The Genoprotective Role of Naringin
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Naringin Electrochemical Response
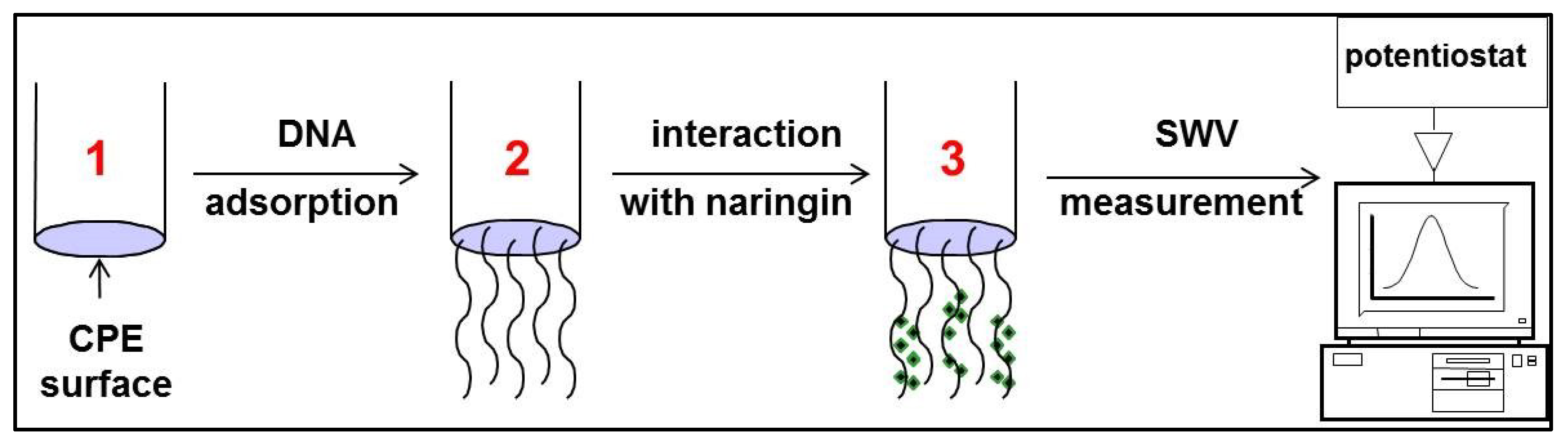
2.2.2. Interaction of Naringin with Synthetic Oligonucleotide and dsDNA
2.2.3. Statistical Analysis
3. Results
3.1. Electrochemical (SWV) Signals of Naringin
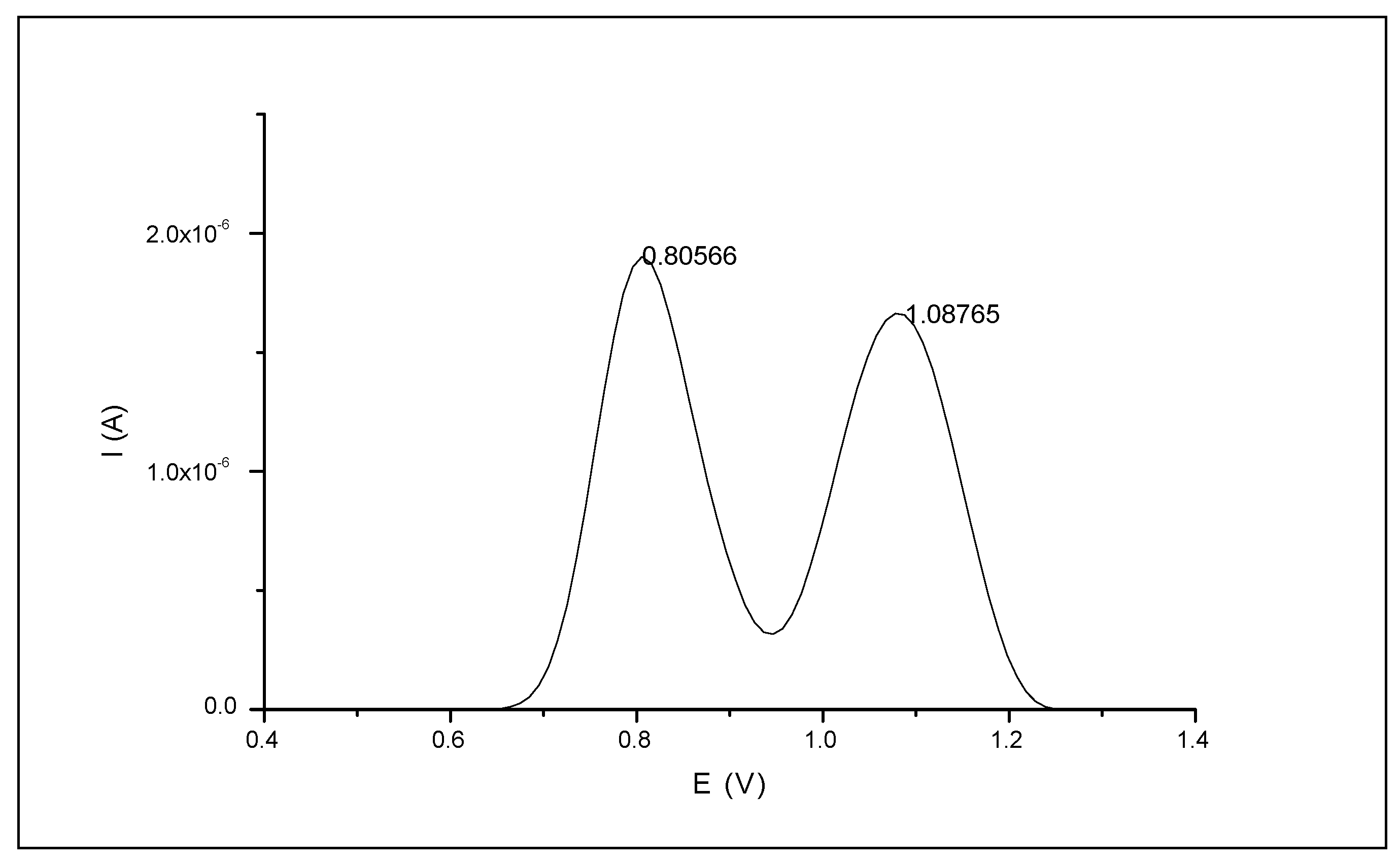
3.1.1. Naringin Electrochemical Signals
3.1.2. Calibration Curve
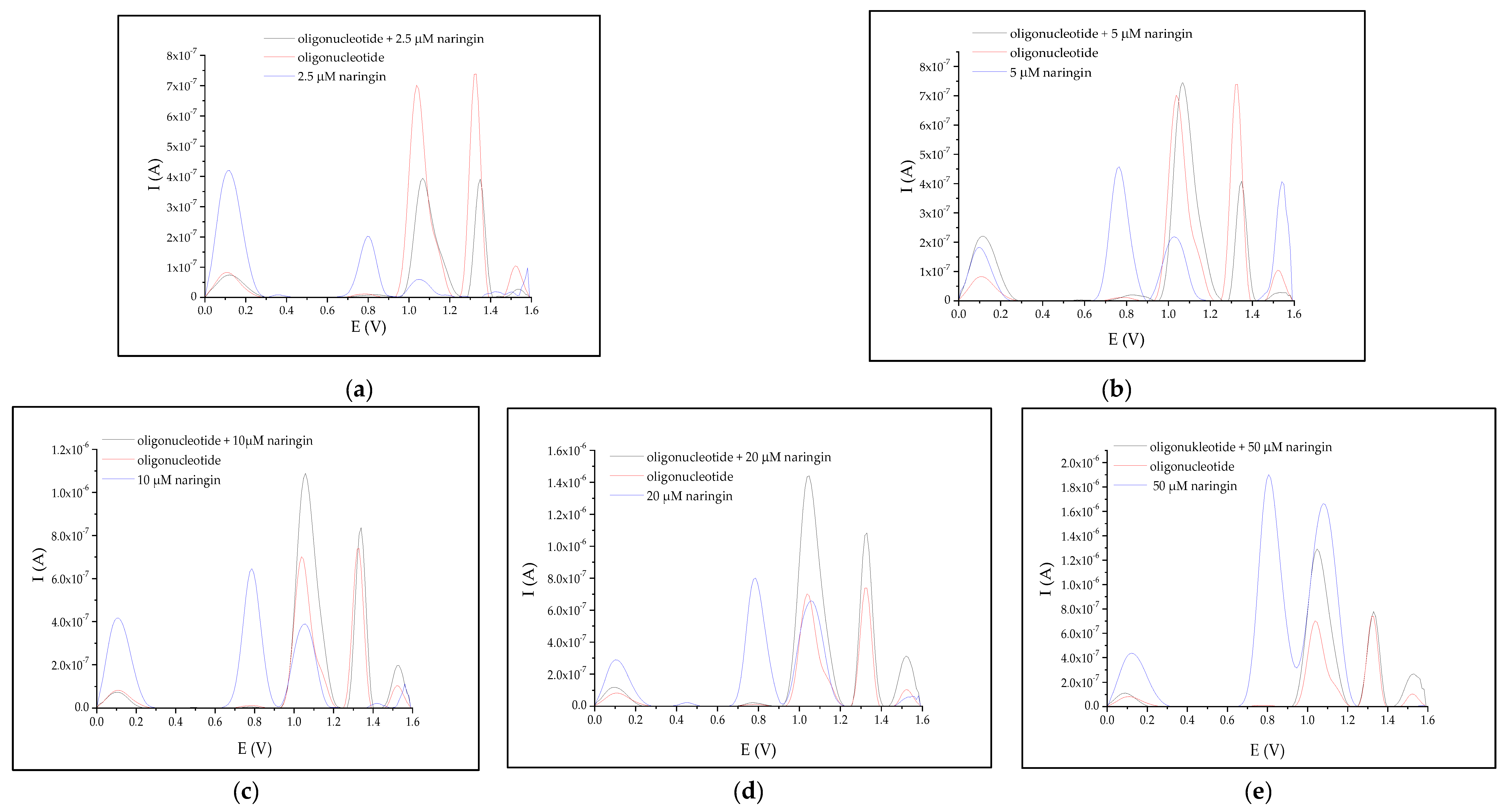
3.2. Interaction between Naringin and Oligonucleotide
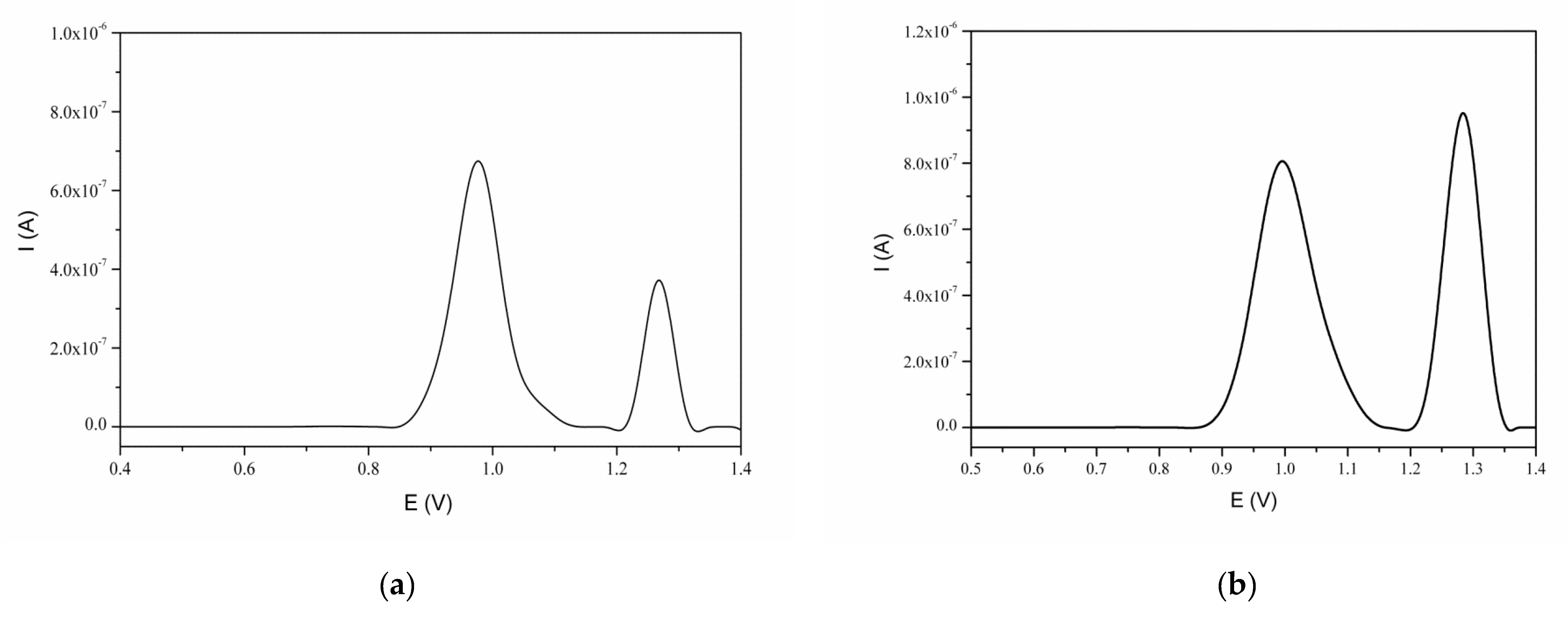
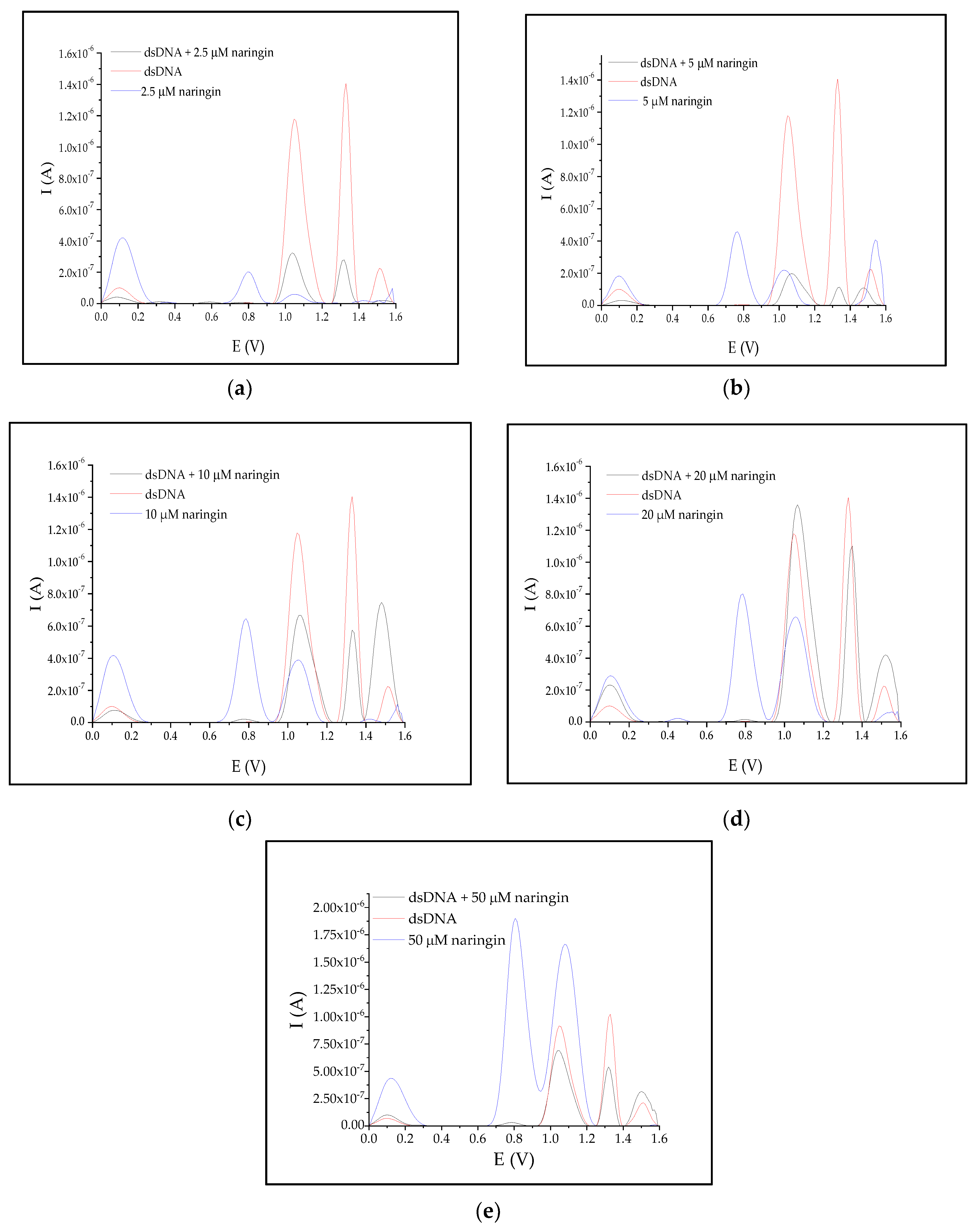
3.3. Interaction of Naringin with dsDNA
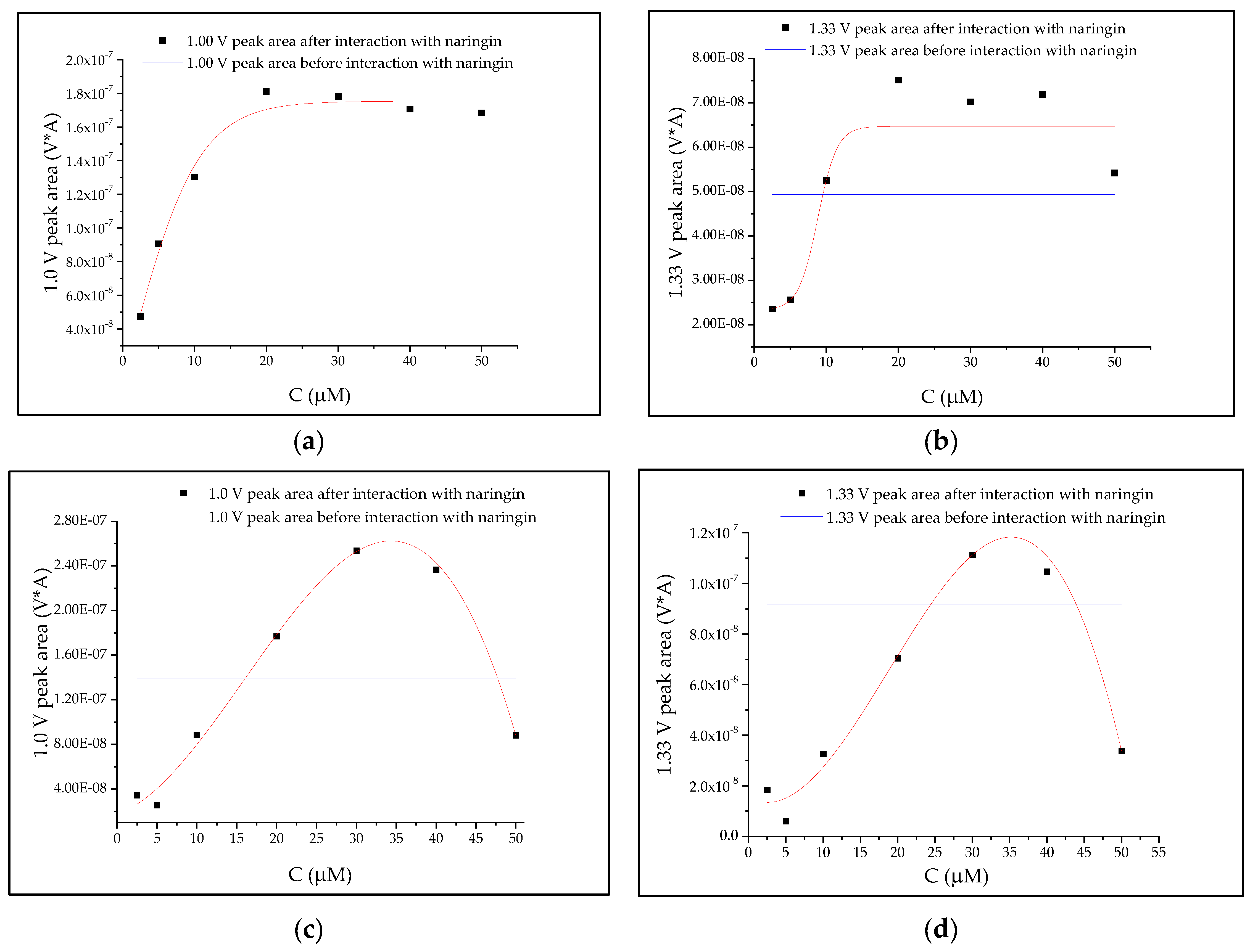
3.4. Correlation Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
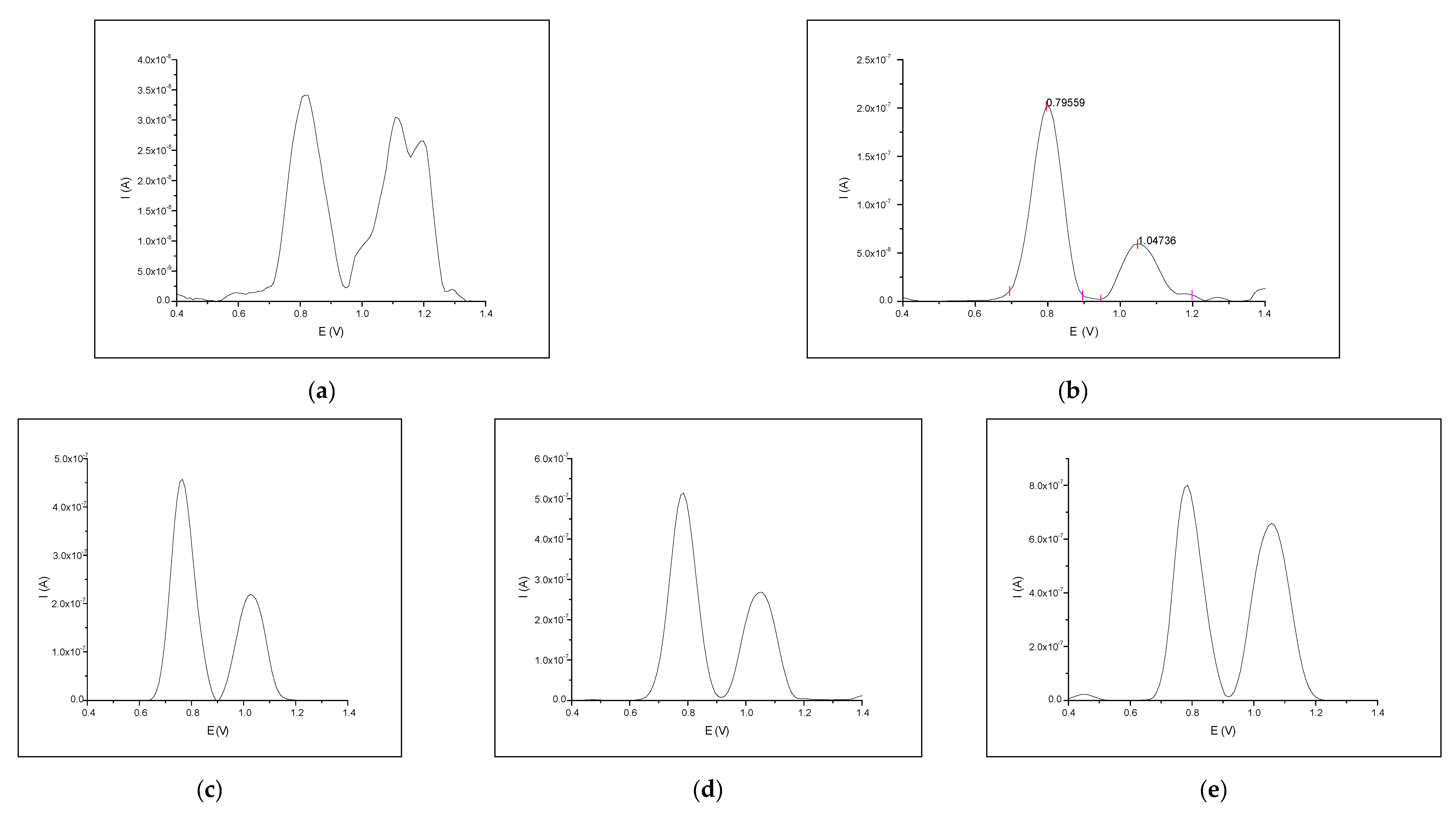
References
- Kim, S.H.; Kim, M.S.; Lee, M.S.; Park, Y.S.; Lee, H.J.; Kang, S.; Lee, H.S.; Lee, K.-E.; Yang, H.J.; Kim, M.J.; et al. Korean diet: Characteristics and historical background. J. Ethn. Foods 2016, 3, 26–31. [Google Scholar] [CrossRef]
- de Lorgeril, M. Mediterranean Diet and Cardiovascular Disease: Historical Perspective and Latest Evidence. Curr. Atheroscler. Rep. 2013, 15, 370. [Google Scholar] [CrossRef] [PubMed]
- Reitsema, L.J.; Kozłowski, T. Diet and society in Poland before the state: Stable isotope evidence from a Wielbark population (2nd c. AD). Anthropol. Rev. 2013, 76, 1–22. [Google Scholar] [CrossRef][Green Version]
- Corella, D.; Ordovás, J.M. How does the Mediterranean diet promote cardiovascular health? Current progress toward molecular mechanisms. BioEssays 2014, 36, 526–537. [Google Scholar] [CrossRef]
- Maâtouk, F.; MounaMaâtouk; Barhoumi, H.; Jaffrezic, N. Electrochemical Study of Naringenin-DNA Interaction on Modified DNA-PAH-MPA Gold Electrode. Sens. Lett. 2017, 15, 492–497. [Google Scholar] [CrossRef]
- Dudczak, J.; Ligaj, M. The Comparison of Functonality of the Methods Used for the Assessment of DNA Samples Quality. In Current Trends in Commodity Science: Development and Assessment of Non-Food Products; Michocka, K., Tichoniuk, M., Eds.; Almat Systemy Zarządzania: Poznań, Poland, 2015; ISBN 978-83-938018-9-3. [Google Scholar]
- Fojta, M. Detecting DNA Damage with Electrodes. In Electrochemistry of Nucleic Acids and Proteins—Towards Electrochemical Sensors for Genomics and Proteomics. Volume 1; Palecek, E., Scheller, F., Wang, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 385–431. ISBN 978-0-444-52150-7. [Google Scholar]
- Szczepaniak, O.M. Jakość handlowych izolatów DNA. In Młodzi Towaroznawcy 2017; Zmudziński, W., Rybicka, I., Włodarska, K., Bińczak, O., Sady, S., Szulczyńska, J., Pawłowski, T., Eds.; Wydział Towaroznawstwa, Uniwersytet Ekonomiczny w Poznaniu: Poznań, Poland, 2017; pp. 165–168. [Google Scholar]
- Ligaj, M.; Dudczak, J.; Mikołajczak, A.; Filipiak, M. Electrochemical detection of heat-induced nucleobases damage and DNA samples aging. In Collection Symposium Series; Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic: Prague, Czech Republic, 2014; pp. 314–317. [Google Scholar]
- Mountzouris, J.A.; Hurley, L.H. Small Molecule-DNA Interaction. In Bioorganic Chemistry: Nucleic Acids; Hecht, S.M., Ed.; Oxford University Press: New York, NY, USA; Oxford, UK, 1996; pp. 288–323. ISBN 0-19-508467-5. [Google Scholar]
- Ligaj, M.; Tichoniuk, M.; Gwiazdowska, D.; Filipiak, M. Electrochemical DNA biosensor for the detection of pathogenic bacteria Aeromonas hydrophila. Electrochim. Acta 2014, 128, 67–74. [Google Scholar] [CrossRef]
- Jasnowska, J.; Ligaj, M.; Stupnicka, B.; Filipiak, M. DNA sensor for o-dianisidine. Bioelectrochemistry 2004, 64, 85–90. [Google Scholar] [CrossRef]
- Panwar, N.; Soehartono, A.M.; Chan, K.K.; Zeng, S.; Xu, G.; Qu, J.; Coquet, P.; Yong, K.T.; Chen, X. Nanocarbons for Biology and Medicine: Sensing, Imaging, and Drug Delivery. Chem. Rev. 2019, 119, 9559–9656. [Google Scholar] [CrossRef]
- Mendoza, S.; Bustos, E.; Manriquez, J.; Godinez, L.A. Voltammetric Techniques. In Agricultural and Food Electroanalysis; Escarpa, A., Gonzalez, M.C., Lopez, M.A., Eds.; John Wiley & Sons: Chichester, UK; Ltd.7yy7: West Sussex, UK, 2015; pp. 23–48. ISBN 9781119961864. [Google Scholar]
- Kundu, M.; Krishnan, P.; Kotnala, R.K.; Sumana, G. Recent developments in biosensors to combat agricultural challenges and their future prospects. Trends Food Sci. Technol. 2019, 88, 157–178. [Google Scholar] [CrossRef]
- Blasco, A.J.; Barrigas, I.; González, M.C.; Escarpa, A. Fast and simultaneous detection of prominent natural antioxidants using analytical microsystems for capillary electrophoresis with a glassy carbon electrode: A new gateway to food environments. Electrophoresis 2005, 26, 4664–4673. [Google Scholar] [CrossRef]
- David, I.G.; Litescu, S.C.; Popa, D.E.; Buleandra, M.; Iordache, L.; Albu, C.; Alecu, A.; Penu, R.L. Voltammetric analysis of naringenin at a disposable pencil graphite electrode—Application to polyphenol content determination in citrus juice. Anal. Methods 2018, 10, 5763–5772. [Google Scholar] [CrossRef]
- Tichoniuk, M. Konstrukcja i Zastosowanie Elektrochemicznego Biosensora DNA do Detekcji Mikroflory Patogennej w Żywności; Uniwersytet Ekonomiczny w Poznaniu: Poznań, Poland, 2010. [Google Scholar]
- Ligaj, M. Bioczujniki do Wykrywania GMO; Wydawnictwo Uniwersytetu Ekonomicznego: Poznań, Poland, 2010; ISBN 978-83-7417-471-8. [Google Scholar]
- Stempkowska, I.; Ligaj, M.; Jasnowska, J.; Langer, J.; Filipiak, M. Electrochemical response of oligonucleotides on carbon paste electrode. Bioelectrochemistry 2007, 70, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Yang, H.-H.; Zhu, C.-L.; Chen, X.; Chen, G.-N. A Graphene Platform for Sensing Biomolecules. Angew. Chem. Int. Ed. 2009, 48, 4785–4787. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, C.D.; Nafisi, S.; Rajabi, M.; Shadaloi, A.; Tarantilis, P.A.; Polissiou, M.G.; Bariyanga, J.; Tajmir-Riahi, H.A. Structural analysis of DNA and RNA interactions with antioxidant flavonoids. Spectroscopy 2009, 23, 29–43. [Google Scholar] [CrossRef]
- Nafisi, S.; Hashemi, M.; Rajabi, M.; Tajmir-Riahi, H.A. DNA Adducts with Antioxidant Flavonoids: Morin, Apigenin, and Naringin. DNA Cell Biol. 2008, 27, 433–442. [Google Scholar] [CrossRef]
- Cavia-Saiz, M.; Busto, M.D.; Pilar-Izquierdo, M.C.; Ortega, N.; Perez-Mateos, M.; Muñiz, P. Antioxidant properties, radical scavenging activity and biomolecule protection capacity of flavonoid naringenin and its glycoside naringin: A comparative study. J. Sci. Food Agric. 2010, 90, 1238–1244. [Google Scholar] [CrossRef]
- Motawi, T.K.; Teleb, Z.A.; El-Boghdady, N.A.; Ibrahim, S.A. Effect of simvastatin and naringenin coadministration on rat liver DNA fragmentation and cytochrome P450 activity: An in vivo and in vitro study. J. Physiol. Biochem. 2014, 70, 225–237. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Chakraborty, S.; Sengupta, P.K.; Bhowmik, S. Exploring the Interactions of the Dietary Plant Flavonoids Fisetin and Naringenin with G-Quadruplex and Duplex DNA, Showing Contrasting Binding Behavior: Spectroscopic and Molecular Modeling Approaches. J. Phys. Chem. B 2016, 120, 8942–8952. [Google Scholar] [CrossRef]
- Ameer, B.; Weintraub, R.A.; Johnson, J.V.; Yost, R.A.; Rouseff, R.L. Flavanone absorption after naringin, hesperidin, and citrus administration. Clin. Pharmacol. Ther. 1996, 60, 34–40. [Google Scholar] [CrossRef]
- Yousuf, S.; Enoch, I.V.M.V. Binding interactions of naringenin and naringin with calf thymus DNA and the role of β-cyclodextrin in the binding. AAPS PharmSciTech 2013, 14, 770–781. [Google Scholar] [CrossRef]
- Mello, L.D.; Pereira, R.M.S.; Sawaya, A.C.H.F.; Eberlin, M.N.; Kubota, L.T. Electrochemical and spectroscopic characterization of the interaction between DNA and Cu(II)–naringin complex. J. Pharm. Biomed. Anal. 2007, 45, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, C.D.; Tarantilis, P.A.; Tajmir-Riahi, H.-A.; Polissiou, M.G. DNA Interaction with Saffron’s Secondary Metabolites Safranal, Crocetin, and Dimethylcrocetin. DNA Cell Biol. 2007, 26, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, F.; Valadbeigi, S.; Sajjadi, S.E.; Shokoohinia, Y.; Azizian, H.; Taheripak, G. Grandivittin as a natural minor groove binder extracted from Ferulago macrocarpa to ct-DNA, experimental and in silico analysis. Chem. Biol. Interact. 2016, 258, 89–101. [Google Scholar] [CrossRef] [PubMed]
| Calibration Curve | Intercept | Slope | r2 | ||||
|---|---|---|---|---|---|---|---|
| Value | Standard Deviation | ||||||
| SN | 0 | 1.014 × 10−8 | 0.029 × 10−8 | 0.99495 | |||
| Variance | Degrees of freedom | Total squares | Mean squares | F value | p > F | ||
| Between samples | 1 | 3.117 × 10−13 | 3.117 × 10−13 | 1184.254 | 3.897 × 10−7 | ||
| Inside sample | 5 | 1.316 × 10−15 | 0.263 × 10−15 | ||||
| Sum of totals | 6 | 3.130 × 10−13 | |||||
| S1 | Intercept | Slope | r2 | ||||
| Value | Standard deviation | ||||||
| 0 | 5.116 × 10−9 | 0.228 × 10−9 | 0.98816 | ||||
| Variance | Degrees of freedom | Total squares | Mean squares | F value | p > F | ||
| Between samples | 1 | 7.934 × 10−14 | 7.934 × 10−14 | 501.800 | 3.294 × 10−6 | ||
| Inside sample | 5 | 7.906 × 10−16 | 1.581 × 10−16 | ||||
| Sum of totals | 6 | 8.013 × 10−14 | |||||
| S2 | Intercept | Slope | r2 | ||||
| Value | Standard deviation | ||||||
| 0 | 5.023 × 10−9 | 0.162 × 10−9 | 0.99379 | ||||
| Variance | Degrees of freedom | Total squares | Mean squares | F value | p > F | ||
| Between samples | 1 | 7.649 × 10−14 | 7.649 × 10−14 | 961.392 | 6.550 × 10−7 | ||
| Inside sample | 5 | 3.978 × 10−16 | 0.796 × 10−16 | ||||
| Sum of totals | 6 | 7.689 × 10−14 | |||||
| Naringin Concentration (μM) | N | Average Peak Area (A × V) | Relative Peak Area (%) | ||||
|---|---|---|---|---|---|---|---|
| +1.0 V after Interaction | +1.3 V after Interaction | +1.0 V before Interaction | +1.3 V before Interaction | +1.0 V after Interaction | +1.3 V after Interaction | ||
| 2.5 | 5 | 4.762 × 10−8 | 2.358 × 10−8 | 6.154 × 10−8 | 4.935 × 10−8 | 77.38 | 47.79 |
| 5.0 | 4 | 9.075 × 10−8 | 2.565 × 10−8 | 147.46 | 51.97 | ||
| 10.0 | 3 | 1.304 × 10−7 | 5.248 × 10−8 | 211.91 | 106.34 | ||
| 20.0 | 4 | 1.810 × 10−7 | 7.518 × 10−8 | 294.12 | 152.33 | ||
| 30.0 | 3 | 1.783 × 10−7 | 7.025 × 10−8 | 289.73 | 142.35 | ||
| 40.0 | 2 | 1.707 × 10−7 | 7.191 × 10−8 | 277.38 | 145.71 | ||
| 50.0 | 2 | 1.685 × 10−7 | 5.424 × 10−8 | 273.73 | 109.90 | ||
| Naringin Concentration (μM) | N | First Peak Maximum Location (V) | Latter Peak Maximum Location (V) | Relative Shift (%) | |||
| After Interaction | Before Interaction | After Interaction | Before Interaction | First Peak | Latter Peak | ||
| 2.5 | 5 | 1.067 | 1.037 | 1.349 | 1.329 | 2.89 | 1.50 |
| 5.0 | 4 | 1.067 | 1.349 | 2.89 | 1.50 | ||
| 10.0 | 3 | 1.057 | 1.339 | 1.93 | 0.75 | ||
| 20.0 | 4 | 1.047 | 1.329 | 0.96 | 0 | ||
| 30.0 | 3 | 1.047 | 1.329 | 0.96 | 0 | ||
| 40.0 | 3 | 1.047 | 1.329 | 0.96 | 0 | ||
| 50.0 | 2 | 1.047 | 1.329 | 0.96 | 0 | ||
| Naringin Concentration (μM) | N | Average Peak Area | Relative Peak Area | ||||
|---|---|---|---|---|---|---|---|
| V × A | % | ||||||
| +1.0 V after Interaction | +1.3 V after Interaction | +1.0 V before Interaction | +1.3 V before Interaction | +1.0 V after Interaction | +1.3 V after Interaction | ||
| 2.5 | 3 | 3.445 × 10−8 | 1.836 × 10−8 | 1.394 × 10−7 | 9.179 × 10−8 | 24.72 | 20.00 |
| 5.0 | 4 | 2.557 × 10−8 | 6.033 × 10−9 | 18.34 | 6.57 | ||
| 10.0 | 3 | 8.825 × 10−8 | 3.253 × 10−8 | 63.31 | 35.44 | ||
| 20.0 | 4 | 1.769 × 10−7 | 7.043 × 10−8 | 126.87 | 76.73 | ||
| 30.0 | 3 | 2.538 × 10−7 | 1.112 × 10−7 | 182.07 | 121.15 | ||
| 40.0 | 3 | 2.367 × 10−7 | 1.047 × 10−7 | 169.80 | 114.06 | ||
| 50.0 | 5 | 8.808 × 10−8 | 3.385 × 10−8 | 63.19 | 36.88 | ||
| Naringin concentration(μM) | N | First peak maximum location (V) | Latter peak maximum location (V) | Relative shift (%) | |||
| After interaction | Before interaction | After interaction | Before interaction | First peak | Latter peak | ||
| 2.5 | 3 | 1.037 | 1.047 | 1.319 | 1.329 | −0.96 | −0.75 |
| 5.0 | 4 | 1.067 | 1.339 | 1.91 | 0.75 | ||
| 10.0 | 3 | 1.067 | 1.329 | 1.91 | 0 | ||
| 20.0 | 4 | 1.067 | 1.349 | 1.91 | 1.50 | ||
| 30.0 | 3 | 1.067 | 1.339 | 1.91 | 0.75 | ||
| 40.0 | 3 | 1.067 | 1.339 | 1.91 | 0.75 | ||
| 50.0 | 5 | 1.047 | 1.319 | 0 | −0.75 | ||
| Oligonucleotide | ||||||
|---|---|---|---|---|---|---|
| Peak Area | A1 | A2 | x0 | dx | Statistics | |
| Χ2 | r2 | |||||
| +1.0 V | −1.126 × 10−7 | 1.754 × 10−7 | 1.330 | 4.621 | 2.345 × 10−16 | 0.9232 |
| +1.3 V | 2.334 × 10−8 | 6.470 × 10−8 | 8.839 | 1.32944 | 2.195 × 10−16 | 0.5324 |
| dsDNA | ||||||
| Peak area | A | B | C | D | Statistics | |
| Χ2 | r2 | |||||
| +1.0 V | 1.750 × 10−8 | 2.423 × 10−9 | 4.822 × 10−10 | −1.005 × 10−11 | 3.573 × 10−16 | 0.9015 |
| +1.3 V | 1.541 × 10−8 | −1.606 × 10−9 | 3.408 × 10−10 | −6.027 × 10−12 | 1.346 × 10−16 | 0.7700 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczepaniak, O.; Ligaj, M.; Kobus-Cisowska, J.; Tichoniuk, M.; Dziedziński, M.; Przeor, M.; Szulc, P. The Genoprotective Role of Naringin. Biomolecules 2020, 10, 700. https://doi.org/10.3390/biom10050700
Szczepaniak O, Ligaj M, Kobus-Cisowska J, Tichoniuk M, Dziedziński M, Przeor M, Szulc P. The Genoprotective Role of Naringin. Biomolecules. 2020; 10(5):700. https://doi.org/10.3390/biom10050700
Chicago/Turabian StyleSzczepaniak, Oskar, Marta Ligaj, Joanna Kobus-Cisowska, Mariusz Tichoniuk, Marcin Dziedziński, Monika Przeor, and Piotr Szulc. 2020. "The Genoprotective Role of Naringin" Biomolecules 10, no. 5: 700. https://doi.org/10.3390/biom10050700
APA StyleSzczepaniak, O., Ligaj, M., Kobus-Cisowska, J., Tichoniuk, M., Dziedziński, M., Przeor, M., & Szulc, P. (2020). The Genoprotective Role of Naringin. Biomolecules, 10(5), 700. https://doi.org/10.3390/biom10050700









