Uranyl Binding to Proteins and Structural-Functional Impacts
Abstract
:1. Introduction
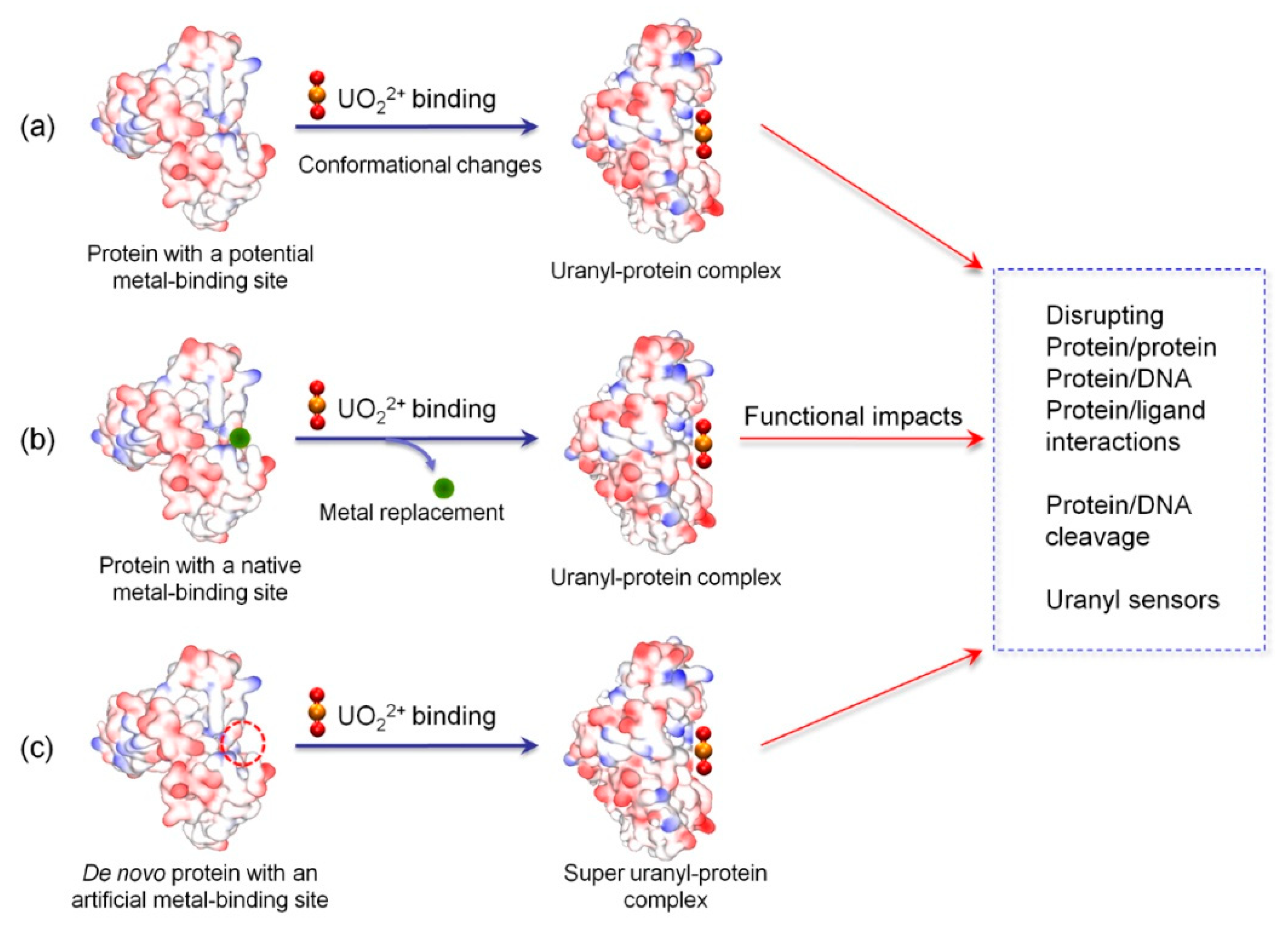
2. Uranyl Binding to Proteins
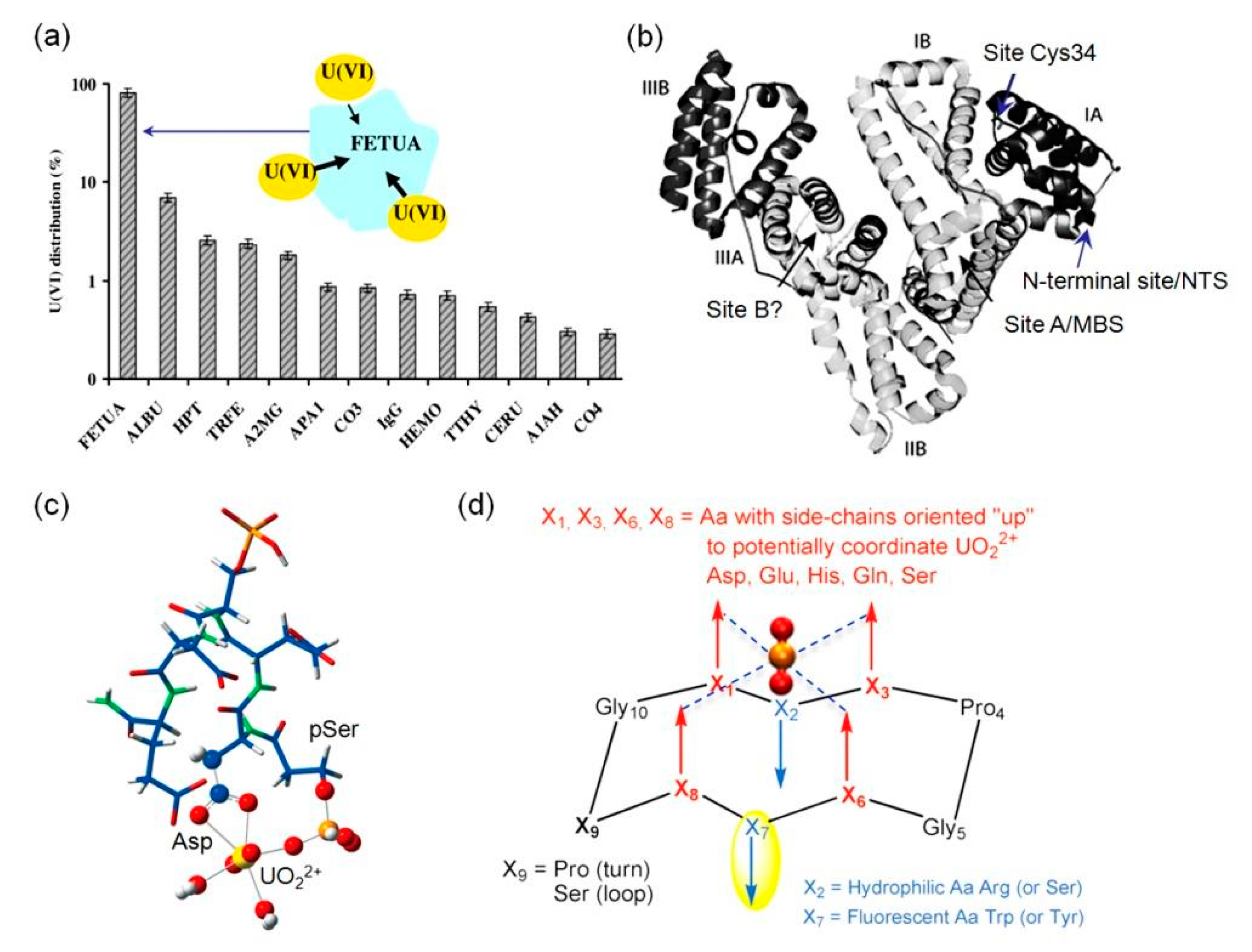
2.1. Binding to Potential Metal-Binding Sites
2.2. Binding to Native Metal-Binding Sites
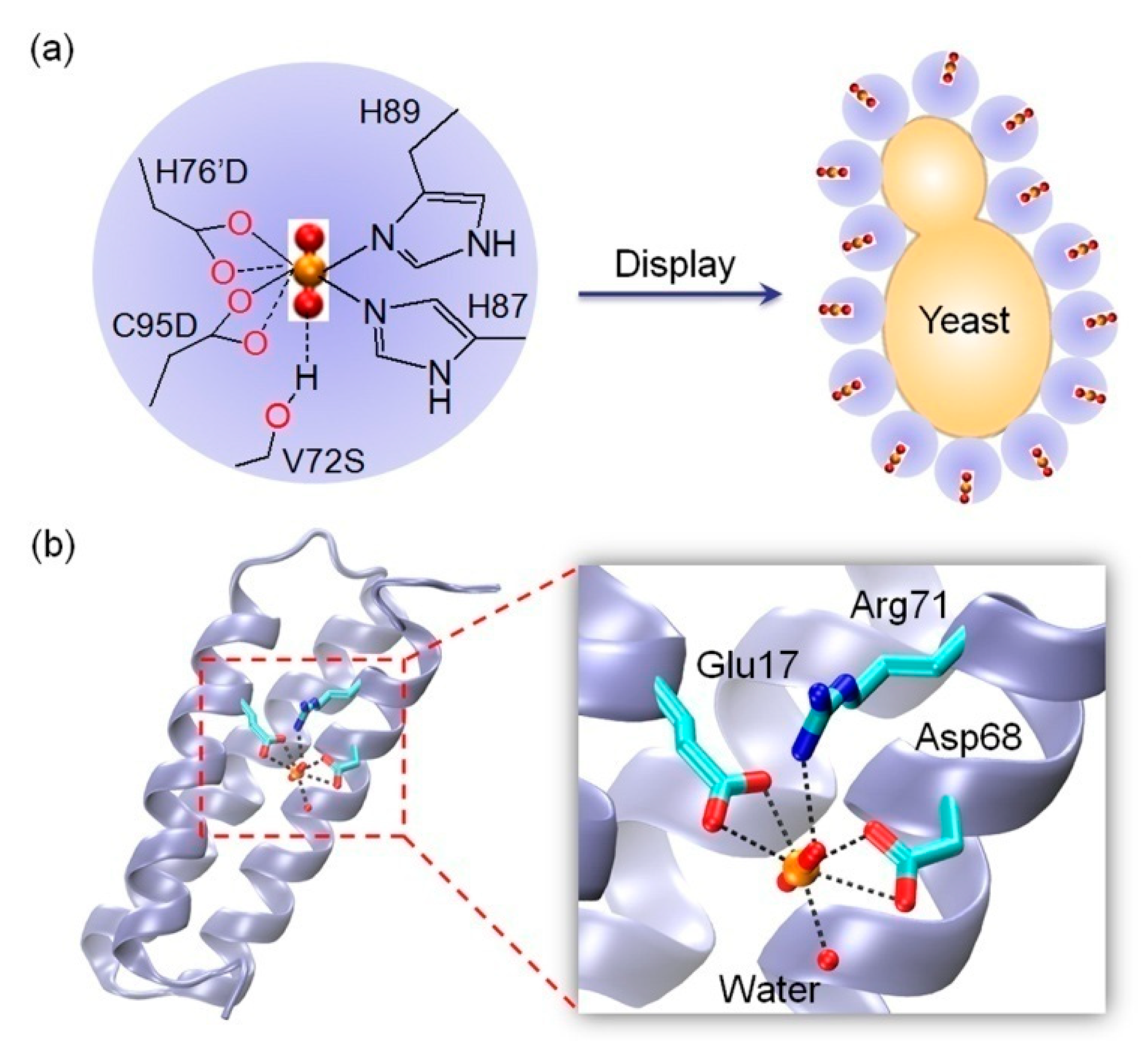
2.3. Binding to Artificial Metal-Binding Sites
3. Structural-Functional Impacts of Uranyl Binding
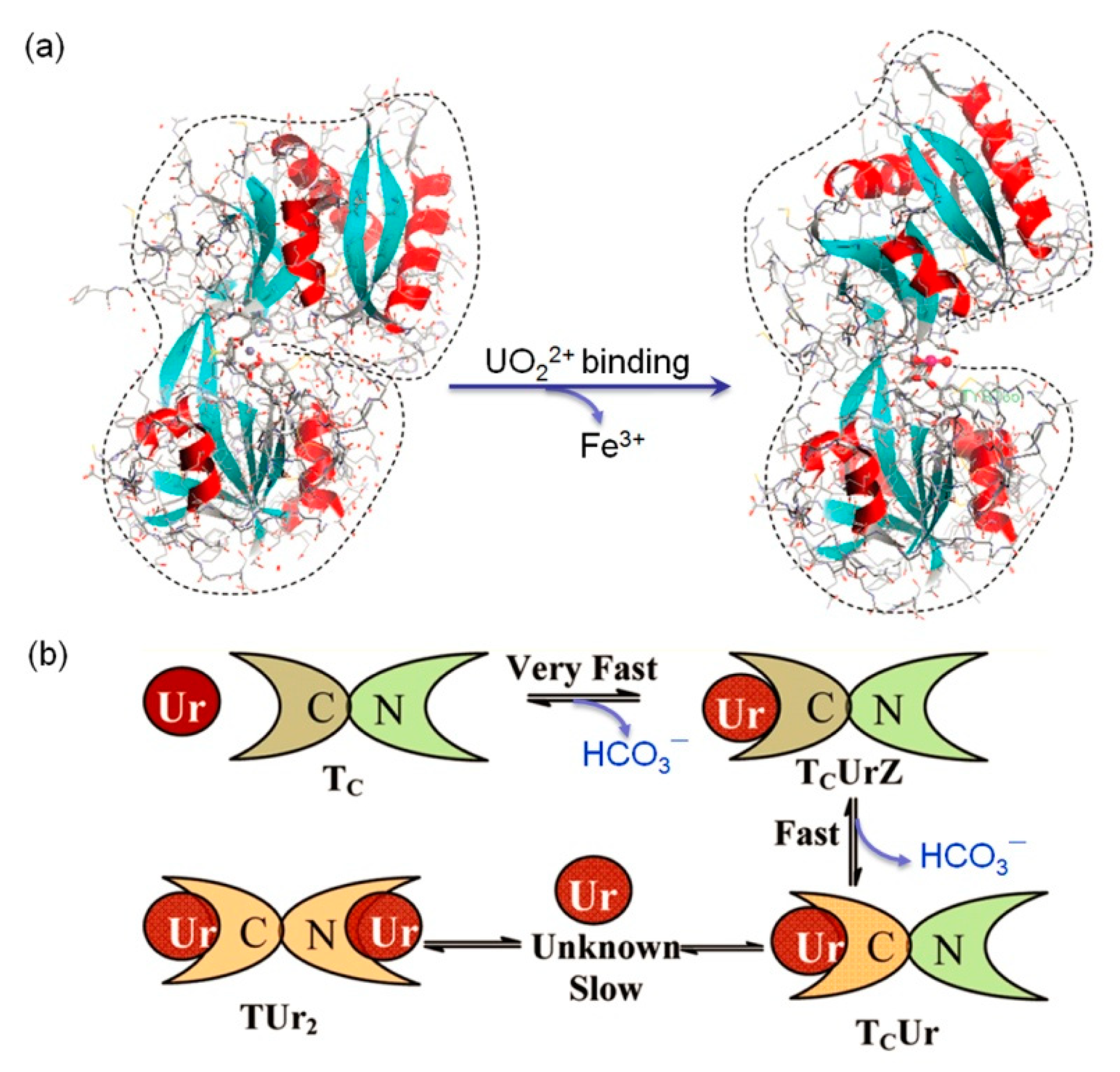
3.1. Inducing Conformational Changes
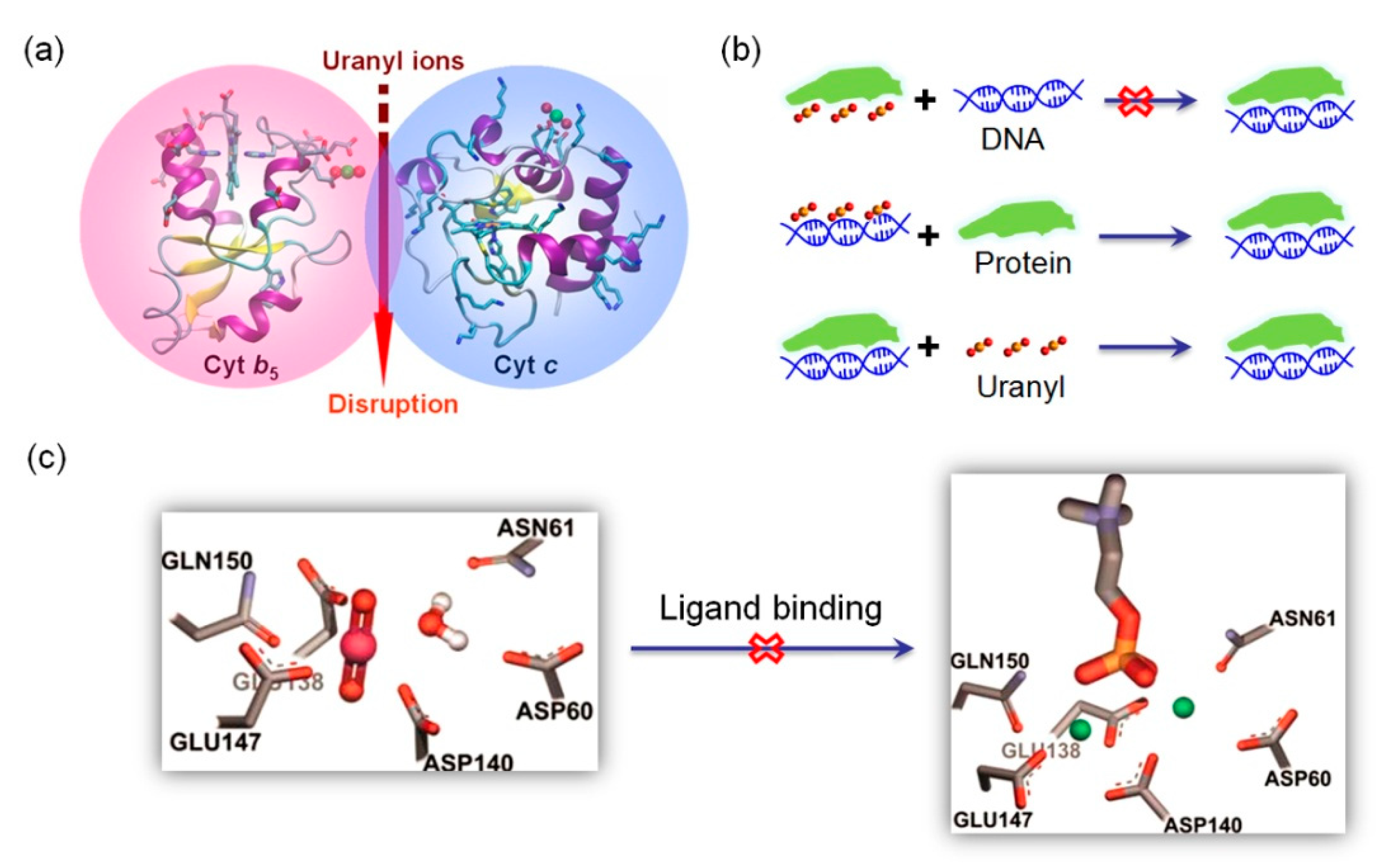
3.2. Disrupting Protein-Protein/DNA/Ligand Interactions
3.3. Protein/DNA Cleavage and Other Impacts
4. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Domingo, J.L. Reproductive and developmental toxicity of natural and depleted uranium: A review. Reprod. Toxicol. 2001, 15, 603–609. [Google Scholar] [CrossRef]
- Craft, E.S.; Abu-Qare, A.W.; Flaherty, M.M.; Garofolo, M.C.; Rincavage, H.L.; Abou-Donia, M.B. Depleted and natural uranium: Chemistry and toxicological effects. J. Toxicol. Environ. Health Part B 2004, 7, 297–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garmash, S.; Smirnova, V.; Karp, O.; Usacheva, A.; Berezhnov, A.; Ivanov, V.; Chernikov, A.; Bruskov, V.; Gudkov, S. Pro-oxidative, genotoxic and cytotoxic properties of uranyl ions. J. Environ. Radioact. 2014, 127, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Hurault, L.; Creff, G.; Hagège, A.; Santucci-Darmanin, S.; Pagnotta, S.; Farlay, D.; Auwer, C.D.; Pierrefite-Carle, V.; Carle, G.F. Uranium Effect on Osteocytic Cells In Vitro. Toxicol. Sci. 2019, 170, 199–209. [Google Scholar] [CrossRef]
- Gao, N.; Huang, Z.; Liu, H.; Hou, J.; Liu, X. Advances on the toxicity of uranium to different organisms. Chemosphere 2019, 237, 124548. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef]
- Van Horn, J.D.; Huang, H. Uranium (VI) bio-coordination chemistry from biochemical, solution and protein structural data. Coord. Chem. Rev. 2006, 250, 765–775. [Google Scholar] [CrossRef]
- Pible, O.; Guilbaud, P.; Pellequer, J.-L.; Vidaud, C.; Quemeneur, E. Structural insights into protein–uranyl interaction: Towards an in silico detection method. Biochimie 2006, 88, 1631–1638. [Google Scholar] [CrossRef]
- Carugo, O. Structural features of uranium-protein complexes. J. Inorg. Biochem. 2018, 189, 1–6. [Google Scholar] [CrossRef]
- Zhou, L.; Bosscher, M.; Zhang, C.; Ozcubukcu, S.; Zhang, L.; Zhang, W.; Li, C.J.; Liu, J.; Jensen, M.; Lai, L.; et al. A protein engineered to bind uranyl selectively and with femtomolar affinity. Nat. Chem. 2014, 6, 236–241. [Google Scholar] [CrossRef]
- Lu, Y.; Yeung, N.; Sieracki, N.; Marshall, N.M. Design of functional metalloproteins. Nature 2009, 460, 855–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef]
- Poulos, T.L. Heme Enzyme Structure and Function. Chem. Rev. 2014, 114, 3919–3962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chakraborty, S.; Hosseinzadeh, P.; Yü, Y.; Tian, S.; Petrík, I.; Bhagi, A.; Lu, Y. Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers. Chem. Rev. 2014, 114, 4366–4469. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W. Rational design of metalloenzymes: From single to multiple active sites. Coord. Chem. Rev. 2017, 336, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Yuan, H.; Liu, C.; He, B.; Gao, S.-Q.; Wen, G.-B.; Tan, X.; Lin, Y.-W. A Rationally Designed Myoglobin Exhibits a Catalytic Dehalogenation Efficiency More than 1000-Fold That of a Native Dehaloperoxidase. ACS Catal. 2018, 8, 9619–9624. [Google Scholar] [CrossRef]
- Zhang, P.; Xu, J.; Wang, X.-J.; He, B.; Gao, S.-Q.; Lin, Y.-W. The Third Generation of Artificial Dye-Decolorizing Peroxidase Rationally Designed in Myoglobin. ACS Catal. 2019, 9, 7888–7893. [Google Scholar] [CrossRef]
- Zhang, P.; Yuan, H.; Xu, J.; Wang, X.-J.; Gao, S.-Q.; Tan, X.; Lin, Y.-W. A Catalytic Binding Site Together with a Distal Tyr in Myoglobin Affords Catalytic Efficiencies Similar to Natural Peroxidases. ACS Catal. 2019, 10, 891–896. [Google Scholar] [CrossRef]
- Götzke, L.; Schaper, G.; März, J.; Kaden, P.; Huittinen, N.; Stumpf, T.; Kammerlander, K.K.; Brunner, E.; Hahn, P.; Mehnert, A.; et al. Coordination chemistry of f-block metal ions with ligands bearing bio-relevant functional groups. Coord. Chem. Rev. 2019, 386, 267–309. [Google Scholar] [CrossRef]
- Creff, G.; Zurita, C.; Jeanson, A.; Carle, G.; Vidaud, C.; Auwer, C.D. What do we know about actinides-proteins interactions? Radiochim. Acta 2019, 107, 993–1009. [Google Scholar] [CrossRef]
- Vidaud, C.; Dedieu, A.; Basset, C.; Plantevin, S.; Dany, I.; Pible, O.; Quéméneur, E. Screening of Human Serum Proteins for Uranium Binding. Chem. Res. Toxicol. 2005, 18, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Basset, C.; Averseng, O.; Ferron, P.-J.; Richaud, N.; Hagège, A.; Pible, O.; Vidaud, C. Revision of the Biodistribution of Uranyl in Serum: Is Fetuin-A the Major Protein Target? Chem. Res. Toxicol. 2013, 26, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.-N.S.; Bourgeois, D.; Basset, C.; Vidaud, C.; Hagège, A. Assessment of CE-ICP/MS hyphenation for the study of uranyl/protein interactions. Electrophoresis 2015, 36, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Bal, W.; Sokołowska, M.; Kurowska, E.; Faller, P. Binding of transition metal ions to albumin: Sites, affinities and rates. Biochim. Biophys. Acta BBA Gen. Subj. 2013, 1830, 5444–5455. [Google Scholar] [CrossRef]
- Montavon, G.; Apostolidis, C.; Bruchertseifer, F.; Repinc, U.; Morgenstern, A. Spectroscopic study of the interaction of U(VI) with transferrin and albumin for speciation of U(VI) under blood serum conditions. J. Inorg. Biochem. 2009, 103, 1609–1616. [Google Scholar] [CrossRef] [Green Version]
- Szyrwiel, L.; Liauchuk, V.; Chavatte, L.; Łobiński, R. In vitro induction and proteomics characterisation of a uranyl–protein interaction network in bovine serum. Metallomics 2015, 7, 1604–1611. [Google Scholar] [CrossRef] [Green Version]
- Bourgeois, D.; Burt-Pichat, B.; Le Goff, X.; Garrevoet, J.; Tack, P.; Falkenberg, G.; Van Hoorebeke, L.; Vincze, L.; Denecke, M.; Meyer, D.; et al. Micro-distribution of uranium in bone after contamination: New insight into its mechanism of accumulation into bone tissue. Anal. Bioanal. Chem. 2015, 407, 6619–6625. [Google Scholar] [CrossRef]
- Safi, S.; Creff, G.; Jeanson, A.; Qi, L.; Basset, C.; Roques, J.; Solari, P.; Simoni, E.; Vidaud, C.; Auwer, C.D. Osteopontin: A Uranium Phosphorylated Binding-Site Characterization. Chem. A Eur. J. 2013, 19, 11261–11269. [Google Scholar] [CrossRef]
- Qi, L.; Basset, C.; Averseng, O.; Quéméneur, E.; Hagège, A.; Vidaud, C. Characterization of UO22+binding to osteopontin, a highly phosphorylated protein: Insights into potential mechanisms of uranyl accumulation in bones. Metallomics 2014, 6, 166–176. [Google Scholar] [CrossRef]
- Huynh, T.-N.S.; Vidaud, C.; Hagège, A. Investigation of uranium interactions with calcium phosphate-binding proteins using ICP/MS and CE-ICP/MS. Metallomics 2016, 8, 1185–1192. [Google Scholar] [CrossRef]
- Lebrun, C.; Starck, M.; Gathu, V.; Chenavier, Y.; Delangle, P. Engineering Short Peptide Sequences for Uranyl Binding. Chem. A Eur. J. 2014, 20, 16566–16573. [Google Scholar] [CrossRef] [PubMed]
- Starck, M.; Sisommay, N.; Laporte, F.A.; Oros, S.; Lebrun, C.; Delangle, P. Preorganized Peptide Scaffolds as Mimics of Phosphorylated Proteins Binding Sites with a High Affinity for Uranyl. Inorg. Chem. 2015, 54, 11557–11562. [Google Scholar] [CrossRef] [PubMed]
- Starck, M.; Laporte, F.A.; Oros, S.; Sisommay, N.; Gathu, V.; Solari, P.; Creff, G.; Roques, J.; Auwer, C.D.; Lebrun, C.; et al. Cyclic Phosphopeptides to Rationalize the Role of Phosphoamino Acids in Uranyl Binding to Biological Targets. Chem. A Eur. J. 2017, 23, 5281–5290. [Google Scholar] [CrossRef] [PubMed]
- Laporte, F.A.; Lebrun, C.; Vidaud, C.; Delangle, P. Phosphate-Rich Biomimetic Peptides Shed Light on High-Affinity Hyperphosphorylated Uranyl Binding Sites in Phosphoproteins. Chem. A Eur. J. 2019, 25, 8570–8578. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-T.; Han, J.; Gu, M.; Liu, J.; Li, Y.; Huang, Z.; Yu, H.; Chu-Ting, Y.; Wang, X. Fluorescent recognition of uranyl ions by a phosphorylated cyclic peptide. Chem. Commun. 2015, 51, 11769–11772. [Google Scholar] [CrossRef]
- Garai, A.; Delangle, P. Recent advances in uranyl binding in proteins thanks to biomimetic peptides. J. Inorg. Biochem. 2020, 203, 110936. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Wang, J. Structure and function of heme proteins in non-native states: A mini-review. J. Inorg. Biochem. 2013, 129, 162–171. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Sawyer, E.; Wang, J. Rational Heme Protein Design: All Roads Lead to Rome. Chem. Asian J. 2013, 8, 2534–2544. [Google Scholar] [CrossRef]
- Lin, Y.-W. Rational design of heme enzymes for biodegradation of pollutants toward a green future. Biotechnol. Appl. Biochem. 2020. [Google Scholar] [CrossRef]
- Kumar, A.; Ali, M.; Ningthoujam, R.; Gaikwad, P.; Kumar, M.; Nath, B.B.; Pandey, B.N. The interaction of actinide and lanthanide ions with hemoglobin and its relevance to human and environmental toxicology. J. Hazard. Mater. 2016, 307, 281–293. [Google Scholar] [CrossRef]
- Chudaev, M.V.; Gilep, A.A.; Usanov, S.A. Site-directed mutagenesis of cytochrome b5 for studies of its interaction with cytochrome P450. Biochem. Mosc. 2001, 66, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-W.; Wang, W.-H.; Zhang, Q.; Lu, H.-J.; Yang, P.-Y.; Xie, Y.; Huang, Z.-X.; Wu, H.-M. Converting Cytochrome b5 into Cytochrome c-Like Protein. ChemBioChem 2005, 6, 1356–1359. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; He, B.; Du, K.; Wang, X.; Gao, S.; Lin, Y.-W. Peroxidase Activity of a c-Type Cytochrome b5 in the Non-Native State is Comparable to that of Native Peroxidases. ChemistryOpen 2017, 6, 325–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, D.; Liao, L.-F.; Zhao, M.-M.; Wu, M.-L.; Wu, Y.-M.; Lin, Y.-W. Interactions of uranyl ion with cytochrome b5 and its His39Ser variant as revealed by molecular simulation in combination with experimental methods. J. Mol. Model. 2011, 18, 1009–1013. [Google Scholar] [CrossRef]
- Sun, M.-H.; Liu, S.-Q.; Du, K.-J.; Nie, C.-M.; Lin, Y.-W. A spectroscopic study of uranyl-cytochrome b5/cytochrome c interactions. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 118, 130–137. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, W.-H.; Wang, Y.-H.; Case, M.; Qian, W.; McLendon, G.; Huang, Z.-X. Mapping the Electron Transfer Interface between Cytochrome b5 and Cytochrome c. Biochemistry 2004, 43, 3527–3536. [Google Scholar] [CrossRef]
- Lin, Y.-W. Rational Design of Artificial Metalloproteins and Metalloenzymes with Metal Clusters. Molecules 2019, 24, 2743. [Google Scholar] [CrossRef] [Green Version]
- Maret, W. Redox biochemistry of mammalian metallothioneins. JBIC J. Biol. Inorg. Chem. 2011, 16, 1079–1086. [Google Scholar] [CrossRef]
- Acharya, C.; Blindauer, C.A. Unexpected Interactions of the Cyanobacterial Metallothionein SmtA with Uranium. Inorg. Chem. 2016, 55, 1505–1515. [Google Scholar] [CrossRef]
- Pickart, C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Arnesano, F.; Belviso, B.D.; Caliandro, R.; Falini, G.; Fermani, S.; Natile, G.; Siliqi, D. Crystallographic Analysis of Metal-Ion Binding to Human Ubiquitin. Chem. A Eur. J. 2010, 17, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Camara-Artigas, A.; Plaza-Garrido, M.; Martinez-Rodriguez, S.; Bacarizo, J. New crystal form of human ubiquitin in the presence of magnesium. Acta Cryst. Sect. F Struct. Biol. Commun. 2016, 72, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-W.; Nie, C.-M.; Liao, L.-F. Insights into Uranyl Ion Binding to Ubiquitin from Molecular Modeling and Dynamics Simulations. Chem. Lett. 2011, 40, 1330–1331. [Google Scholar] [CrossRef]
- Malard, V.; Prat, O.; Darrouzet, E.; Berenguer, F.; Sage, N.; Quéméneur, E. Proteomic analysis of the response of human lung cells to uranium. Proteomics 2005, 5, 4568–4580. [Google Scholar] [CrossRef]
- Lestaevel, P.; Houpert, P.; Bussy, C.; Dhieux, B.; Gourmelon, P.; Paquet, F. The brain is a target organ after acute exposure to depleted uranium. Toxicology 2005, 212, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 2003, 40, 427–446. [Google Scholar] [CrossRef] [Green Version]
- Eom, H.; Song, W.J. Emergence of metal selectivity and promiscuity in metalloenzymes. JBIC J. Biol. Inorg. Chem. 2019, 24, 517–531. [Google Scholar] [CrossRef]
- Mudgal, V.; Madaan, N.; Mudgal, A.; Singh, R.B.; Mishra, S. Effect of Toxic Metals on Human Health. Open Nutraceuticals J. 2010, 3, 94–99. [Google Scholar] [CrossRef] [Green Version]
- Wally, J.; Halbrooks, P.J.; Vonrhein, C.; Rould, M.A.; Everse, S.J.; Mason, A.B.; Buchanan, S.K. The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding. J. Biol. Chem. 2006, 281, 24934–24944. [Google Scholar] [CrossRef] [Green Version]
- MacGillivray, R.T.A.; Moore, S.A.; Chen, J.; Anderson, B.F.; Baker, H.; Luo, Y.; Bewley, M.; Smith, C.A.; Murphy, M.E.P.; Wang, Y.; et al. Two High-Resolution Crystal Structures of the Recombinant N-Lobe of Human Transferrin Reveal a Structural Change Implicated in Iron Release. Biochemistry 1998, 37, 7919–7928. [Google Scholar] [CrossRef]
- Vidaud, C.; Gourion-Arsiquaud, S.; Rollin-Genetet, F.; Torne-Celer, C.; Plantevin, S.; Pible, O.; Berthomieu, C.; Quéméneur, E. Structural Consequences of Binding of UO22+ to Apotransferrin: Can This Protein Account for Entry of Uranium into Human Cells? Biochemistry 2007, 46, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Benavides-Garcia, M.G.; Balasubramanian, K. Structural Insights into the Binding of Uranyl with Human Serum Protein Apotransferrin Structure and Spectra of Protein−Uranyl Interactions. Chem. Res. Toxicol. 2009, 22, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Brunger, A.T. The 1.0 Å crystal structure of Ca2+-bound calmodulin: An analysis of disorder and implications for functionally relevant plasticity. J. Mol. Biol. 2000, 301, 1237–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardoux, R.; Sauge-Merle, S.; Lemaire, D.; Delangle, P.; Guilloreau, L.; Adriano, J.-M.; Berthomieu, C. Modulating Uranium Binding Affinity in Engineered Calmodulin EF-Hand Peptides: Effect of Phosphorylation. PLoS ONE 2012, 7, 41922. [Google Scholar] [CrossRef] [Green Version]
- Brulfert, F.; Safi, S.; Jeanson, A.; Martinez-Baez, E.; Roques, J.; Berthomieu, C.; Solari, P.-L.; Sauge-Merle, S. Simoni, Éric Structural Environment and Stability of the Complexes Formed Between Calmodulin and Actinyl Ions. Inorg. Chem. 2016, 55, 2728–2736. [Google Scholar] [CrossRef]
- Sauge-Merle, S.; Brulfert, F.; Pardoux, R.; Solari, P.L.; Lemaire, D.; Safi, S.; Guilbaud, P.; Simoni, E.; Merroun, M.L.; Berthomieu, C. Structural Analysis of Uranyl Complexation by the EF-Hand Motif of Calmodulin: Effect of Phosphorylation. Chem. A Eur. J. 2017, 23, 15505–15517. [Google Scholar] [CrossRef] [Green Version]
- Wegner, S.V.; Boyaci, H.; Chen, H.; Jensen, M.P.; He, C. Engineering a uranyl-specific binding protein from NikR. Angew. Chem. Int. Ed. Engl. 2009, 48, 2339–2341. [Google Scholar] [CrossRef]
- Kuroda, K.; Ebisutani, K.; Iida, K.; Nishitani, T.; Ueda, M. Enhanced Adsorption and Recovery of Uranyl Ions by NikR Mutant-Displaying Yeast. Biomolecules 2014, 4, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Stellato, C.C.; Lai, R.Y. Engineering uranyl-chelating peptides from NikR for electrochemical peptide-based sensing applications. J. Electroanal. Chem. 2020, 858, 113698. [Google Scholar] [CrossRef]
- Weinhold, B. Unknown Quantity: Regulating Radionuclides in Tap Water. Environ. Health Perspect. 2012, 120, 350–356. [Google Scholar] [CrossRef] [Green Version]
- Kou, S.; Yang, Z.; Sun, F. Protein Hydrogel Microbeads for Selective Uranium Mining from Seawater. ACS Appl. Mater. Interfaces 2017, 9, 2035–2039. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wei, J.; Wang, Y.; Yang, C.; Zhao, S.; Li, C.; Dong, Y.; Bai, K.; Li, Y.; Teng, H.; et al. A Genetically Encoded Protein Polymer for Uranyl Binding and Extraction Based on the SpyTag–SpyCatcher Chemistry. ACS Synth. Biol. 2018, 7, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Yu, Q.; Wen, J.; Li, C.; Guo, Z.; Wang, X.; Wang, N. Ultrafast and Highly Selective Uranium Extraction from Seawater by Hydrogel-like Spidroin-based Protein Fiber. Angew. Chem. Int. Ed. 2019, 58, 11785–11790. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, J.S.; Lindquist, S. Mechanisms of protein-folding diseases at a glance. Dis. Model. Mech. 2014, 7, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.E.; Møllegaard, N.E.; Jeppesen, C. DNA Conformational analysis in solution by uranyl mediated photocleavage. Nucleic Acids Res. 1990, 18, 3847–3851. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Feng, Y.; Wang, Y.; Wang, L.; Shi, W.-Q. Interactions between U(VI) and bovine serum albumin. J. Radioanal. Nucl. Chem. 2013, 298, 903–908. [Google Scholar] [CrossRef]
- Hoarau, M.; Koebke, K.J.; Chen, Z.; Marsh, E.N.G. Probing Metal Ion Discrimination in a Protein Designed to Bind Uranyl Cation With Femtomolar Affinity. Front. Mol. Biosci. 2019, 6, 73. [Google Scholar] [CrossRef] [Green Version]
- Dautry-Varsat, A.; Ciechanover, A.; Lodish, H.F. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA 1983, 80, 2258–2262. [Google Scholar] [CrossRef] [Green Version]
- Hémadi, M.; Ha-Duong, N.-T.; Chahine, J.-M.E.H. Can Uranium Be Transported by the Iron-Acquisition Pathway? Ur Uptake by Transferrin. J. Phys. Chem. B 2011, 115, 4206–4215. [Google Scholar]
- Hémadi, M.; Ha-Duong, N.-T.; Plantevin, S.; Vidaud, C.; Chahine, J.-M.E.H. Can uranium follow the iron-acquisition pathway? Interaction of uranyl-loaded transferrin with receptor 1. JBIC J. Biol. Inorg. Chem. 2009, 15, 497–504. [Google Scholar] [CrossRef]
- Periyakaruppan, A.; Sarkar, S.; Ravichandran, P.; Sadanandan, B.; Sharma, C.S.; Ramesh, V.; Hall, J.C.; Thomas, R.; Wilson, B.L.; Ramesh, G.T. Uranium induces apoptosis in lung epithelial cells. Arch. Toxicol. 2008, 83, 595–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Du, K.-J.; Fang, Z.; You, Y.; Wen, G.-B.; Lin, Y.-W. Chemical and biological insights into uranium-induced apoptosis of rat hepatic cell line. Radiat. Environ. Biophys. 2015, 54, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A. Zinc Finger Proteins as Potential Targets for Toxic Metal Ions: Differential Effects on Structure and Function. Antioxid. Redox Signal. 2001, 3, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Hartsock, W.J.; Cohen, J.D.; Segal, D.J. Uranyl Acetate as a Direct Inhibitor of DNA-Binding Proteins. Chem. Res. Toxicol. 2007, 20, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Kushner, I.; Samols, D. C-reactive Protein. J. Biol. Chem. 2004, 279, 48487–48490. [Google Scholar] [CrossRef] [Green Version]
- Vilahur, G.; Badimon, L. Biological actions of pentraxins. Vasc. Pharm. 2015, 73, 38–44. [Google Scholar] [CrossRef]
- Pible, O.; Vidaud, C.; Plantevin, S.; Pellequer, J.-L.; Quéméneur, E. Predicting the disruption by UO2 2+ of a protein-ligand interaction. Protein Sci. 2010, 19, 2219–2230. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Lin, Y.-W.; Yu, D.-H. Uranyl photocatalysis: Precisely controlled oxidation of sulfides with ground-state oxygen. Sci. China Ser. B Chem. 2020, 63, 291–293. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Jeppesen, C.; Buchardt, O. Uranyl salts as photochemical agents for cleavage of DNA and probing of protein-DNA contacts. Febs Lett. 1988, 235, 122–124. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, L.H.; Nielsen, P.E.; Jørgensen, C.I.; Kragelund, B.B.; Møllegaard, N.E. Phosphate Selective Uranyl Photo-Affinity Cleavage of Proteins. Determination of Phosphorylation Sites. ChemBioChem 2008, 9, 2377–2381. [Google Scholar] [CrossRef]
- Duff, M.R., Jr.; Kumar, C.V. Site-selective photocleavage of proteins by uranyl ions. Angew. Chem. Int. Ed. Engl. 2005, 45, 137–139. [Google Scholar] [CrossRef]
- Zhang, Q.; Jørgensen, T.J.; Nielsen, P.E.; Møllegaard, N.E. A Phosphorylation Tag for Uranyl Mediated Protein Purification and Photo Assisted Tag Removal. PLoS ONE 2014, 9, 91138. [Google Scholar] [CrossRef]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, N.M.; Istok, J.D.; Watson, D.; Lu, Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.K.; Liu, J.; He, Y.; Lu, Y. Biochemical Characterization of a Uranyl Ion-Specific DNAzyme. ChemBioChem 2009, 10, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Wang, Z.; Liu, J.; Lu, Y. Highly Sensitive and Selective Colorimetric Sensors for Uranyl (UO22+): Development and Comparison of Labeled and Label-Free DNAzyme-Gold Nanoparticle Systems. J. Am. Chem. Soc. 2008, 130, 14217–14226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Huang, Q.; Mao, Y.; Wang, X.; Wang, Y.; Hu, Q.; Wang, H.; Wang, X. Sensors for determination of uranium: A review. Trac. Trends Anal. Chem. 2019, 118, 89–111. [Google Scholar] [CrossRef]
- Carriere, M.; Proux, O.; Milgram, S.; Thiebault, C.; Avoscan, L.; Barré, N.; Auwer, C.D.; Gouget, B. Transmission electron microscopic and X-ray absorption fine structure spectroscopic investigation of U repartition and speciation after accumulation in renal cells. JBIC J. Biol. Inorg. Chem. 2008, 13, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-F.; Chen, Y.; Song, L.-P.; Fang, Z.; Zhang, J.; Shi, F.; Lin, Y.-W.; Sun, Y.; Zhang, Y.-B.; Rocha, J. Cooperative Capture of Uranyl Ions by a Carbonyl-Bearing Hierarchical-Porous Cu-Organic Framework. Angew. Chem. Int. Ed. 2019, 58, 18808–18812. [Google Scholar] [CrossRef]
- Koohi-Moghadam, M.; Wang, H.; Wang, Y.; Yang, X.; Li, H.; Wang, J.; Sun, H. Predicting disease-associated mutation of metal-binding sites in proteins using a deep learning approach. Nat. Mach. Intell. 2019, 1, 561–567. [Google Scholar] [CrossRef]


| Protein/Peptide | Binding Site | Affinity (KD) | Structural and Functional Impacts | Refs. |
|---|---|---|---|---|
| Fetuin-A | 3 binding sites | ~30 nM–10 µM | The secondary structure was slightly modified upon binding of 3 eq. of UO22+ | [22] |
| HSA | 4 potential binding sites | 1.7 µM | Conformational changes for the secondary structure | [22,24] |
| Cyclic peptides | 1-2 binding sites | logK = 8.2~11.3 | Design of uranyl sensors | [35,36] |
| Phosphorylated OPN | 9 binding sites | 3.6 nM | Structural rearrangements | [29] |
| Cyt b5 | 1 binding site E37, E43, Wat | 10 µM | Slight conformational alterations of both the heme-binding domain and the hydrophobic core. | [44] |
| Cyt c | 1 binding site E66, E69, Wat | 87 µM | Induces conformational changes and decreases the peroxidase activity | [45] |
| Cyt b5- Cyt c complex | Not determined | 30 µM | Interference of the interactions between Cyt b5 and Cyt c | [45] |
| SmtA | 1 binding site E34, D38, Water | ~10-10 M | Very minor adjustments of either backbone or side chains | [49] |
| Ub | 1-binding site E18, D21, Water | Not determined | Slight conformational changes and a different dynamic property | [53] |
| Tf | 2 binding sites D63, Y95, Y188 | 2.8 µM | Large conformational changes and interference of the protein-receptor interactions | [22,61] |
| CaM | 4 binding sites D1, D3, D5, E12 | 32 nM (pH 6) | Conformational changes for the EF-hand binding motif | [64] |
| Engineered CaM | 4 binding sites D1, D3, D5, Y7, pThr9, E12 | 5 nM (pH 6) 0.25 nM (pH 7) | Conformational changes for the EF-hand binding motif | [64] |
| CRP | 1 binding site E138, D140, E147 | 0.68 µM | Disrupts ligand binding to the protein | [87] |
| Engineered NikR | 1 binding site D76, D95, H87, H89, S72 (H-bond) | 53 nM | High uranyl selectivity; DNA-binding ability | [67] |
| De novo SUP | 1 binding site E17, D68, Water, R71 (H-bond) | 7.4 fM | Conformational changes for the secondary structure | [10,77] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-W. Uranyl Binding to Proteins and Structural-Functional Impacts. Biomolecules 2020, 10, 457. https://doi.org/10.3390/biom10030457
Lin Y-W. Uranyl Binding to Proteins and Structural-Functional Impacts. Biomolecules. 2020; 10(3):457. https://doi.org/10.3390/biom10030457
Chicago/Turabian StyleLin, Ying-Wu. 2020. "Uranyl Binding to Proteins and Structural-Functional Impacts" Biomolecules 10, no. 3: 457. https://doi.org/10.3390/biom10030457
APA StyleLin, Y.-W. (2020). Uranyl Binding to Proteins and Structural-Functional Impacts. Biomolecules, 10(3), 457. https://doi.org/10.3390/biom10030457




