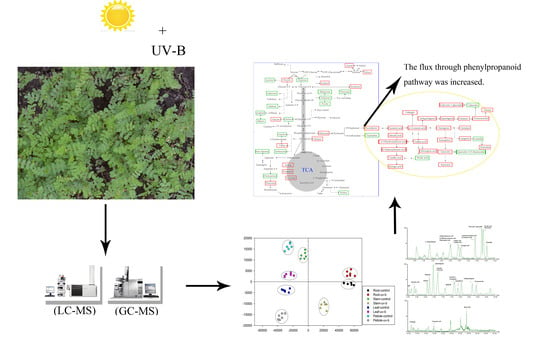
UV-B Radiation Largely Promoted the Transformation of Primary Metabolites to Phenols in Astragalus mongholicus Seedlings
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions and Treatments
2.2. Primary Metabolite Extraction and Gas Chromatography-Mass Spectrometry Analysis
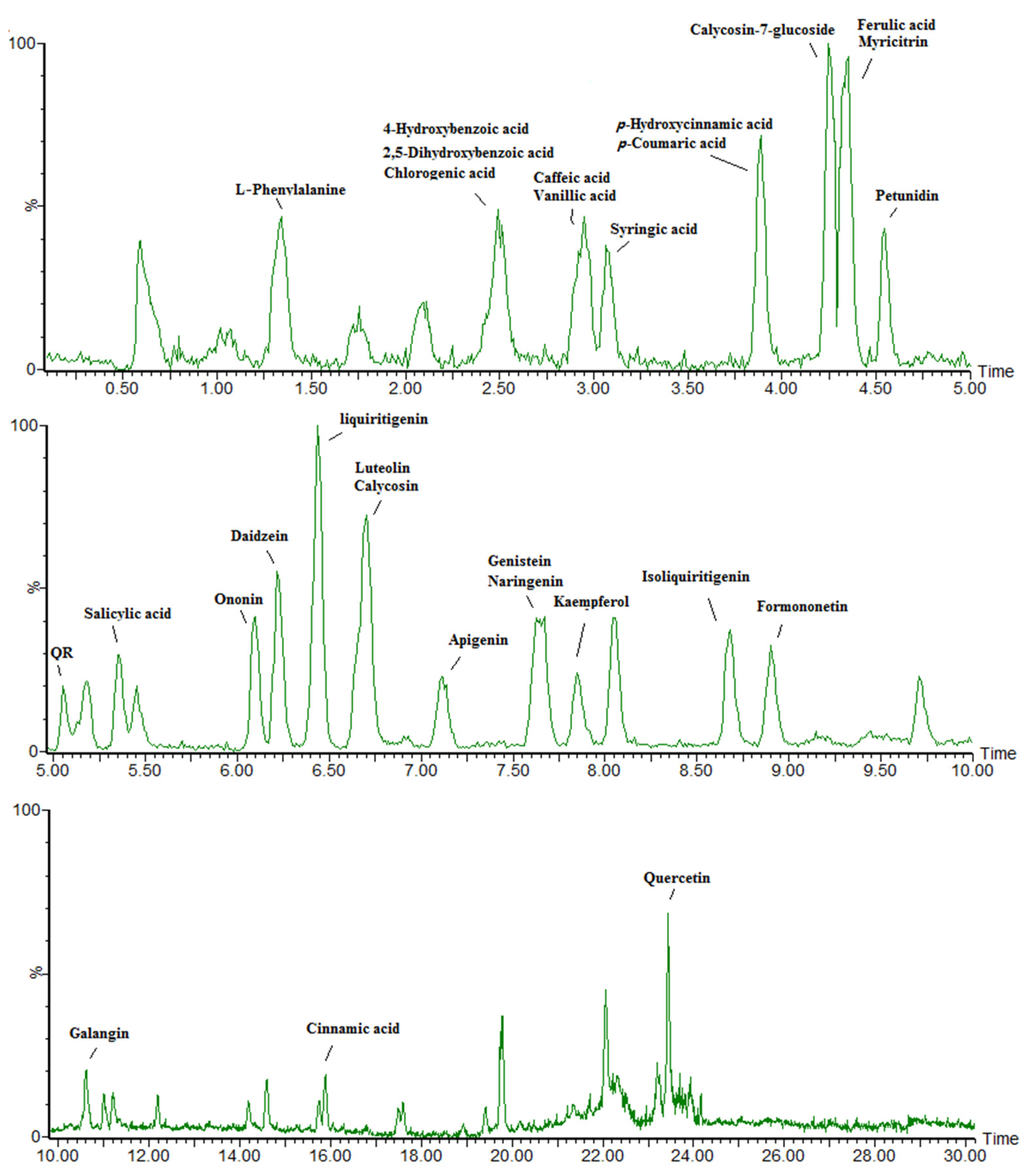
2.3. Phenolic Metabolite Extraction and Liquid Chromatography-Mass Spectrometry Targeted Analysis
2.4. Multivariate Analysis
2.5. RNA Extraction and Real-Time Polymerase Chain Reaction Analysis
3. Results
3.1. Morphological and Physiological Changes Induced by Ultraviolet-B Radiation
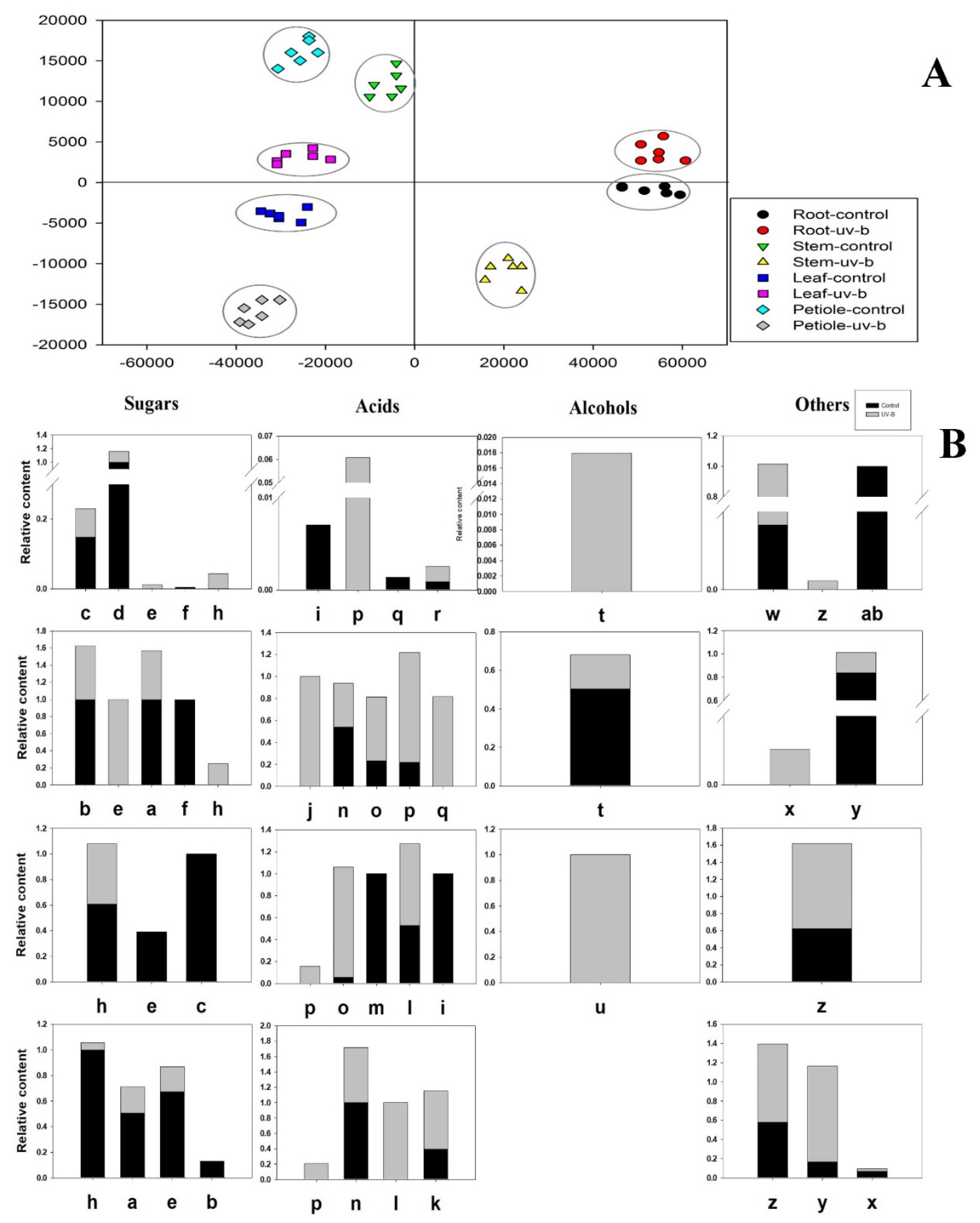
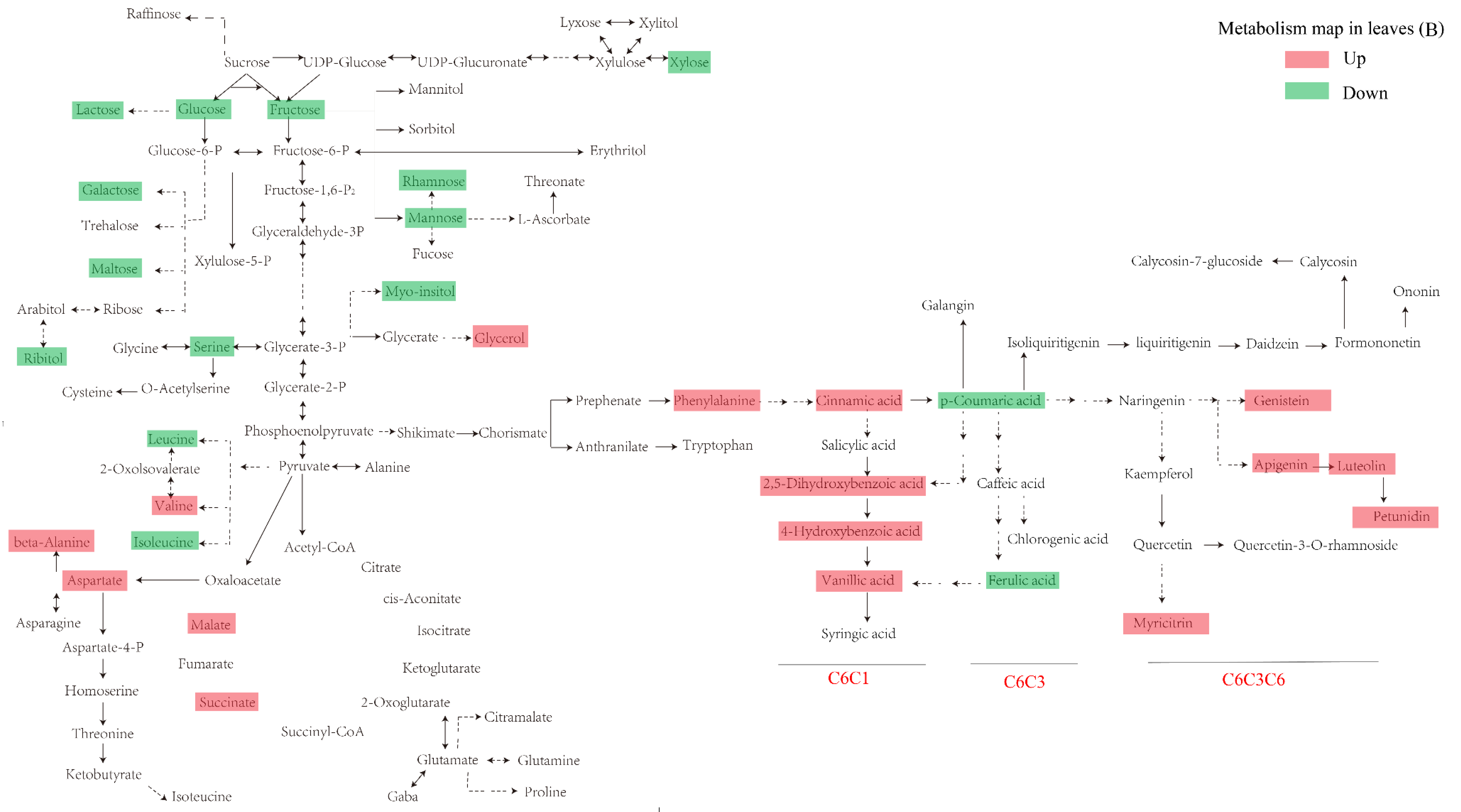
3.2. Primary Metabolism Reprogramming between the below- and Aboveground Organs in Response to Ultraviolet-B Radiation
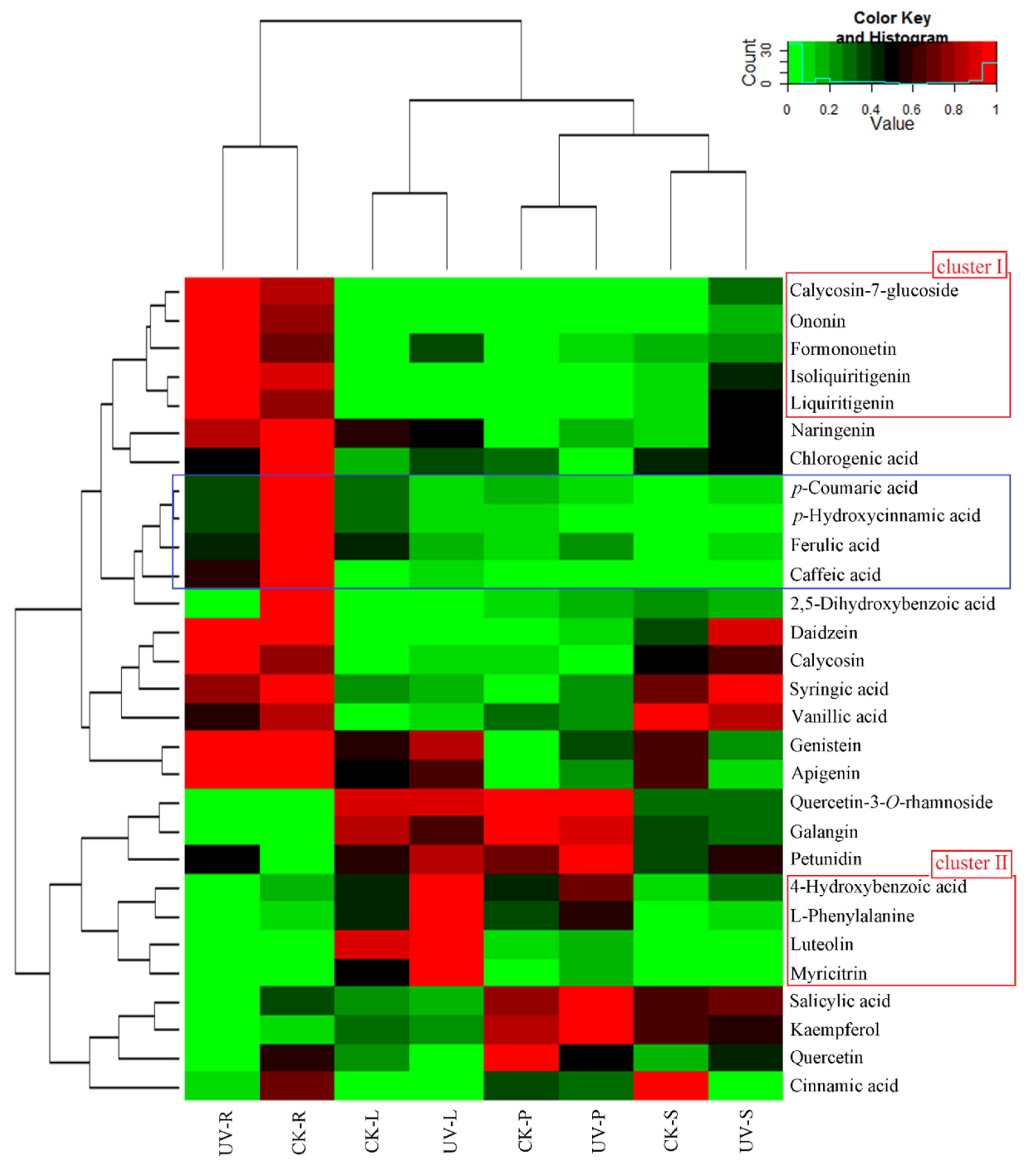
3.3. Phenolic Compounds Were Concentrated in Leaves Compared to Roots under Ultraviolet-B Radiation
3.4. Construction of an Integrative View of the Primary and Phenolic Metabolite Network for Specific Tissues and Growth Conditions
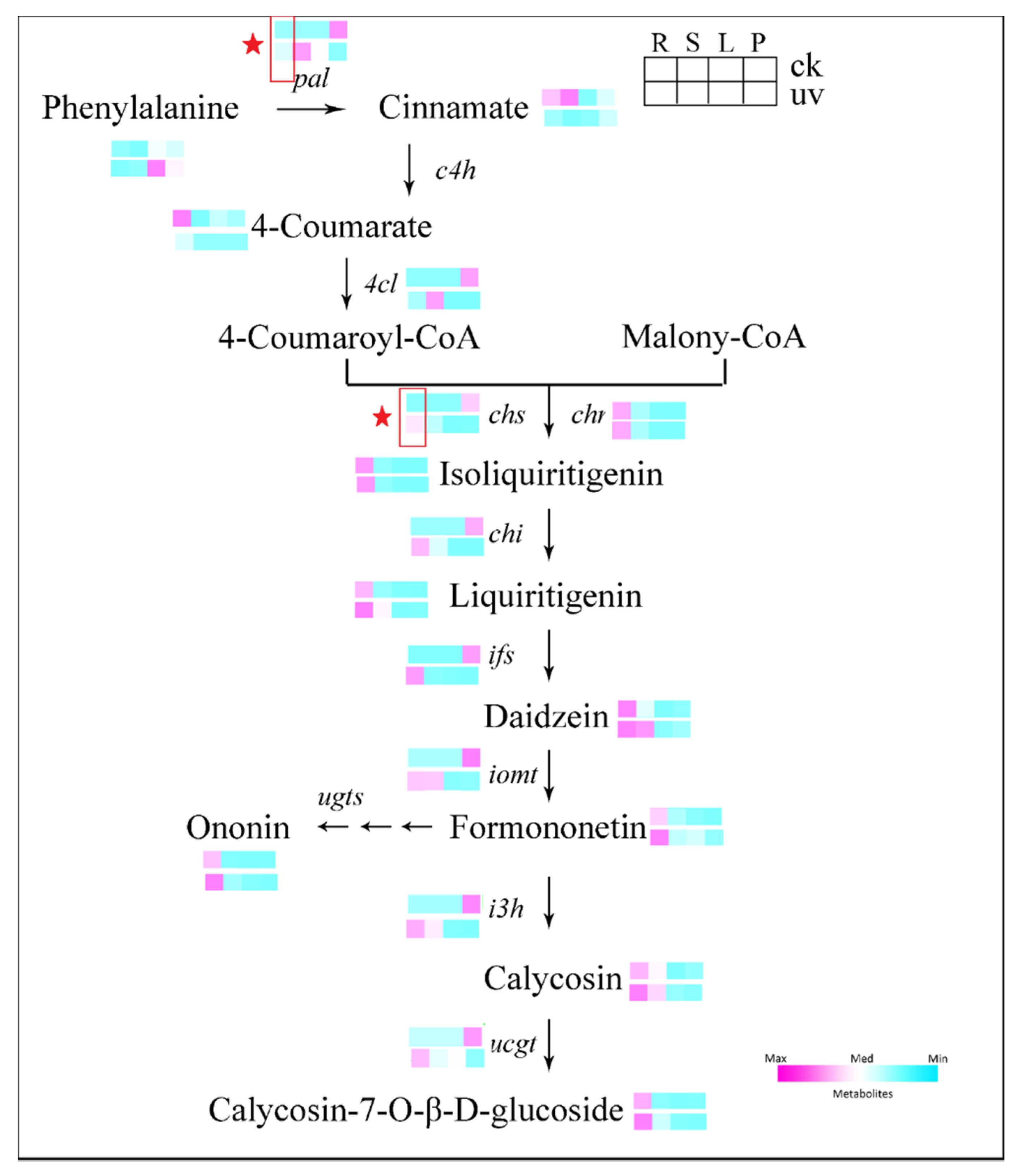
3.5. Expression of Genes Involved in Isoflavonoids Pathway in Leaves and Roots
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pan, H.; Fang, C.; Zhou, T.; Wang, Q.; Chen, J. Accumulation of calycosin and its 7-O-beta-D-glucoside and related gene expression in seedlings of Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao induced by low temperature stress. Plant Cell Rep. 2007, 26, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.; Akay, S.; Mari, A.; Bedir, E.; Pizza, C.; Piacente, S. An analytical approach based on ESI-MS, LC–MS and PCA for the quali–quantitative analysis of cycloartane derivatives in Astragalus spp. J. Pharm. Biomed. Anal. 2013, 85, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-H.; Tsai, W.-J.; Loke, S.-H.; Wu, T.-S.; Chiou, W.-F. Astragalus membranaceus flavonoids (AMF) ameliorate chronic fatigue syndrome induced by food intake restriction plus forced swimming. J. Ethnopharmacol. 2009, 122, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-H.; Li, Q.; Wu, M.-Y.; Guo, D.-J.; Chen, H.-L.; Chen, S.-L.; Seto, S.W.; Au, A.L.; Poon, C.C.; Leung, G.P.-H. Formononetin, an isoflavone, relaxes rat isolated aorta through endothelium-dependent and endothelium-independent pathways. J. Nutr. Biochem. 2010, 21, 613–620. [Google Scholar] [CrossRef]
- Fan, Y.; Wu, D.-Z.; Gong, Y.-Q.; Zhou, J.-Y.; Hu, Z.-B. Effects of calycosin on the impairment of barrier function induced by hypoxia in human umbilical vein endothelial cells. Eur. J. Pharmacol. 2003, 481, 33–40. [Google Scholar] [CrossRef]
- Cho, W.C.S.; Leung, K.N. In vitro and in vivo immunomodulating and immunorestorative effects of Astragalus membranaceus. J. Ethnopharmacol. 2007, 113, 132–141. [Google Scholar] [CrossRef]
- Cho, W.C.; Leung, K.N. In vitro and in vivo anti-tumor effects of Astragalus membranaceus. Cancer Lett. 2007, 252, 43–54. [Google Scholar] [CrossRef]
- Ma, X.; Tu, P.; Chen, Y.; Zhang, T.; Wei, Y.; Ito, Y. Preparative isolation and purification of two isoflavones from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao by high-speed counter-current chromatography. J. Chromatogr. A 2003, 992, 193–197. [Google Scholar] [CrossRef]
- Nakamura, T.; Hashimoto, A.; Nishi, H.; Kokusenya, Y. Investigation on the Marker Substances of Crude Drugs in Formulations. I.: Marker Substances for the Identification of Astragali Radix in Kampo and Drinkable Preparations. Yakugaku Zasshi 1999, 119, 391–400. [Google Scholar] [CrossRef][Green Version]
- Yu, O.; Shi, J.; Hession, A.O.; Maxwell, C.A.; Mcgonigle, B.; Odell, J.T. Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochem. 2003, 63, 753–763. [Google Scholar] [CrossRef]
- Frohnmeyer, H.; Staiger, D. Ultraviolet-B Radiation-Mediated Responses in Plants. Balancing Damage and Protection. Plant Physiol. 2003, 133, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.A.K.; Gaba, V.; Greenberg, B.M. Higher plants and UV-B radiation: Balancing damage, repair and acclimation. Trends Plant Sci. 1998, 3, 131–135. [Google Scholar] [CrossRef]
- Rozema, J.; Van De Staaij, J.; Björn, L.O.; Caldwell, M. UV-B as an environmental factor in plant life: Stress and regulation. Trends Ecol. Evol. 1997, 12, 22–28. [Google Scholar] [CrossRef]
- Kusano, M.; Tohge, T.; Fukushima, A.; Kobayashi, M.; Hayashi, N.; Otsuki, H.; Kondou, Y.; Goto, H.; Kawashima, M.; Matsuda, F.; et al. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 2011, 67, 354–369. [Google Scholar] [CrossRef]
- Matsuura, H.N.; De Costa, F.; Yendo, A.C.A.; Fett-Neto, A.G. Photoelicitation of Bioactive Secondary Metabolites by Ultraviolet Radiation: Mechanisms, Strategies, and Applications. Biotechnol. Med. Plants 2012, 171–190. [Google Scholar]
- Hahlbrock, K.; Scheel, D. Physiology and Molecular Biology of Phenylpropanoid Metabolism. Annu. Rev. Plant Biol. 1989, 40, 347–369. [Google Scholar] [CrossRef]
- Frolov, A.; Henning, A.; Böttcher, C.; Tissier, A.; Strack, D. An UPLC-MS/MS Method for the Simultaneous Identification and Quantitation of Cell Wall Phenolics in Brassica napus Seeds. J. Agric. Food Chem. 2013, 61, 1219–1227. [Google Scholar] [CrossRef]
- Parr, A.J.; Bolwell, G.P. Phenols in the plant and in man. The potential for possible nutritional enhancement of the diet by modifying the phenols content or profile. J. Sci. Food Agric. 2000, 80, 985–1012. [Google Scholar] [CrossRef]
- Beckman, C.H. Phenolic-storing cells: Keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 2000, 57, 101–110. [Google Scholar] [CrossRef]
- Valcarcel, J.; Reilly, K.; Gaffney, M.; O’Brien, N.M. Antioxidant Activity, Total Phenolic and Total Flavonoid Content in Sixty Varieties of Potato (Solanum tuberosum L.) Grown in Ireland. Potato Res. 2015, 58, 221–244. [Google Scholar] [CrossRef]
- García, B.A.; Berrueta, L.A.; Garmón-Lobato, S.; Gallo, B.; Vicente, F. A general analytical strategy for the characterization of phenolic compounds in fruit juices by high-performance liquid chromatography with diode array detection coupled to electrospray ionization and triple quadrupole mass spectrometry. J. Chromatogr. A 2009, 1216, 5398–5415. [Google Scholar] [CrossRef] [PubMed]
- Caravaca, G.; López-Cobo, A.; Verardo, V.; Segura-Carretero, A.; Gutierrez, A.F. HPLC-DAD-Q-TOF-MS as a powerful platform for the determination of phenolic and other polar compounds in the edible part of mango and its by-products (peel, seed and seed husk). Electrophoresis 2016, 37, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.M.; Weaver, L.M. The shikimate pathway. Annu. Rev. Plant Boil. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Boil. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Babst, B.A.; Harding, S.A.; Tsai, C.-J. Biosynthesis of Phenolic Glycosides from Phenylpropanoid and Benzenoid Precursors in Populus. J. Chem. Ecol. 2010, 36, 286–297. [Google Scholar] [CrossRef]
- Wink, M. Biochemistry of Plant Secondary Metabolism, 2nd ed.; Wiley Blackwell: Hoboken, NJ, USA, 2010; Volume 40, pp. 484–485. [Google Scholar]
- Marković, S.; Tošović, J. Application of Time-Dependent Density Functional and Natural Bond Orbital Theories to the UV–vis Absorption Spectra of Some Phenolic Compounds. J. Phys. Chem. A 2015, 119, 9352–9362. [Google Scholar] [CrossRef]
- Song, J.-Z.; Mo, S.-F.; Yip, Y.-K.; Qiao, C.-F.; Han, Q.-B.; Xu, H. Development of microwave assisted extraction for the simultaneous determination of isoflavonoids and saponins in radix astragali by high performance liquid chromatography. J. Sep. Sci. 2007, 30, 819–824. [Google Scholar] [CrossRef]
- Wu, T.; Bligh, S.A.; Gu, L.-H.; Wang, Z.-T.; Liu, H.-P.; Cheng, X.-M.; Branford-White, C.J.; Hu, Z.-B. Simultaneous determination of six isoflavonoids in commercial Radix Astragali by HPLC-UV. Fitoterapia 2005, 76, 157–165. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Wang, Y.; Abozeid, A.; Yuan-Gang, Z.; Tang, Z.-H. The integration of GC–MS and LC–MS to assay the metabolomics profiling in Panax ginseng and Panax quinquefolius reveals a tissue- and species-specific connectivity of primary metabolites and ginsenosides accumulation. J. Pharm. Biomed. Anal. 2017, 135, 176–185. [Google Scholar] [CrossRef]
- Park, H.-W.; In, G.; Kim, J.-H.; Cho, B.-G.; Han, G.-H.; Chang, I.-M. Metabolomic approach for discrimination of processed ginseng genus (Panax ginseng and Panax quinquefolius) using UPLC-QTOF MS. J. Ginseng Res. 2013, 38, 59–65. [Google Scholar] [CrossRef]
- Li, L.; Luo, G.-A.; Liang, Q.; Hu, P.; Wang, Y.-M. Rapid qualitative and quantitative analyses of Asian ginseng in adulterated American ginseng preparations by UPLC/Q-TOF-MS. J. Pharm. Biomed. Anal. 2010, 52, 66–72. [Google Scholar] [CrossRef]
- Fernie, A.R.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Boil. 2004, 5, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Combining Genomics, Metabolome Analysis, and Biochemical Modelling to Understand Metabolic Networks. Comp. Funct. Genom. 2001, 2, 155–168. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Zhu, B.; Yao, Z.; Yin, P.; Lu, X.; Kong, H.; Fan, F.; Jiao, B.; Xu, G. Analysis of Urinary Metabolic Signatures of Early Hepatocellular Carcinoma Recurrence after Surgical Removal Using Gas Chromatography–Mass Spectrometry. J. Proteome Res. 2012, 11, 4361–4372. [Google Scholar] [CrossRef]
- Li, Y.; Ruan, Q.; Li, Y.; Ye, G.; Lü, X.; Lin, X.; Xu, G. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. J. Chromatogr. A 2012, 1255, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Corrigendum: Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2015, 10, 1457. [Google Scholar] [CrossRef]
- Schauer, N.; Steinhauser, D.; Strelkov, S.; Schomburg, D.; Allison, G.; Moritz, T.; Lundgren, K.; Roessner, U.; Forbes, M.G.; Willmitzer, L.; et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005, 579, 1332–1337. [Google Scholar] [CrossRef]
- Ye, G.; Liu, Y.; Yin, P.; Zeng, Z.; Huang, Q.; Kong, H.; Lu, X.; Zhong, L.; Zhang, Z.; Xu, G. Study of Induction Chemotherapy Efficacy in Oral Squamous Cell Carcinoma Using Pseudotargeted Metabolomics. J. Proteome Res. 2014, 13, 1994–2004. [Google Scholar] [CrossRef] [PubMed]
- Imperlini, E.; Santorelli, L.; Orrù, S.; Scolamiero, E.; Ruoppolo, M.; Caterino, M. Mass Spectrometry-Based Metabolomic and Proteomic Strategies in Organic Acidemias. BioMed Res. Int. 2016, 2016, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Goodacre, R. An introduction to liquid chromatography–mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem. Anal. 2010, 21, 33–47. [Google Scholar] [CrossRef]
- Farag, M.A.; Huhman, D.; Lei, Z.; Sumner, L. Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC–UV–ESI–MS and GC–MS. Phytochemistry 2007, 68, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Rogachev, I.; Aharoni, A. UPLC-MS-Based Metabolite Analysis in Tomato. Adv. Struct. Saf. Stud. 2011, 860, 129–144. [Google Scholar]
- Doerfler, H.; Lyon, D.; Nägele, T.; Sun, X.; Fragner, L.; Hadaček, F.; Egelhofer, V.; Weckwerth, W. Granger causality in integrated GC–MS and LC–MS metabolomics data reveals the interface of primary and secondary metabolism. Metabolomics 2012, 9, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Han, J.S.; Lee, S.; Kim, H.Y.; Lee, C.H. MS-Based Metabolite Profiling of Aboveground and Root Components of Zingiber mioga and Officinale. Molecules 2015, 20, 16170–16185. [Google Scholar] [CrossRef]
- Cuadros-Inostroza, A.; Ruiz-Lara, S.; González, E.; Eckardt, A.; Willmitzer, L.; Peña-Cortés, H. GC–MS metabolic profiling of Cabernet Sauvignon and Merlot cultivars during grapevine berry development and network analysis reveals a stage- and cultivar-dependent connectivity of primary metabolites. Metabolomics 2016, 12, 39. [Google Scholar] [CrossRef]
- Moing, A.; Aharoni, A.; Biais, B.; Rogachev, I.; Meir, S.; Brodsky, L.; Allwood, J.W.; Erban, A.; Dunn, W.; Kay, L.; et al. Extensive metabolic cross-talk in melon fruit revealed by spatial and developmental combinatorial metabolomics. New Phytol. 2011, 190, 683–696. [Google Scholar] [CrossRef]
- Logemann, E.; Tavernaro, A.; Schulz, W.; Somssich, I.; Hahlbrock, K. UV light selectively coinduces supply pathways from primary metabolism and flavonoid secondary product formation in parsley. Proc. Natl. Acad. Sci. USA 2000, 97, 1903–1907. [Google Scholar] [CrossRef]
- Liu, C.-J.; Deavours, B.E.; Richard, S.B.; Ferrer, J.-L.; Blount, J.W.; Huhman, D.; Dixon, R.A.; Noel, J.P. Structural Basis for Dual Functionality of Isoflavonoid O-Methyltransferases in the Evolution of Plant Defense Responses[OA]. Plant Cell 2006, 18, 3656–3669. [Google Scholar] [CrossRef]
- Liu, C.-J.; Huhman, D.; Sumner, L.; Dixon, R.A. Regiospecific hydroxylation of isoflavones by cytochrome p450 81E enzymes from Medicago truncatula. Plant J. 2003, 36, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Dhaubhadel, S.; McGarvey, B.D.; Williams, R.; Gijzen, M. Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol. Boil. 2003, 53, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.Y. The Diphenylether Herbicide Lactofen Induces Cell Death and Expression of Defense-Related Genes in Soybean1. Plant Physiol. 2005, 139, 1784–1794. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Gai, Q.-Y.; Wang, W.; Luo, M.; Gu, C.-B.; Fu, Y.-J.; Ma, W. Ultraviolet Radiation-Elicited Enhancement of Isoflavonoid Accumulation, Biosynthetic Gene Expression, and Antioxidant Activity in Astragalus membranaceus Hairy Root Cultures. J. Agric. Food Chem. 2015, 63, 8216–8224. [Google Scholar] [CrossRef]
- Caldwell, M.M.; Ballare, C.L.; Bornman, J.F.; Flint, S.D.; Bjorn, L.O.; Teramura, A.H.; Kulandaivelu, G.; Tevini, M. Terrestrial ecosystems, increased solar ultraviolet radiation and interactions with other climatic change factors. Photochem. Photobiol. Sci. 2003, 2, 29–38. [Google Scholar]
- Xiong, F.S.; Day, T.A. Effect of solar ultraviolet-B radiation during springtime ozone depletion on photosynthesis and biomass production of Antarctic vascular plants. Plant Physiol. 2001, 125, 738–751. [Google Scholar]
- Xu, R.Y.; Nan, P.; Yang, Y.; Pan, H.; Zhou, T.; Chen, J. Ultraviolet irradiation induces accumulation of isoflavonoids and transcription of genes of enzymes involved in the calycosin-7-O-beta-d-glucoside pathway in Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao. Physiol. Plant 2011, 142, 265–273. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Wang, Y.; Abozeid, A.; Tian, D.; Zhang, X.-N.; Tang, Z. The Different Resistance of Two Astragalus Plants to UV-B Stress is Tightly Associated with the Organ-specific Isoflavone Metabolism. Photochem. Photobiol. 2017, 94, 115–125. [Google Scholar] [CrossRef]
- Li, J.; Ou-Lee, T.M.; Raba, R.; Last, A.R.L. Arabidopsis flavonoid mutants are hypersensitive to UB-B irradiation. Plant Cell. 1993, 5, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; McClure, J.W. Effects of UV-B on activities of enzymes of secondary phenolic metabolism in barley primary leaves. Physiol. Plant. 1995, 93, 734–739. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Aiamla-Or, S.; Shigyo, M.; Ito, S.-I.; Yamauchi, N. Involvement of chloroplast peroxidase on chlorophyll degradation in postharvest broccoli florets and its control by UV-B treatment. Food Chem. 2014, 165, 224–231. [Google Scholar] [CrossRef]
- Cuadros-Inostroza, Á.; Caldana, C.; Redestig, H.; Kusano, M.; Lisec, J.; Peña-Cortés, H.; Willmitzer, L.; Hannah, M. TargetSearch-a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinform. 2009, 10, 428. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Dileo, M.V.; Strahan, G.D.; Bakker, M.D.; Hoekenga, O. Weighted Correlation Network Analysis (WGCNA) Applied to the Tomato Fruit Metabolome. PLoS ONE 2011, 6, e26683. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Wang, Y.; Zhang, Z.-H.; Zu, Y.-G.; Efferth, T.; Tang, Z.-H. The Combined Effects of Ethylene and MeJA on Metabolic Profiling of Phenolic Compounds in Catharanthus roseus Revealed by Metabolomics Analysis. Front. Physiol. 2016, 7, 2555. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.-G.; Pang, H.-H.; Yu, J.-H.; Li, D.-W.; Wei, X.-X.; Gao, Y.-X.; Tong, L. Responses in the morphology, physiology and biochemistry of Taxus chinensis var. mairei grown under supplementary UV-B radiation. J. Photochem. Photobiol. B Boil. 2010, 98, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Soriano-Melgar, L.D.A.A.; Alcaraz-Meléndez, L.; Méndez-Rodríguez, L.C.; Puente, M.E.; Rivera-Cabrera, F.; Zenteno-Savin, T. Antioxidant responses of damiana (Turnera diffusa Willd) to exposure to artificial ultraviolet (UV) radiation in an in vitro model; part ii; UV-B radiation. Nutr. Hosp. 2014, 29, 1116–1122. [Google Scholar]
- Dias, D.A.; Hill, C.B.; Jayasinghe, N.S.; Atieno, J.; Sutton, T.; Roessner, U. Quantitative profiling of polar primary metabolites of two chickpea cultivars with contrasting responses to salinity. J. Chromatogr. B 2015, 1000, 1–13. [Google Scholar] [CrossRef]
- Harborne, J.B.; A Williams, C. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Peer, W.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Boil. 2005, 8, 317–323. [Google Scholar] [CrossRef]
- Ryan, K.G.; Swinny, E.E.; Markham, K.R.; Winefield, C. Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry 2002, 59, 23–32. [Google Scholar] [CrossRef]
- Ryan, K.G.; Swinny, E.E.; Winefield, C.; Markham, K.R. Flavonoids and UV photoprotection in Arabidopsis mutants. Z. Nat. C 2001, 56, 745–754. [Google Scholar] [CrossRef]
- Gibon, Y.; Pyl, E.-T.; Sulpice, R.; Lunn, J.; Höhne, M.; Günther, M.; Stitt, M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsisis grown in very short photoperiods. Plant Cell Environ. 2009, 32, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Robbins, M.P. Biochemistry of plant secondary metabolism. Annual Plant Reviews, Volume 2. Edited by Michael Wink. Eur. J. Plant Pathol. 2000, 106, 487. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef]
- Agati, G.; Cerovic, Z.G.; Pinelli, P.; Tattini, M. Light-induced accumulation of ortho-dihydroxylated flavonoids as non-destructively monitored by chlorophyll fluorescence excitation techniques. Environ. Exp. Bot. 2011, 73, 3–9. [Google Scholar] [CrossRef]
- Zhang, Z.-Z.; Che, X.-N.; Pan, Q.-H.; Li, X.-X.; Duan, C.-Q. Transcriptional activation of flavan-3-ols biosynthesis in grape berries by UV irradiation depending on developmental stage. Plant Sci. 2013, 208, 64–74. [Google Scholar] [CrossRef]
- Kanazawa, K.; Hashimoto, T.; Yoshida, S.; Sungwon, P.; Fukuda, S. Short Photoirradiation Induces Flavonoid Synthesis and Increases Its Production in Postharvest Vegetables. J. Agric. Food Chem. 2012, 60, 4359–4368. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, S.; Rani, A.; Gulati, A.; Ahuja, P.S. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct. Integr. Genom. 2008, 9, 125–134. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence (5′ to 3′) | |
|---|---|---|
| CHS | Forward | CCTTCTTTGGATGCTAGACAAGACA |
| Reverse | CGAAGACCCAAGAGTTTGGTTAGTT | |
| PAL | Forward | CATCAAATCTCTCTGGCAGTAGGAA |
| Reverse | AGTTCACATCTTGGTTATGCTGCTC | |
| C4H | Forward | AACAAAGTGAGGGATGAAATTGACA |
| Reverse | GGATTGCCATTCTTAGCCTTAGTGT | |
| 4CL | Forward | TGTCCCTCCTATTGTTTTGGCTATT |
| Reverse | CTTTGGGGAATTTAGCTCTGACAGT | |
| CHR | Forward | AAACAAGGTTACAGGCATTTTGACA |
| Reverse | GGAAGAACGAGATGAGGATGATTTT | |
| CHI | Forward | ATCGAGTTTTTCCACCAGGATCTAC |
| Reverse | ATCATAGTCTCCAACACAGCCTCAG | |
| IFS | Forward | CCTTCACCTATTGGACAAACCTCTT |
| Reverse | CCTGGTATTAAAGGAAGAAGCCTCA | |
| IOMT | Forward | GCACAAAACACAAGATCAAACTTC |
| Reverse | GCATTACGGCCATTGATTG | |
| I3′H | Forward | GGATGTTAAAGAAGCGAAGCAATTT |
| Reverse | ATCAAACAATCTCAACAAAGGCAAA | |
| UCGT | Forward | AGGTTTTGAAGATTTATGCACCA |
| Reverse | TCCTTTCTGAGTTCCAGGACA | |
| 18S | Forward | TGCAGAATCCCGTGAACCATC |
| Reverse | AGGCATCGGGCAACGATATG |
| Height (cm) | Root/Whole Fresh Weight (FW) (%) | Leaf Area (cm2) | Chlorophyll (mg·g−1) | |
|---|---|---|---|---|
| Control | 24.2 ± 0.6 | 19.07 ± 0.31 | 0.292 ± 0.004 | 1.242 ± 0.008 |
| UV-treated | 23.4 ± 0.6 | 20.7 ± 0.67 | 0.230 ± 0.006 | 0.799 ± 0.011 |
| CAT (U·g−1·min−1, FW) | POD (U·g−1·min−1, FW) | APX (U·g−1·min−1, FW) | H2O2 (μmoL·g−1) | |
| Control | 26.25 ± 0.01 | 1800 ± 1 | 0.06 ± 0.001 | 3.59 ± 0.01 |
| UV-treated | 36.75 ± 0.01 | 7050 ± 1 | 0.15 ± 0.003 | 3.79 ± 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liu, J.; Abozeid, A.; Wu, K.-X.; Guo, X.-R.; Mu, L.-Q.; Tang, Z.-H. UV-B Radiation Largely Promoted the Transformation of Primary Metabolites to Phenols in Astragalus mongholicus Seedlings. Biomolecules 2020, 10, 504. https://doi.org/10.3390/biom10040504
Liu Y, Liu J, Abozeid A, Wu K-X, Guo X-R, Mu L-Q, Tang Z-H. UV-B Radiation Largely Promoted the Transformation of Primary Metabolites to Phenols in Astragalus mongholicus Seedlings. Biomolecules. 2020; 10(4):504. https://doi.org/10.3390/biom10040504
Chicago/Turabian StyleLiu, Yang, Jia Liu, Ann Abozeid, Ke-Xin Wu, Xiao-Rui Guo, Li-Qiang Mu, and Zhong-Hua Tang. 2020. "UV-B Radiation Largely Promoted the Transformation of Primary Metabolites to Phenols in Astragalus mongholicus Seedlings" Biomolecules 10, no. 4: 504. https://doi.org/10.3390/biom10040504
APA StyleLiu, Y., Liu, J., Abozeid, A., Wu, K.-X., Guo, X.-R., Mu, L.-Q., & Tang, Z.-H. (2020). UV-B Radiation Largely Promoted the Transformation of Primary Metabolites to Phenols in Astragalus mongholicus Seedlings. Biomolecules, 10(4), 504. https://doi.org/10.3390/biom10040504