet A Contribute to Heparin Binding and Activation of Vaspin (Serpin A12). J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.Q.; Agata, J.; Chao, L.; Chao, J. Kallistatin is a new inhibitor of angiogenesis and tumor growth. Blood 2002, 100, 3245–3252. [Google Scholar] [CrossRef]
- Chao, J.; Stallone, J.N.; Liang, Y.M.; Chen, L.M.; Wang, D.Z.; Chao, L. Kallistatin is a potent new. vasodilator. J. Clin. Invest. 1997, 100, 11–17. [Google Scholar] [CrossRef] [PubMed]

| Crystal | PDB 6F4U | PDB 6F4V |
|---|---|---|
| Kallistatin | Kallistatin-heparin | |
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c (Å) | 113.51, 113.51, 76.17 | 113.77, 113.77, 76.56 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Solvent content (%) | 44 | 44 |
| Data processing statistics | ||
| Wavelength (Å) | 1.196 | 1.196 |
| Resolution (Å) | 35.67–1.9; 2.0–1.9 | 56.88–1.80; 1.84–1.8 |
| Total reflections | 185,915; 26,532 | 366,297; 13,230 |
| Unique reflections | 44,414; 6433 | 52,290; 3094 |
| Multiplicity | 4.2; 4.1 | 7.0; 4.3 |
| Mean I/sd(I) | 14; 2.5 | 15.3; 1.5 |
| Completeness (%) | 99.9; 99.7 | 99.9; 99.3 |
| Rmerge | 0.043; 0.326 | 0.061; 0.648 |
| Rmeas | 0.049; 0.374 | 0.071; 0.832 |
| Model | ||
| Number of atoms modeled | 3293 | 3364 |
| Protein | 3058 | 3067 |
| Water | 185 | 230 |
| Refinement statistics (Å) | ||
| Reflections in working/free set | 42,206/2208 | 49,688/2602 |
| R-factor/R-free (%) | 0.168/0.193 | 0.173/0.196 |
| a r.m.s. deviation of bonds (Å)/angles (°) from ideality | 0.0068/1.12 | 0.0114/1.46 |
| Ramachandran plot (favored/outliers, %) | 98.40/0 | 95.89/0 |
| Wilson B-factor (Å2) | 30.1 | 28.1 |
| MolProbity score | 0.98, b 100th percentile (n = 12,147, 1.9 ± 0.25 Å) | 1.04, b 100th percentile (n = 11,444, 1.80 ± 0.25 Å) |
| Protein | kapp (M−1s−1) | SI | kapp × SI (M−1s−1) |
|---|---|---|---|
| rKAL-WT | 2.4 ± 0.1 × 103 | 1.4 ± 0.1 | 3.4 × 103 |
| rKAL-R259D | 2.2 ± 0. 1 × 103 | 1.6 ± 0.1 | 3.5 × 103 |
| rKAL-R300D | 1.6 ± 0.1 × 103 | 1.8 ± 0.1 | 2.9 × 103 |
| rKAL-R394D | 1.0 ± 0.1 × 103 | 1.8 ± 0.1 | 1.8 × 103 |
| rKAL-307/308D | 1.3 ± 0.1 × 103 | 1.5 ± 0.1 | 2.0 × 103 |
| rKAL-312/313D | 1.0 ± 0.1 × 103 | 1.5 ± 0.1 | 1.5 × 103 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; Wu, J.; Zheng, Y.; Shu, Z.; Wei, Z.; Sun, Y.; Carrell, R.W.; Zhou, A. Heparin Blocks the Inhibition of Tissue Kallikrein 1 by Kallistatin through Electrostatic Repulsion. Biomolecules 2020, 10, 828. https://doi.org/10.3390/biom10060828
Ma L, Wu J, Zheng Y, Shu Z, Wei Z, Sun Y, Carrell RW, Zhou A. Heparin Blocks the Inhibition of Tissue Kallikrein 1 by Kallistatin through Electrostatic Repulsion. Biomolecules. 2020; 10(6):828. https://doi.org/10.3390/biom10060828
Chicago/Turabian StyleMa, Lina, Jiawei Wu, Ying Zheng, Zimei Shu, Zhenquan Wei, Yinbiao Sun, Robin W. Carrell, and Aiwu Zhou. 2020. "Heparin Blocks the Inhibition of Tissue Kallikrein 1 by Kallistatin through Electrostatic Repulsion" Biomolecules 10, no. 6: 828. https://doi.org/10.3390/biom10060828
APA StyleMa, L., Wu, J., Zheng, Y., Shu, Z., Wei, Z., Sun, Y., Carrell, R. W., & Zhou, A. (2020). Heparin Blocks the Inhibition of Tissue Kallikrein 1 by Kallistatin through Electrostatic Repulsion. Biomolecules, 10(6), 828. https://doi.org/10.3390/biom10060828






