Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Description
2.2. Sample Collection and Preparation
2.3. LC–MS/MS Analysis
2.4. Protein Identification and Data Processing
2.5. Quantitative Label-Free Proteomic Analysis
2.6. Western Blot Analysis
2.7. Bioinformatic Analysis
3. Results
3.1. SDS–PAGE and Protein Identification in Brain Samples from MPS IIIB and WT Mice
3.2. Quantitative Analysis of Differentially Expressed Proteins in Brain Samples from MPS IIIB and WT Mice
3.3. Bioinformatic Analysis of Differentially Abundant Proteins
4. Discussion
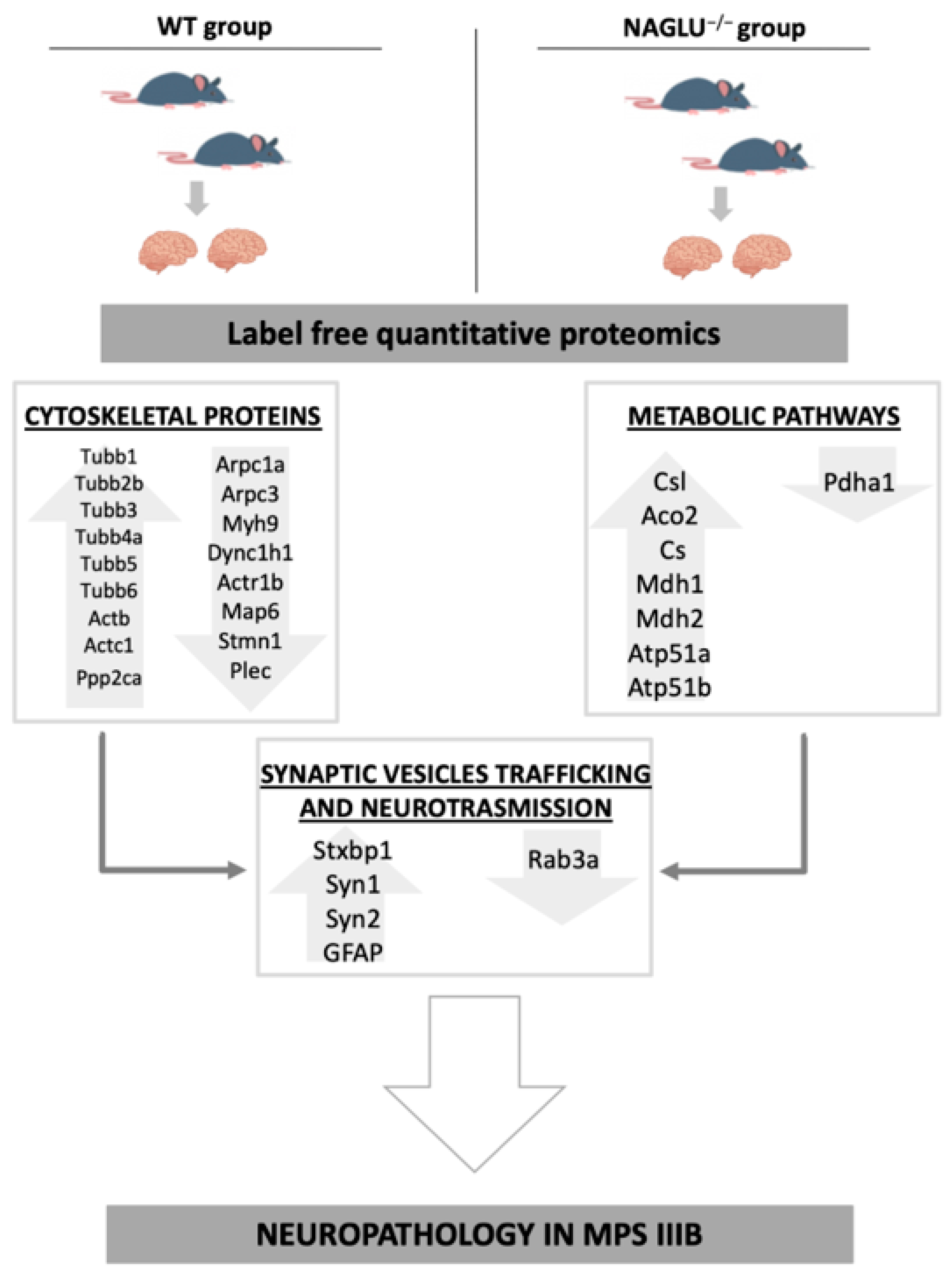
4.1. Deregulated Proteins Involved in Cytoskeleton Organization
4.2. Deregulated Proteins Involved in Metabolic Pathways
4.3. Deregulated Proteins Involved in Synaptic Vesicle Trafficking and Neurotransmission
4.4. Other Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Neufeld, E.F.; Muenzer, J. The mucopolysaccharidoses. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 2001; pp. 3421–3452. [Google Scholar]
- De Pasquale, V.; Pavone, L.M. Heparan sulfate proteoglycans: The sweet side of development turns sour in mucopolysaccharidoses. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 165539. [Google Scholar] [CrossRef] [PubMed]
- Gilkes, J.A.; Heldermon, C.D. Mucopolysaccharidosis III (Sanfilippo Syndrome) disease presentation and experimental therapies. Pediatr. Endocrinol. Rev. 2014, 12, 133–140. [Google Scholar] [PubMed]
- De Pasquale, V.; Sarogni, P.; Pistorio, V.; Cerulo, G.; Paladino, S.; Pavone, L.M. Targeting Heparan Sulfate Proteoglycans as a Novel Therapeutic Strategy for Mucopolysaccharidoses. Mol. Ther. Methods Clin. Dev. 2018, 10, 8–16. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, V.; Pezone, A.; Sarogni, P.; Tramontano, A.; Schiattarella, G.G.; Avvedimento, V.E.; Paladino, S.; Pavone, L.M. EGFR activation triggers cellular hypertrophy and lysosomal disease in NAGLU-depleted cardiomyoblasts; mimicking the hallmarks of mucopolysaccharidosis IIIB. Cell Death Dis. 2018, 9, 40. [Google Scholar] [CrossRef]
- Shapiro, E.G.; Jones, S.A.; Escolar, M.L. Developmental and behavioral aspects of mucopolysaccharidoses with brain manifestations—Neurological signs and symptoms. Mol. Genet. Metab. 2017, 122, 1–7. [Google Scholar] [CrossRef]
- Whitley, C.B.; Cleary, M.; Eugen Mengel, K.; Harmatz, P.; Shapiro, E.; Nestrasil, I.; Haslett, P.; Whiteman, D.; Alexanderian, D. Observational prospective natural history of patients with Sanfilippo syndrome type B. J. Pediatr. 2018, 197, 198–206. [Google Scholar] [CrossRef]
- Heldermon, C.D.; Hennig, A.K.; Ohlemiller, K.K.; Ogilvie, J.M.; Herzog, E.D.; Breidenbach, A.; Vogler, C.; Wozniak, D.F.; Sands, M.S. Development of sensory, motor and behavioral deficits in the murine model of Sanfilippo syndrome type B. PLoS ONE 2007, 2, 772. [Google Scholar] [CrossRef]
- Bigger, B.W.; Begley, D.J.; Virgintino, D.; Pshezhetsky, A.V. Anatomical changes and pathophysiology of the brain in mucopolysaccharidosis disorders. Mol. Genet. Metab. 2018, 125, 322–331. [Google Scholar] [CrossRef]
- Zafeiriou, D.I.; Savvopoulou-Augoustidou, P.A.; Sewell, A.; Papadopoulou, F.; Badouraki, M.; Vargiami, E.; Gombakis, N.P.; Katzos, G.S. Serial magnetic resonance imaging findings in mucopolysaccharidosis IIIB (Sanfilippo’s syndrome B). Brain Dev. 2001, 23, 385–389. [Google Scholar] [CrossRef]
- Barone, R.; Nigro, F.; Triulzi, F.; Musumeci, S.; Fiumara, A.; Pavone, L. Clinical and neuroradiological follow-up in mucopolysaccharidosis type III (Sanfilippo syndrome). Neuropediatrics 1999, 30, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Cusi, V.; Pineda, M.; Galofre, E.; Vila, J. Focal dendritic swellings in Purkinje cells in mucopolysaccharidoses types I, II and III. A Golgi and ultrastructural study. Neuropathol. Appl. Neurobiol. 1988, 14, 315–323. [Google Scholar] [CrossRef]
- Ellinwood, N.M.; Wang, P.; Skeen, T.; Sharp, N.J.; Cesta, M.; Decker, S.; Edwards, N.J.; Bublot, I.; Thompson, J.N.; Bush, W.; et al. A model of mucopolysaccharidosis IIIB (Sanfilippo syndrome type IIIB): N-acetyl-alpha-D-glucosaminidase deficiency in Schipperke dogs. J. Inherit. Metab. Dis. 2003, 26, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Lavery, M.A.; Green, W.R.; Jabs, E.W.; Luckenbach, M.W.; Cox, J.L. Ocular histopathology and ultrastructure of Sanfilippo’s syndrome, type III-B. Arch. Ophthalmol. 1983, 101, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- McCarty, D.M.; DiRosario, J.; Gulaid, K.; Killedar, S.; Oosterhof, A.; van Kuppevelt, T.H.; Martin, P.T.; Fu, H. Differential distribution of heparan sulfate glycoforms and elevated expression of heparan sulfate biosynthetic enzyme genes in the brain of mucopolysaccharidosis IIIB mice. Metab. Brain Dis. 2011, 26, 9–19. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, R.; Dobrenis, K.; Walkley, S.U. Differential subcellular localization of cholesterol, gangliosides, and glycosaminoglycans in murine models of mucopolysaccharide storage disorders. J. Comp. Neurol. 2004, 480, 415–426. [Google Scholar] [CrossRef]
- Hara, A.; Kitazawa, N.; Taketomi, T. Abnormalities of glycosphingolipids in mucopolysaccharidosis type III B. J. Lipid Res. 1984, 25, 175–184. [Google Scholar]
- Ohmi, K.; Greenberg, D.S.; Rajavel, K.S.; Ryazantsev, S.; Li, H.H.; Neufeld, E.F. Activated microglia in cortex of mouse models of mucopolysaccharidoses I and IIIB. Proc. Natl. Acad. Sci. USA 2003, 100, 1902–1907. [Google Scholar] [CrossRef]
- Fu, H.; Bartz, J.D.; Stephens, R.L., Jr.; McCarty, D.M. Peripheral nervous system neuropathology and progressive sensory impairments in a mouse model of Mucopolysaccharidosis IIIB. PLoS ONE 2012, 7, 45992. [Google Scholar] [CrossRef]
- Ausseil, J.; Desmaris, N.; Bigou, S.; Attali, R.; Corbineau, S.; Vitry, S.; Parent, M.; Cheillan, D.; Fuller, M.; Maire, I.; et al. Early neurodegeneration progresses independently of microglial activation by heparan sulfate in the brain of mucopolysaccharidosis IIIB mice. PLoS ONE 2008, 3, 2296. [Google Scholar] [CrossRef]
- Li, H.H.; Zhao, H.Z.; Neufeld, E.F.; Cai, Y.; Gómez-Pinilla, F. Attenuated plasticity in neurons and astrocytes in the mouse model of Sanfilippo syndrome type B. J. Neurosci. Res. 2002, 69, 30–38. [Google Scholar] [CrossRef]
- Caterino, M.; Corbo, C.; Imperlini, E.; Armiraglio, M.; Pavesi, E.; Aspesi, A.; Loreni, F.; Dianzani, I.; Ruoppolo, M. Differential proteomic analysis in human cells subjected to ribosomal stress. Proteomics 2013, 13, 1220–1227. [Google Scholar] [CrossRef]
- Imperlini, E.; Santorelli, L.; Orrù, S.; Scolamiero, E.; Ruoppolo, M.; Caterino, M. Mass spectrometry-based metabolomic and proteomic strategies in organic acidemias. BioMed Res. Int. 2016, 2016, 9210408. [Google Scholar] [CrossRef] [PubMed]
- Barbarani, G.; Ronchi, A.; Ruoppolo, M.; Santorelli, L.; Steinfelder, R.; Elangovan, S.; Fugazza, C.; Caterino, M. Unravelling pathways downstream Sox6 induction in K562 erythroid cells by proteomic analysis. Sci. Rep. 2017, 7, 14088. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Cevenini, A.; Marchese, E.; Imperlini, E.; Raia, M.; Del Vecchio, L.; Caterino, M.; Ruoppolo, M. Label-free quantitative proteomics in a methylmalonyl-CoA mutase-silenced neuroblastoma cell line. Int. J. Mol. Sci. 2018, 19, 3580. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, 183–189. [Google Scholar] [CrossRef]
- Armentano, M.F.; Caterino, M.; Miglionico, R.; Ostuni, A.; Pace, M.C.; Cozzolino, F.; Monti, M.; Milella, L.; Carmosino, M.; Pucci, P.; et al. New insights on the functional role of URG7 in the cellular response to ER stress. Biol. Cell 2018, 110, 147–158. [Google Scholar] [CrossRef]
- Bianchi, L.; Gagliardi, A.; Landi, C.; Focarelli, R.; De Leo, V.; Luddi, A.; Bini, L.; Piomboni, P. Protein pathways working in human follicular fluid: The future for tailored IVF? Expert Rev. Mol. Med. 2016, 8, 9. [Google Scholar] [CrossRef]
- Li, H.H.; Yu, W.H.; Rozengurt, N.; Zhao, H.Z.; Lyons, K.M.; Anagnostaras, S.; Fanselow, M.S.; Suzuki, K.; Vanier, M.T.; Neufeld, E.F. Mouse model of Sanfilippo syndrome type B produced by targeted disruption of the gene encoding alpha-N-acetylglucosaminidase. Proc. Natl. Acad. Sci. USA 1999, 96, 14505–14510. [Google Scholar] [CrossRef]
- Schiattarella, G.G.; Cerulo, G.; De Pasquale, V.; Cocchiaro, P.; Paciello, O.; Avallone, L.; Belfiore, M.P.; Iacobellis, F.; Di Napoli, D.; Magliulo, F.; et al. The Murine Model of Mucopolysaccharidosis IIIB Develops Cardiopathies over Time Leading to Heart Failure. PLoS ONE 2015, 10, e0131662. [Google Scholar] [CrossRef]
- Pavone, L.M.; Rea, S.; Trapani, F.; De Pasquale, V.; Tafuri, S.; Papparella, S.; Paciello, O. Role of serotonergic system in the pathogenesis of fibrosis in canine idiopathic inflammatory myopathies. Neuromuscul. Disord. 2012, 22, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Spina, A.; Rea, S.; De Pasquale, V.; Mastellone, V.; Avallone, L.; Pavone, L.M. Fate map of serotonin transporter-expressing cells in developing mouse thyroid. Anat. Rec. (Hoboken) 2011, 294, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Cerulo, G.; Tafuri, S.; De Pasquale, V.; Rea, S.; Romano, S.; Costagliola, A.; Della Morte, R.; Avallone, L.; Pavone, L.M. Serotonin activates cell survival and apoptotic death responses in cultured epithelial thyroid cells. Biochimie 2014, 105, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Colavita, I.; Esposito, N.; Martinelli, R.; Catanzano, F.; Melo, J.V.; Pane, F.; Ruoppolo, M.; Salvatore, F. Gaining insights into the Bcr-Abl activity-independent mechanisms of resistance to imatinib mesylate in KCL22 cells: A comparative proteomic approach. Biochim. Biophys. Acta 2010, 1804, 1974–1987. [Google Scholar] [CrossRef]
- Caterino, M.; Chandler, R.J.; Sloan, J.L.; Dorko, K.; Cusmano-Ozog, K.; Ingenito, L.; Strom, S.C.; Imperlini, E.; Scolamiero, E.; Venditti, C.P.; et al. The proteome of methylmalonic acidemia (MMA): The elucidation of altered pathways in patient livers. Mol. Biosyst. 2016, 12, 566–574. [Google Scholar] [CrossRef]
- Caterino, M.; Pastore, A.; Strozziero, M.G.; Di Giovamberardino, G.; Imperlini, E.; Scolamiero, E.; Ingenito, L.; Boenzi, S.; Ceravolo, F.; Martinelli, D.; et al. The proteome of cblC defect: In vivo elucidation of altered cellular pathways in humans. J. Inherit. Metab. Dis. 2015, 38, 969–979. [Google Scholar] [CrossRef]
- Caterino, M.; Zacchia, M.; Costanzo, M.; Bruno, G.; Arcaniolo, D.; Trepiccione, F.; Siciliano, R.A.; Mazzeo, M.F.; Ruoppolo, M.; Capasso, G. Urine proteomics revealed a significant correlation between urine-fibronectin abundance and estimated-GFR decline in patients with Bardet-Biedl Syndrome. Kidney Blood Press. Res. 2018, 43, 389–405. [Google Scholar] [CrossRef]
- Alberio, T.; Pieroni, L.; Ronci, M.; Banfi, C.; Bongarzone, I.; Bottoni, P.; Brioschi, M.; Caterino, M.; Chinello, C.; Cormio, A.; et al. Toward the Standardization of Mitochondrial Proteomics: The Italian Mitochondrial Human Proteome Project Initiative. J. Proteome Res. 2017, 16, 4319–4329. [Google Scholar] [CrossRef]
- Caterino, M.; Aspesi, A.; Pavesi, E.; Imperlini, E.; Pagnozzi, D.; Ingenito, L.; Santoro, C.; Dianzani, I.; Ruoppolo, M. Analysis of the interactome of ribosomal protein S19 mutants. Proteomics 2014, 14, 2286–2296. [Google Scholar] [CrossRef]
- Capobianco, V.; Caterino, M.; Iaffaldano, L.; Nardelli, C.; Sirico, A.; Del Vecchio, L.; Martinelli, P.; Pastore, L.; Pucci, P.; Sacchetti, L. Proteome analysis of human amniotic mesenchymal stem cells (hA-MSCs) reveals impaired antioxidant ability, cytoskeleton and metabolic functionality in maternal obesity. Sci. Rep. 2016, 6, 25270. [Google Scholar] [CrossRef]
- Di Pasquale, P.; Caterino, M.; Di Somma, A.; Squillace, M.; Rossi, E.; Landini, P.; Iebba, V.; Schippa, S.; Papa, R.; Selan, L.; et al. Exposure of E. coli to DNA-methylating agents impairs biofilm formation and invasion of eukaryotic cells via down regulation of the N-Acetylneuraminate Lyase NanA. Front. Microbiol. 2016, 7, 147. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Cevenini, A.; Salvatore, F. Biomarker discovery by proteomics-based approaches for early detection and personalized medicine in colorectal cancer. Proteom. Clin. Appl. 2017, 11, 5–6. [Google Scholar]
- Croisé, P.; Estay-Ahumada, C.; Gasman, S.; Ory, S. Rho GTPases; phosphoinositides, and actin: A tripartite framework for efficient vesicular trafficking. Small GTPases 2014, 5, 29469. [Google Scholar] [CrossRef] [PubMed]
- Vemu, A.; Atherton, J.; Spector, J.O.; Moores, C.A.; Roll-Mecak, A. Tubulin isoform composition tunes microtubule dynamics. Mol. Biol. Cell 2017, 28, 3564–3572. [Google Scholar] [CrossRef] [PubMed]
- Tischfield, M.A.; Engle, E.C. Distinct alpha- and beta-tubulin isotypes are required for the positioning, differentiation and survival of neurons: New support for the ‘multi-tubulin’ hypothesis. Biosci. Rep. 2010, 30, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Engle, E.C. Human genetic disorders of axon guidance. Cold Spring Harb. Perspect. Biol. 2010, 2, a001784. [Google Scholar] [CrossRef]
- Romaniello, R.; Arrigoni, F.; Fry, A.E.; Bassi, M.T.; Rees, M.I.; Borgatti, R.; Pilz, D.T.; Cushion, T.D. Tubulin genes and malformations of cortical development. Eur. J. Med. Genet. 2018, 61, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.; Przybilla, M.J.; Whitley, C.B. Proteomic analysis of mucopolysaccharidosis I mouse brain with two-dimensional polyacrylamide gel electrophoresis. Mol. Genet. Metab. 2017, 120, 101–110. [Google Scholar] [CrossRef][Green Version]
- Cheon, M.S.; Fountoulakis, M.; Cairns, N.J.; Dierssen, M.; Herkner, K.; Lubec, G. Decreased protein levels of stathmin in adult brains with Down syndrome and Alzheimer’s disease. J. Neural Transm. Suppl. 2001, 61, 281–288. [Google Scholar]
- Howell, B.; Larsson, N.; Gullberg, M.; Cassimeris, L. Dissociation of the tubulin-sequestering and microtubule catastrophe-promoting activities of oncoprotein 18/stathmin. Mol. Biol. Cell 1999, 10, 105–118. [Google Scholar] [CrossRef]
- Ohkawa, N.; Fujitani, K.; Tokunaga, E.; Furuya, S.; Inokuchi, K. The microtubule destabilizer stathmin mediates the development of dendritic arbors in neuronal cells. J. Cell Sci. 2007, 120, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Tortosa, E.; Adolfs, Y.; Fukata, M.; Pasterkamp, R.J.; Kapitein, L.C.; Hoogenraad, C.C. Dynamic Palmitoylation Targets MAP6 to the Axon to Promote Microtubule Stabilization during Neuronal Polarization. Neuron 2017, 17, 809–825. [Google Scholar] [CrossRef]
- Schwenk, B.M.; Lang, C.M.; Hogl, S.; Tahirovic, S.; Orozco, D.; Rentzsch, K.; Lichtenthaler, S.F.; Hoogenraad, C.C.; Capell, A.; Haass, C.; et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 2014, 33, 450–467. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef]
- Overly, C.C.; Hollenbeck, P.J. Dynamic organization of endocytic pathways in axons of cultured sympathetic neurons. J. Neurosci. 1996, 16, 6056–6064. [Google Scholar] [CrossRef] [PubMed]
- McKenney, R.J.; Huynh, W.; Tanenbaum, M.E.; Bhabha, G.; Vale, R.D. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 2014, 354, 337–341. [Google Scholar] [CrossRef]
- Lie, P.P.Y.; Nixon, R.A. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 2019, 122, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998, 111, 2477–2486. [Google Scholar] [PubMed]
- Leung, C.L.; Green, K.J.; Liem, R.K. Plakins: A family of versatile cytolinker proteins. Trends Cell Biol. 2002, 12, 37–45. [Google Scholar] [CrossRef]
- Redmann, M.; Benavides, G.A.; Berryhill, T.F.; Wani, W.Y.; Ouyang, X.; Johnson, M.S.; Ravi, S.; Barnes, S.; Darley-Usmar, V.M.; Zhang, J. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2017, 11, 73–81. [Google Scholar] [CrossRef]
- Ryazantsev, S.; Yu, W.H.; Zhao, H.Z.; Neufeld, E.F.; Ohmi, K. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol. Genet. Metab. 2007, 90, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Knecht, E.; Martinez-Ramón, A.; Grisolia, S. Autophagy of mitochondria in rat liver assessed by immunogold procedures. J. Histoch. Cytochem. 1988, 36, 1433–1440. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Di Malta, C.; Fryer, J.D.; Settembre, C.; Ballabio, A. Autophagy in astrocytes: A novel culprit in lysosomal storage disorders. Autophagy 2012, 8, 1871–1872. [Google Scholar] [CrossRef]
- Fraldi, A.; Klein, A.D.; Medina, D.L.; Settembre, C. Brain disorders due to lysosomal dysfunction. Ann. Rev. Neurosci. 2016, 39, 277–295. [Google Scholar] [CrossRef]
- Kim, S.G.; Hoffman, G.R.; Poulogiannis, G.; Buel, G.R.; Jang, Y.J.; Lee, K.W.; Kim, B.Y.; Erikson, R.L.; Cantley, L.C.; Choo, A.Y.; et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell 2013, 49, 172–185. [Google Scholar] [CrossRef]
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 129, 2475–2481. [Google Scholar] [CrossRef]
- Camandola, S.; Mattson, M.P. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 2017, 36, 1474–1492. [Google Scholar] [CrossRef]
- García-Cazorla, À.; Saudubray, J.M. Cellular neurometabolism: A tentative to connect cell biology and metabolism in neurology. J. Inherit. Metab. Dis. 2018, 41, 1043–1054. [Google Scholar] [CrossRef]
- Oyarzabal, A.; Marin-Valencia, I. Synaptic energy metabolism and neuronal excitability, in sickness and health. J. Inherit. Metab. Dis. 2019, 42, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Rizo, J. Mechanism of neurotransmitter release coming into focus. Protein Sci. 2018, 27, 1364–1391. [Google Scholar] [CrossRef] [PubMed]
- Guiberson, N.G.L.; Pineda, A.; Abramov, D.; Kharel, P.; Carnazza, K.E.; Wragg, R.T.; Dittman, J.S.; Burré, J. Mechanism-based rescue of Munc18-1 dysfunction in varied encephalopathies by chemical chaperones. Nat. Commun. 2018, 9, 3986. [Google Scholar] [CrossRef] [PubMed]
- Hilfiker, S.; Pieribone, V.A.; Czernik, A.J.; Kao, H.T.; Augustine, G.J.; Greengard, P. Synapsins as regulators of neurotransmitter release. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 269–279. [Google Scholar] [CrossRef]
- Hosaka, M.; Hammer, R.E.; Südhof, T.C. A phospho-switch controls the dynamic association of synapsins with synaptic vesicles. Neuron 1999, 24, 377–387. [Google Scholar] [CrossRef]
- Shin, O.H. Exocytosis and synaptic vesicle function. Compr. Physiol. 2014, 4, 149–175. [Google Scholar] [CrossRef]
- Mirza, F.J.; Zahid, S. The Role of Synapsins in Neurological Disorders. Neurosci. Bull. 2018, 34, 349–358. [Google Scholar] [CrossRef]
- Zahid, S.; Oellerich, M.; Asif, A.R.; Ahmed, N. Differential expression of proteins in brain regions of Alzheimer’s disease patients. Neurochem. Res. 2014, 39, 208–215. [Google Scholar] [CrossRef]
- Orlando, M.; Lignani, G.; Maragliano, L.; Fassio, A.; Onofri, F.; Baldelli, P.; Giovedí, S.; Benfenati, F. Functional role of ATP binding to synapsin I in synaptic vesicle trafficking and release dynamics. J. Neurosci. 2014, 34, 14752–14768. [Google Scholar] [CrossRef]
- Vieira, O.V. Rab3a and Rab10 are regulators of lysosome exocytosis and plasma membrane repair. Small GTPases 2018, 9, 349–351. [Google Scholar] [CrossRef]
- Zhen, Y.; Stenmark, H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015, 128, 3171–3176. [Google Scholar] [CrossRef] [PubMed]
- Encarnacao, M.; Espada, L.; Escrevente, C.; Mateus, D.; Ramalho, J.; Michelet, X.; Santarino, I.; Hsu, V.W.; Brenner, M.B.; Barral, D. A Rab3a-dependent complex essential for lysosome positioning and plasma membrane repair. J. Cell Biol. 2016, 213, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Ginns, E.I.; Mak, S.K.; Ko, N.; Karlgren, J.; Akbarian, S.; Chou, V.P.; Guo, Y.; Lim, A.; Samuelsson, S.; LaMarca, M.L.; et al. Neuroinflammation and α-synuclein accumulation in response to glucocerebrosidase deficiency are accompanied by synaptic dysfunction. Mol. Genet. Metab. 2014, 111, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Tomic, S. Dopamine transport system imaging is pathologic in Niemann-Pick type C-case report. Neurol. Sci. 2018, 39, 1139–1140. [Google Scholar] [CrossRef]
- Terbeek, J.; Latour, P.; Van Laere, K.; Vandenberghe, W. Abnormal dopamine transporter imaging in adult-onset Niemann-Pick disease type C. Park. Relat. Disord. 2017, 36, 107–108. [Google Scholar] [CrossRef]
- Xu, S.; Sleat, D.E.; Jadot, M.; Lobel, P. Glial fibrillary acidic protein is elevated in the lysosomal storage disease classical late-infantile neuronal ceroid lipofuscinosis, but is not a component of the storage material. Biochem. J. 2010, 428, 355–362. [Google Scholar] [CrossRef]
- Baldo, G.; Mayer, F.Q.; Martinelli, B.; Dilda, A.; Meyer, F.; Ponder, K.P.; Giugliani, R.; Matte, U. Evidence of a progressive motor dysfunction in Mucopolysaccharidosis type I mice. Behav. Brain Res. 2012, 233, 169–175. [Google Scholar] [CrossRef]
- Pressey, S.N.; Smith, D.A.; Wong, A.M.; Platt, F.M.; Cooper, J.D. Early glial activation, synaptic changes and axonal pathology in the thalamocortical system of Niemann-Pick type C1 mice. Neurobiol. Dis. 2012, 45, 1086–1100. [Google Scholar] [CrossRef]
- Wilkinson, F.L.; Holley, R.J.; Langford-Smith, K.J.; Badrinath, S.; Liao, A.; Langford-Smith, A.; Cooper, J.D.; Jones, S.A.; Wraith, J.E.; Wynn, R.F.; et al. Neuropathology in mouse models of mucopolysaccharidosis type I, IIIA and IIIB. PLoS ONE 2012, 7, e35787. [Google Scholar] [CrossRef]
- Vitner, E.B.; Futerman, A.H. Neuronal forms of Gaucher disease. Handb. Exp. Pharmacol. 2013, 216, 405–419. [Google Scholar] [CrossRef]
- Tsuji, D. Molecular pathogenesis and therapeutic approach of GM2 gangliosidosis. Yakugaku Zasshi 2013, 133, 269–274. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hordeaux, J.; Dubreil, L.; Robveille, C.; Deniaud, J.; Pascal, Q.; Dequéant, B.; Pailloux, J.; Lagalice, L.; Ledevin, M.; Babarit, C.; et al. Long-term neurologic and cardiac correction by intrathecal gene therapy in Pompe disease. Acta Neuropathol. Commun. 2017, 5, 1–66. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, P.; Tse, D.Y.; Di Ronza, A.; Seymour, M.L.; Martano, G.; Cooper, J.D.; Pereira, F.A.; Passafaro, M.; Wu, S.M.; Sardiello, M. Trehalose reduces retinal degeneration, neuroinflammation and storage burden caused by a lysosomal hydrolase deficiency. Autophagy 2018, 14, 1419–1434. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.; Bigger, B.W. The role of innate immunity in mucopolysaccharide diseases. J. Neurochem. 2019, 148, 639–651. [Google Scholar] [CrossRef]
- Arfi, A.; Richard, M.; Gandolphe, C.; Bonnefont-Rousselot, D.; Therond, P.; Scherman, D. Neuroinflammatory and oxidative stress phenomena in MPS IIIA mouse model: The positive effect of long-term aspirin treatment. Mol. Genet. Metab. 2011, 103, 18–25. [Google Scholar] [CrossRef]
- Martins, C.; Hůlková, H.; Dridi, L.; Dormoy-Raclet, V.; Grigoryeva, L.; Choi, Y.; Langford-Smith, A.; Wilkinson, F.L.; Ohmi, K.; DiCristo, G.; et al. Neuroinflammation, mitochondrial defects and neurodegeneration in mucopolysaccharidosis III type C mouse model. Brain 2015, 138, 336–355. [Google Scholar] [CrossRef]
- Vitner, E.B.; Farfel-Becker, T.; Ferreira, N.S.; Leshkowitz, D.; Sharma, P.; Lang, K.S.; Futerman, A.H. Induction of the type I interferon response in neurological forms of Gaucher disease. J. Neuroinflamm. 2016, 13, 104. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef]
- Holley, R.J.; Ellison, S.M.; Fil, D.; O’Leary, C.; McDermott, J.; Senthivel, N.; Langford-Smith, A.W.W.; Wilkinson, F.L.; D’Souza, Z.; Parker, H.; et al. Macrophage enzyme and reduced inflammation drive brain correction of mucopolysaccharidosis IIIB by stem cell gene therapy. Brain 2018, 141, 99–116. [Google Scholar] [CrossRef]
- Trudel, S.; Trecherel, E.; Gomila, C.; Peltier, M.; Aubignat, M.; Gubler, B.; Morliere, P.; Heard, J.M.; Ausseil, J. Oxidative stress is independent of inflammation in the neurodegenerative Sanfilippo syndrome type B. J. Neurosci. Res. 2015, 93, 424–432. [Google Scholar] [CrossRef]
- Simonaro, C.M.; Ge, Y.; Eliyahu, E.; He, X.; Jepsen, K.J.; Schuchman, E.H. Involvement of the Toll-like receptor 4 pathway and use of TNF-alpha antagonists for treatment of the mucopolysaccharidoses. Proc. Natl. Acad. Sci. USA 2010, 107, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Simonaro, C.M.; D’Angelo, M.; He, X.; Eliyahu, E.; Shtraizent, N.; Haskins, M.E.; Schuchman, E.H. Mechanism of glycosaminoglycan-mediated bone and joint disease: Implications for the mucopolysaccharidoses and other connective tissue diseases. Am. J. Pathol. 2008, 172, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Simonaro, C.M.; Haskins, M.E.; Schuchman, E.H. Articular chondrocytes from animals with a dermatan sulfate storage disease undergo a high rate of apoptosis and release nitric oxide and inflammatory cytokines: A possible mechanism underlying degenerative joint disease in the mucopolysaccharidoses. Lab. Investig. 2001, 81, 1319–1328. [Google Scholar] [CrossRef]

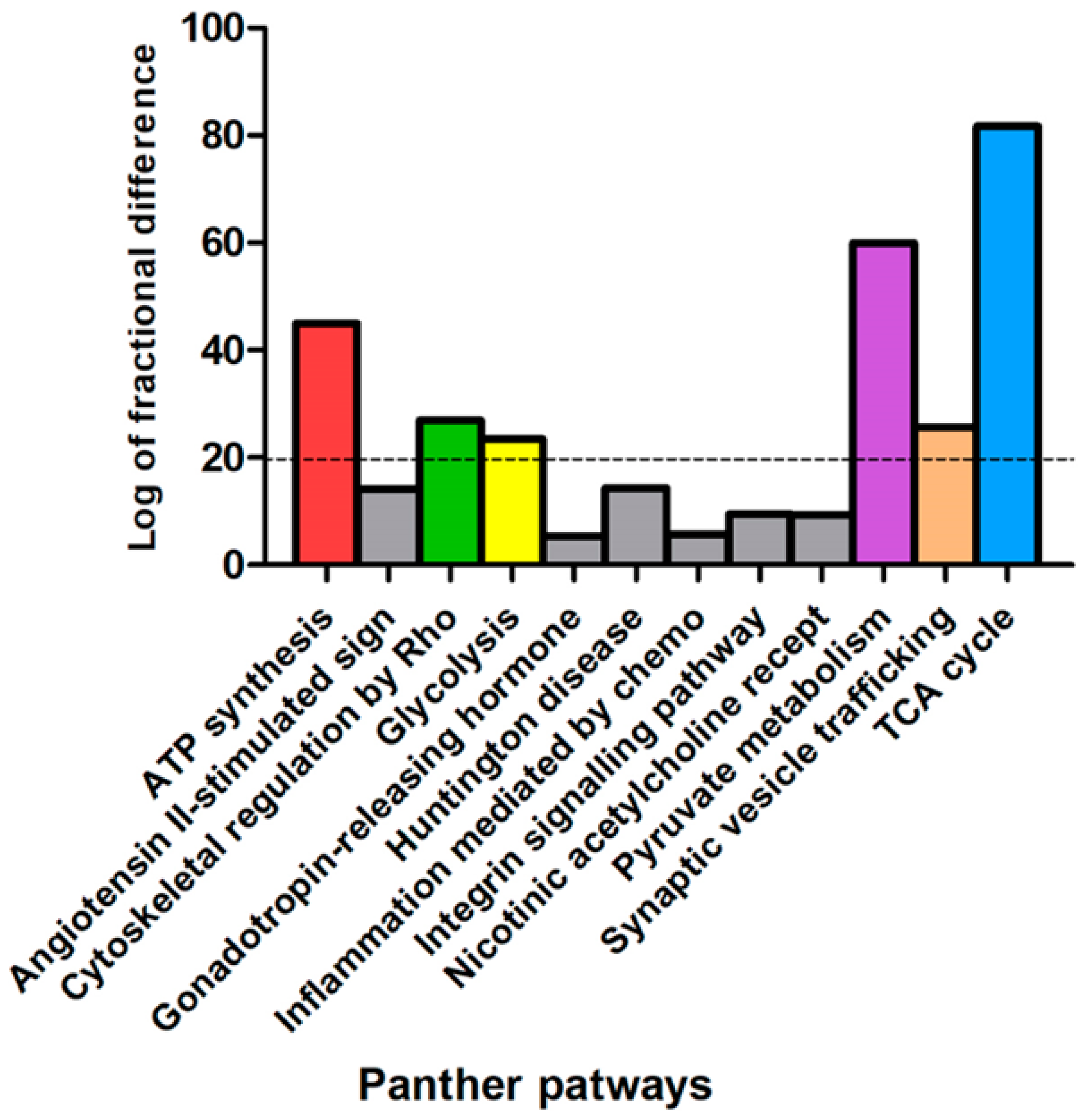
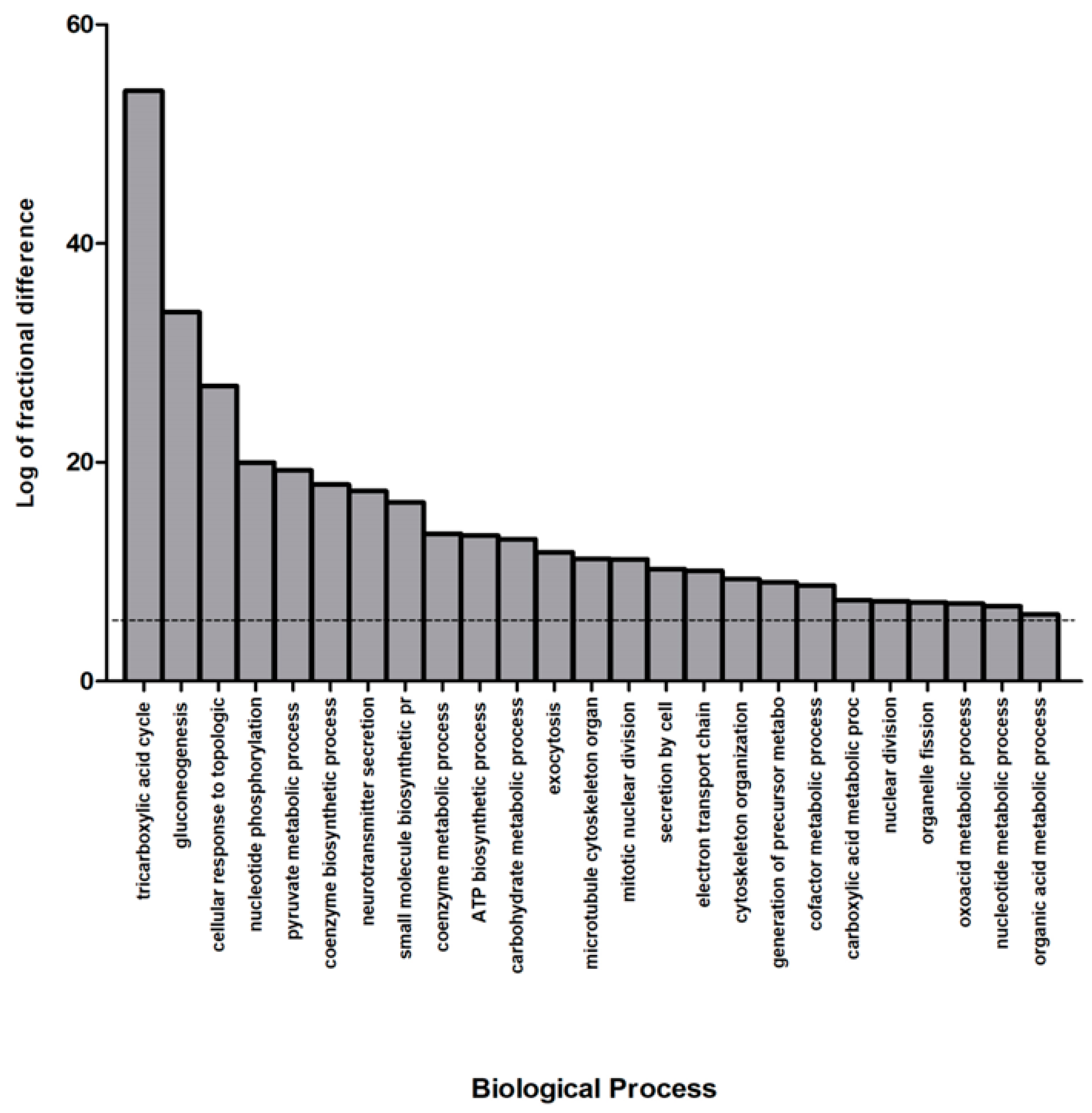
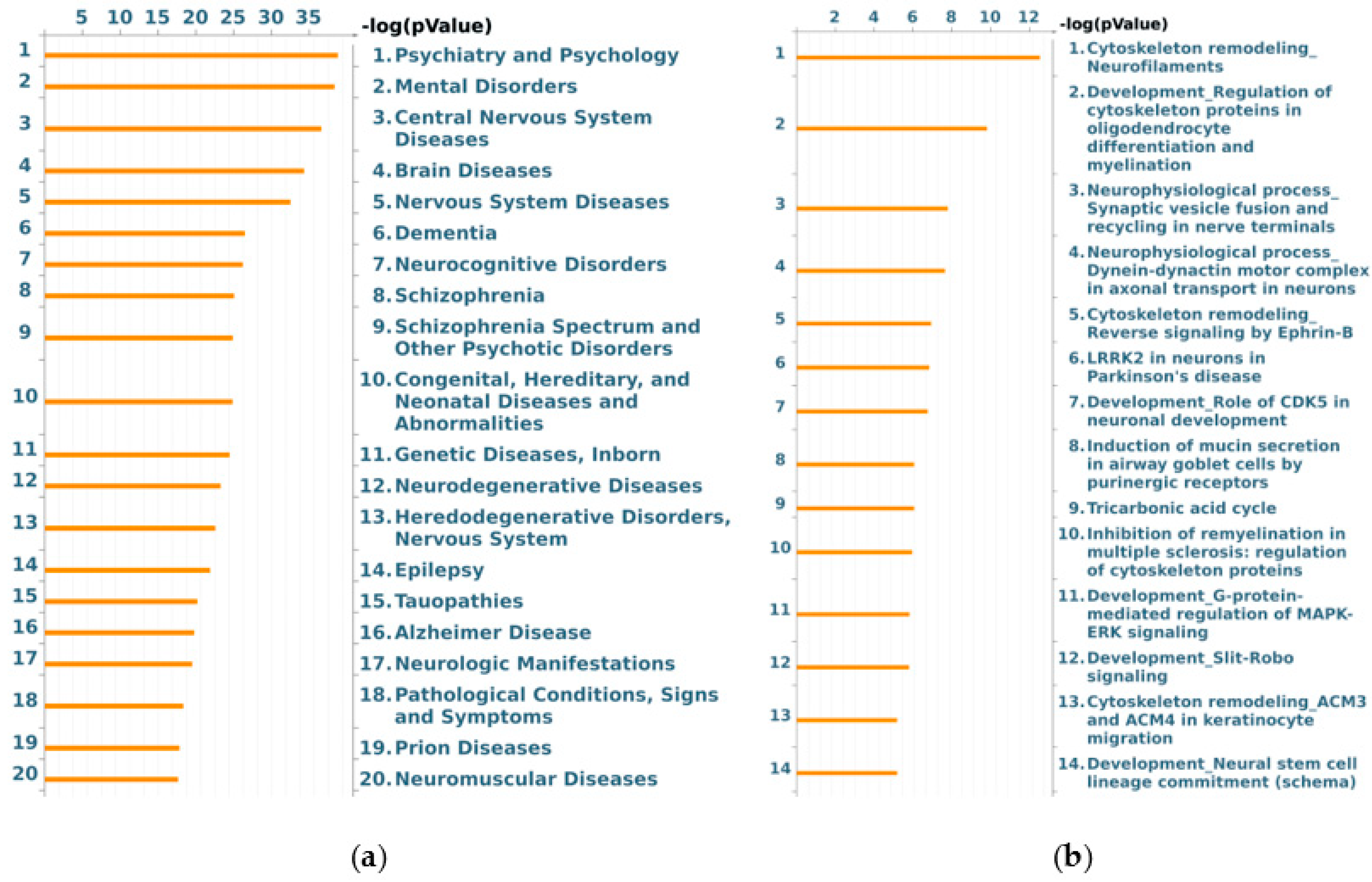
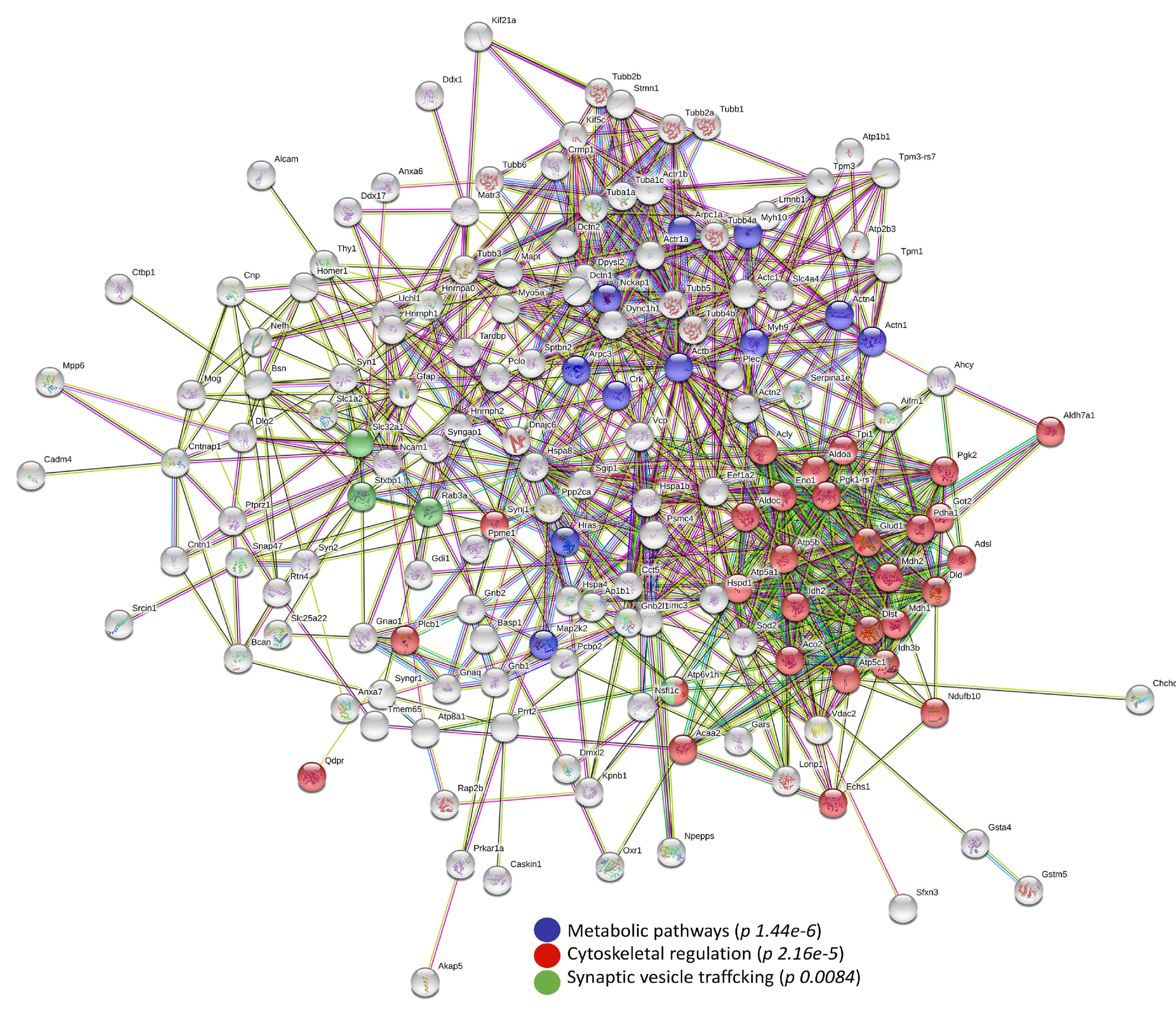
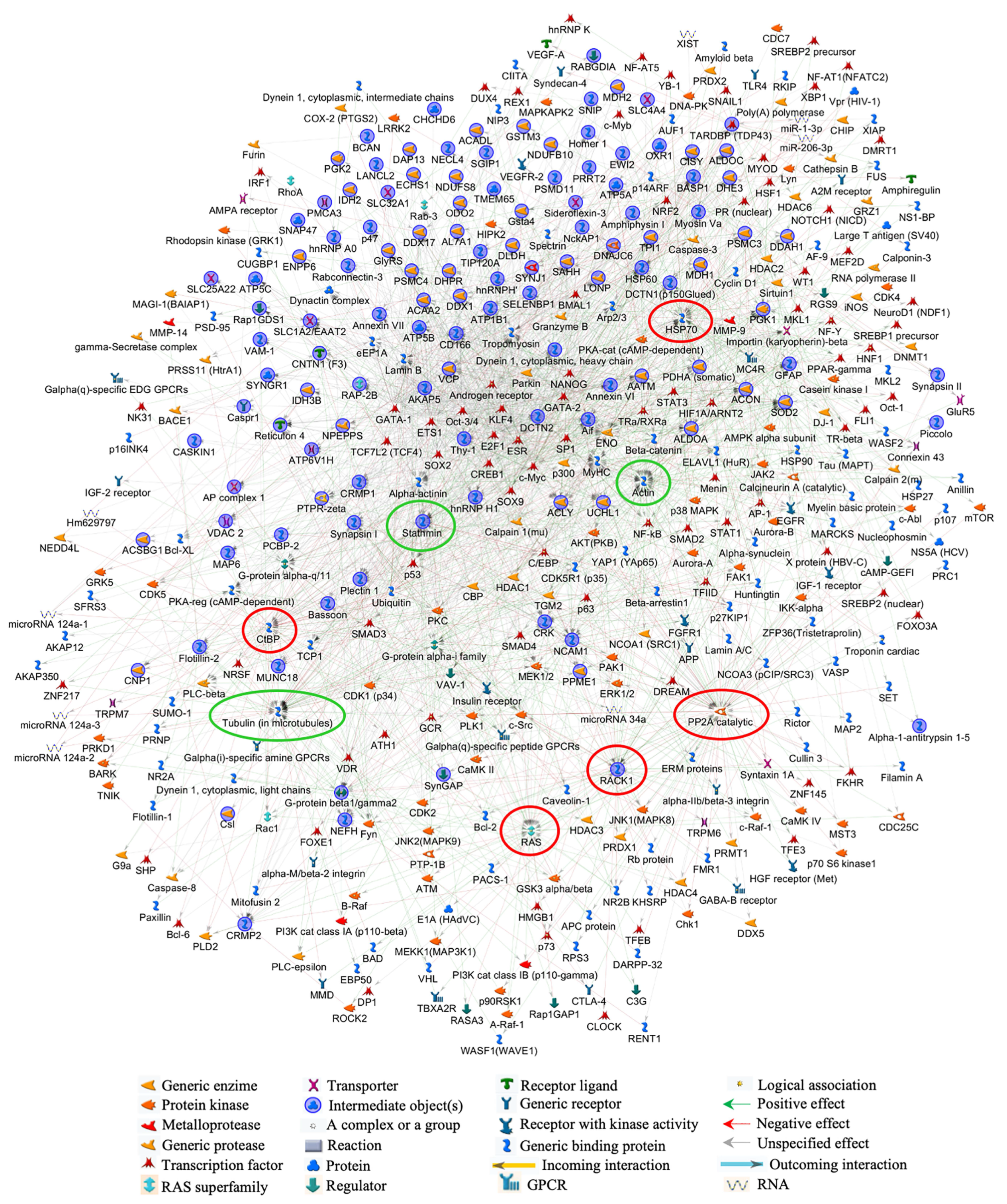
| Swiss-Prot Code | Gene Name | Protein Description | Fold NSAF | p Value | Subcellular Localization |
|---|---|---|---|---|---|
| Q545B6 | Stmn1 | Stathmin | −12.6 | 0.00002 | CK |
| Q9QYX7 | Pclo | Protein piccolo | −8.4 | 0.00101 | CK, GA |
| Q6P9K8 | Caskin1 | Caskin-1 | −3.8 | 0.00262 | CK |
| A8DUK2 | Hbbt1 | Beta-globin | −3.4 | 0.00008 | C |
| Q4VAE3 | Tmem65 | Transmembrane protein 65 | −3.3 | 0.00374 | M, PM |
| Q8CHF1 | mKIAA0531 | Kinesin-like protein (Fragment) | −3.2 | 0.00232 | CK |
| P17563 | Selenbp1 | Selenium-binding protein 1 | −3.2 | 0.00046 | CK, N |
| Q4KMM3 | Oxr1 | Oxidation resistance protein 1 | −3 | 0.00036 | M, N |
| A2AS98 | Nckap1 | Nck-associated protein 1 | −2.9 | 0.00209 | CK, PM |
| C7G3P1 | Ppfia3 | MKIAA0654 protein (Fragment) | −2.6 | 0.00328 | CK |
| Q8BVQ5 | Ppme1 | Protein phosphatase methylesterase 1 | −2.5 | 0.00345 | N |
| Q3UVN5 | Nsfl1c | Putative uncharacterized protein | −2.5 | 0.00002 | CK, GA, N |
| Q9R0Q6 | Arpc1a | Actin-related protein 2/3 complex subunit 1A | −2.4 | 0.00036 | CK, N, PM |
| Q3TF14 | Ahcy | Adenosylhomocysteinase | −2.4 | 0.00329 | CK, N |
| Q9CX86 | Hnrnpa0 | Heterogeneous nuclear ribonucleoprotein A0 | −2.4 | 0.00165 | N |
| Q9Z1B3 | Plcb1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | −2.4 | 0.00671 | CK, N, PM |
| O54991 | Cntnap1 | Contactin-associated protein 1 | −2.3 | 0.00626 | PM |
| B0V2P5 | Dmxl2 | DmX-like protein 2 | −2.3 | 0.01109 | PM |
| E9Q8N8 | Slc4a4 | Anion exchange protein | −2.3 | 0.0011 | PM |
| B1AQX9 | Srcin1 | SRC kinase-signaling inhibitor 1 | −2.3 | 0.0004 | CK |
| P24472 | Gsta4 | Glutathione S-transferase A4 | −2.3 | 0.00125 | M |
| Q3UK83 | Hnrnpa1 | Putative uncharacterized protein | −2.2 | 0.00047 | ERS, N |
| D3Z4J3 | Myo5a | Unconventional myosin-Va | −2.2 | 0.00636 | CK, C, ER, En, GA, Ly, Pe |
| Q61411 | Hras | GTPase HRas | −2.2 | 0.00583 | C, GA, N, PM |
| Q9Z0X1 | Aifm1 | Apoptosis-inducing factor 1, mitochondrial | −2.1 | 0.00398 | CK, M, N |
| B2L107 | Vsnl1 | Visinin-like protein 1 | −2.1 | 0.00018 | C |
| Q9DBF1 | Aldh7a | Alpha-aminoadipic semialdehyde dehydrogenase | −2.1 | 0.02264 | C, M, N |
| Q3TJF2 | Ola1 | Obg-like ATPase 1 | −2.1 | 0.00001 | CK, C, N |
| Q8VDD5 | Myh9 | Myosin-9 | −2 | 0.01358 | C, CK, N, PM |
| Q9JJK2 | Lancl2 | LanC-like protein 2 | −2 | 0.00232 | C, CK, N, PM |
| E9Q2L2 | Dlg2 | Disks large homolog 2 | −2 | 0.00218 | PM |
| Q91VR5 | Ddx1 | ATP-dependent RNA helicase DDX1 | −2 | 0.00216 | C, M, N |
| Q9JHU4 | Dync1h1 | Cytoplasmic dynein 1 heavy chain 1 | −2 | 0.00407 | CK, N |
| Q99PU5 | Acsbg1 | Long-chain-fatty-acid--CoA ligase ACSBG1 | −1.9 | 0.01942 | ER, PM |
| Q64010 | Crk | Adapter molecule crk | −1.9 | 0.0031 | CK, PM |
| P14733 | Lmnb1 | Lamin-B1 | −1.9 | 0.02293 | CK, N |
| P54775 | Psmc4 | 26S protease regulatory subunit 6B | −1.9 | 0.03099 | CK, N |
| A0A1S6GWI0 | Ndufa8 | NADH dehydrogenase (Ubiquinone) 1 alpha subcomplex, 8 | −1.8 | 0.00156 | M |
| Q3U741 | Ddx17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17, isoform CRA_a | −1.8 | 0.00085 | C, N |
| Q3TIQ2 | Rpl12 | Putative uncharacterized protein | −1.8 | 0.01139 | C, N |
| P70168 | Kpnb1 | Importin subunit beta-1 | −1.7 | 0.00011 | C, M, N |
| Q80TZ3 | Dnajc6 | Putative tyrosine-protein phosphatase auxilin | −1.7 | 0.00164 | C |
| Q5DTG0 | Atp8a1 | Phospholipid-transporting ATPase (Fragment) | −1.7 | 0.00428 | ER, GA, PM |
| Q9QXS1 | Plec | Plectin | −1.7 | 0.00377 | C, CK, PM |
| Q921F2 | Tardbp | TAR DNA-binding protein 43 | −1.7 | 0.00786 | N |
| B2RX08 | Sptb | Spectrin beta chain | −1.7 | 0.00319 | C, CK, PM |
| Q3TKG4 | Psmc3 | Putative uncharacterized protein (Fragment) | −1.6 | 0.00396 | N |
| Q3UH59 | Myh10 | Myosin-10 | −1.6 | 0.02527 | C, CK, PM |
| Q9JM76 | Arpc3 | Actin-related protein 2/3 complex subunit 3 | −1.6 | 0.01437 | C, N |
| A0A1S6GWJ8 | Uncharacterized protein | –1,5 | 0.02713 | ||
| Q3UN60 | Mpp6 | Membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6), isoform CRA_b | −1.5 | 0.00085 | PM |
| Q91XV3 | Basp1 | Brain acid soluble protein 1 | −1.5 | 0.00001 | N, PM |
| Q07076 | Anxa7 | Annexin A7 | −1.5 | 0.01183 | C, ER, ERS, N, PM |
| Q3UY05 | Ndufs8 | Putative uncharacterized protein | −1.4 | 0.00008 | M |
| A0A0R4J0Q5 | Lmnb2 | Lamin-B2 | −1.4 | 0.03275 | CK, N |
| Q4FJX9 | Sod2 | Superoxide dismutase | −1.4 | 0.01379 | M |
| Q91VN4 | Chchd6 | MICOS complex subunit Mic25 | −1.4 | 0.01573 | C, M |
| O88737 | Bsn | Protein bassoon | −1.3 | 0.00915 | C, GA |
| Q3UWW9 | Psmd11 | Putative uncharacterized protein | −1.3 | 0.01386 | C, N |
| O08788 | Dctn1 | Dynactin subunit 1 | −1.3 | 0.01243 | CK, C, ERS, N |
| A0A0R4J275 | Ndufa12 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | −1.3 | 0.0197 | C, M |
| Q63932 | Map2k2 | Dual specificity mitogen-activated protein kinase kinase 2 | −1.3 | 0.01951 | CK, C, ER, En, GA, M, N, PM |
| Q61361 | Bcan | Brevican core protein | −1.3 | 0.01264 | ERS |
| Q3V117 | Acly | ATP-citrate synthase | −1.3 | 0.0078 | C, M, N, PM |
| Q8R570 | Snap47 | Synaptosomal-associated protein 47 | −1.3 | 0.00248 | C, PM |
| Q9DCS9 | Ndufb10 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | −1.2 | 0.01688 | M |
| P14824 | Anxa6 | Annexin A6 | −1.2 | 0.01378 | C, En, ERS, Ly, M, N, PM |
| P54071 | Idh2 | Isocitrate dehydrogenase [NADP], mitochondrial | −1.2 | 0.00039 | C, M, Pe |
| Q7TSJ2 | Map6 | Microtubule-associated protein 6 | −1.2 | 0.00159 | CK, GA |
| Q8BGN3 | Enpp6 | Ectonucleotide pyrophosphatase/phosphodiesterase family member 6 | −1.2 | 0.03099 | ERS, PM |
| Q9CZD3 | Gars | Glycine--tRNA ligase | −1.2 | 0.01474 | C, ERS, M |
| A0A0R4J083 | Acadl | Long-chain-specific acyl-CoA dehydrogenase, mitochondrial | −1.2 | 0.03611 | M |
| Q8CGK3 | Lonp1 | Lon protease homolog, mitochondrial | −1.1 | 0.01936 | C, M, N |
| E9Q7Q3 | Tpm3 | Tropomyosin alpha-3 chain | −1.1 | 0.00355 | CK, C |
| Q8BWT1 | Acaa2 | 3-ketoacyl-CoA thiolase, mitochondrial | −1.1 | 0.04478 | M |
| P61226 | Rap2b | Ras-related protein Rap-2b | −1.1 | 0.03025 | C, En, ERS, PM |
| Q8R5C5 | Actr1b | Beta-centractin | −1.1 | 0.00172 | CK |
| E9Q0J5 | Kif21a | Kinesin-like protein KIF21A | −1.1 | 0.04345 | CK, C, PM |
| B9EKR1 | Ptprz1 | Receptor-type tyrosine-protein phosphatase zeta | −1.1 | 0.01214 | ERS, PM |
| Q6ZQ38 | Cand1 | Cullin-associated NEDD8-dissociated protein 1 | −1 | 0.00177 | C, GA, N |
| Q9JI91 | Actn2 | Alpha-actinin-2 | −1 | 0.00286 | CK, PM |
| P68040 | Rack1 | Receptor of activated protein C kinase 1 | −1 | 0.03823 | CK, M, N, PM |
| E9PUL5 | Prrt2 | Proline-rich transmembrane protein 2 | −1 | 0.0411 | PM |
| F6SEU4 | Syngap1 | Ras/Rap GTPase-activating protein SynGAP | −1 | 0.03268 | PM |
| Q8CHG1 | Dclk1 | MKIAA0369 protein (Fragment) | −1 | 0.02306 | PM |
| Q61490 | Alcam | CD166 antigen | −1 | 0.03414 | PM |
| D3Z656 | Synj1 | Synaptojanin-1 | −1 | 0.02126 | PM |
| H3BIV5 | Akap5 | A-kinase anchor protein 5 | −1 | 0.01709 | CK, PM |
| Q8VD37 | Sgip1 | SH3-containing GRB2-like protein 3-interacting protein 1 | −1 | 0.04017 | CK, PM |
| W6PPR4 | Ank3 | 480-kDa ankyrinG | −1 | 0.00579 | CK, C, PM |
| Q9CWS0 | Ddah1 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | −0.9 | 0.04649 | M |
| Q11011 | Npepps | Puromycin-sensitive aminopeptidase | −0.9 | 0.00505 | C, N |
| Q3TXE5 | Canx | Putative uncharacterized protein | −0.9 | 0.00488 | ER, PM |
| Q3UAG2 | Pgd | 6-phosphogluconate dehydrogenase, decarboxylating | −0.9 | 0.02403 | C |
| Q99JX6 | Anxa6 | Annexin | −0.9 | 0.04313 | C, En, ERS, Ly, M, N, PM |
| A0A0G2JEG8 | Amph | Amphiphysin | −0.9 | 0.00912 | CK, PM |
| Q99P72 | Rtn4 | Reticulon-4 | −0.9 | 0.01061 | ER, N, PM |
| Q49S98 | Slc32a1 | Putative uncharacterized protein | −0.9 | 0.04098 | PM |
| B2RQQ5 | Map1b | Microtubule-associated protein 1B | −0.9 | 0.03443 | CK, C, PM |
| D3Z2H9 | Tpm3 | Uncharacterized protein | −0.9 | 0.02982 | CK, C |
| Q6ZQ61 | Matr3 | MCG121979, isoform CRA_c (Fragment) | −0.8 | 0.01811 | N |
| Q3TE45 | Sdhb | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | −0.8 | 0.01667 | M, N, PM |
| Q0VF55 | Atp2b3 | Calcium-transporting ATPase | −0.8 | 0.03416 | PM |
| Q3UEG9 | Flot2 | Putative uncharacterized protein | −0.8 | 0.02632 | CK, En, PM |
| Q91V61 | Sfxn3 | Sideroflexin-3 | −0.8 | 0.00937 | M |
| P61164 | Actr1a | Alpha-centractin | −0.8 | 0.03531 | CK |
| Q571M2 | Hspa4 | MKIAA4025 protein (Fragment) | −0.7 | 0.00487 | C, ERS, N |
| Q9Z2Y3 | Homer1 | Homer protein homolog 1 | −0.7 | 0.01584 | C, PM |
| E9Q455 | Tpm1 | Tropomyosin alpha-1 chain | −0.7 | 0.04225 | C |
| A2A5Y6 | Mapt | Microtubule-associated protein | −0.7 | 0.04838 | CK, C, N, PM |
| A0A1S6GWH1 | Uncharacterized protein | –0,7 | 0.02404 | ||
| P35486 | Pdha1 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, mitochondrial | −0.6 | 0.00552 | M, N |
| Q8CC13 | Ap1b1 | AP complex subunit beta | −0.6 | 0.03216 | C, GA |
| E9Q2W9 | Actn4 | Alpha-actinin-4 (Fragment) | −0.6 | 0.00847 | CK, C, N |
| Q8BVE3 | Atp6v1h | V-type proton ATPase subunit H | −0.6 | 0.02175 | C, Ly |
| A0A0A6YY91 | Ncam1 | Neural cell adhesion molecule 1 (Fragment) | −0.6 | 0.0182 | CK, PM |
| Q3TYK4 | Prkar1a | Putative uncharacterized protein | −0.6 | 0.01177 | CK, C, PM |
| P12960 | Cntn1 | Contactin-1 | −0.6 | 0.02931 | PM |
| Q3TPZ5 | Dctn2 | Dynactin 2 | −0.6 | 0.00054 | CK, C |
| A0A1L1SV25 | Actn4 | Alpha-actinin-4 | −0.6 | 0.0286 | CK, C, N |
| P19246 | Nefh | Neurofilament heavy polypeptide | −0.5 | 0.04106 | CK, M, N, PM |
| A0A0R4J117 | Igsf8 | Immunoglobulin superfamily member 8 | −0.5 | 0.04908 | PM |
| Q68FG2 | Sptbn2 | Spectrin beta chain | −0.5 | 0.04372 | CK, C, ER, En, GA |
| Q80YU5 | Mog | Myelin oligodendrocyte glycoprotein (Fragment) | −0.5 | 0.02477 | C, ER, M, PM |
| E9QB01 | Ncam1 | Neural cell adhesion molecule 1 | −0.5 | 0.03648 | C, PM |
| Q01853 | Vcp | Transitional endoplasmic reticulum ATPase | −0.5 | 0.03249 | C, ER, N |
| P09041 | Pgk2 | Phosphoglycerate kinase 2 | −0.4 | 0.03696 | C |
| Q7TPR4 | Actn1 | Alpha-actinin-1 | −0.4 | 0.01557 | CK, N, PM |
| P63011 | Rab3a | Ras-related protein Rab-3A | −0.4 | 0.0162 | C, En, Ly, PM |
| P09411 | Pgk1 | Phosphoglycerate kinase 1 | −0.3 | 0.01617 | C, ERS, PM |
| O08599 | Stxbp1 | Syntaxin-binding protein 1 | 0.1 | 0.03134 | CK, C, M, PM |
| A0A0A0MQA5 | Tuba4a | Tubulin alpha chain (Fragment) | 0.2 | 0.01472 | CK |
| P68369 | Tuba1a | Tubulin alpha-1A chain | 0.2 | 0.02299 | CK, En |
| Q52L87 | Tuba1c | Tubulin alpha chain | 0.3 | 0.0146 | CK, N |
| P50396 | Gdi1 | Rab GDP dissociation inhibitor alpha | 0.3 | 0.03464 | GA |
| P60710 | Actb | Actin, cytoplasmic 1 | 0.3 | 0.02793 | CK, C, N, PM |
| Q9CZU6 | Cs | Citrate synthase, mitochondrial | 0.3 | 0.00075 | M |
| P05202 | Got2 | Aspartate aminotransferase, mitochondrial | 0.3 | 0.03387 | M, PM |
| P63038 | Hspd1 | 60 kDa heat shock protein, mitochondrial | 0.3 | 0.02215 | CK, ER, En, ERS, GA, M, PM |
| E9Q912 | Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 | 0.3 | 0.00467 | CK, ER, M |
| B2CSK2 | Heat shock protein 1-like protein | 0.3 | 0.00172 | ||
| O88935 | Syn1 | Synapsin-1 | 0.3 | 0.03624 | CK, C, GA, N |
| P68033 | Actc1 | Actin, alpha cardiac muscle 1 | 0.3 | 0.00986 | CK |
| P08249 | Mdh2 | Malate dehydrogenase, mitochondrial | 0.3 | 0.00731 | M |
| Q03265 | Atp5a1 | ATP synthase subunit alpha, mitochondrial | 0.3 | 0.00169 | M, N, PM |
| Q99KI0 | Aco2 | Aconitate hydratase, mitochondrial | 0.3 | 0.00087 | C, M |
| P17879 | Hspa1b | Heat shock 70 kDa protein 1B | 0.4 | 0.00362 | CK, C, M, N, PM |
| O08553 | Dpysl2 | Dihydropyrimidinase-related protein 2 | 0.4 | 0.00004 | CK, C, M, PM |
| Q5FW97 | EG433182 | Enolase 1, alpha non-neuron | 0.4 | 0.00346 | C |
| Q64332 | Syn2 | Synapsin-2 | 0.4 | 0.01708 | PM |
| Q3TQ70 | Gnb1 | Beta1 subnuit of GTP-binding protein | 0.4 | 0.01897 | PM |
| P62631 | Eef1a2 | Elongation factor 1-alpha 2 | 0.4 | 0.00011 | N |
| P80316 | Cct5 | T-complex protein 1 subunit epsilon | 0.4 | 0.04942 | CK, C |
| Q8CE19 | Syn2 | Putative uncharacterized protein | 0.4 | 0.02198 | PM |
| Q3UA81 | Eef1a1 | Elongation factor 1-alpha | 0.4 | 0.00003 | CK, C, M, PM |
| Q8C2Q7 | Hnrnph1 | Heterogeneous nuclear ribonucleoprotein H | 0.4 | 0.01261 | C, N |
| Q9D6F9 | Tubb4a | Tubulin beta-4A chain | 0.4 | 0.00031 | CK |
| P17751 | Tpi1 | Triosephosphate isomerase | 0.4 | 0.00008 | C |
| P48774 | Gstm5 | Glutathione S-transferase Mu 5 | 0.4 | 0.01045 | C, ERS |
| P18872 | Gnao1 | Guanine nucleotide-binding protein G(o) subunit alpha | 0.4 | 0.00334 | PM |
| Q3UYK6 | Slc1a2 | Amino acid transporter | 0.4 | 0.03711 | PM |
| Q8BVI4 | Qdpr | Dihydropteridine reductase | 0.4 | 0.02048 | C, M |
| Q9D2G2 | Dlst | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | 0.4 | 0.00639 | M, N, PM |
| Q3UD06 | Atp5c1 | ATP synthase subunit gamma | 0.4 | 0.00728 | M |
| P68372 | Tubb4b | Tubulin beta-4B chain | 0.4 | 0.00008 | CK |
| P99024 | Tubb5 | Tubulin beta-5 chain | 0.4 | 0.00005 | CK, C, M |
| P56480 | Atp5b | ATP synthase subunit beta, mitochondrial | 0.4 | 0.00166 | M, PM |
| E9QKR0 | Gnb2 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-2 | 0.4 | 0.0162 | PM |
| B2RSN3 | Tubb2b | Tubulin beta chain | 0.4 | 0.00013 | CK |
| Q7TMM9 | Tubb2a | Tubulin beta-2A chain | 0.4 | 0.0001 | CK |
| P63017 | Hspa8 | Heat shock cognate 71 kDa protein | 0.4 | 0.0001 | CK, C, En, ERS, Ly, N, PM |
| Q3TIC8 | Uqcrc1 | Putative uncharacterized protein | 0.4 | 0.00014 | C, M |
| P26443 | Glud1 | Glutamate dehydrogenase 1, mitochondrial | 0.4 | 0.00008 | M |
| Q3TYV5 | Cnp | 2′,3′-cyclic-nucleotide 3′-phosphodiesterase | 0.5 | 0.00001 | ERS |
| B7U582 | Heat shock protein 70-2 | 0.5 | 0.00001 | ||
| Q8BH95 | Echs1 | Enoyl-CoA hydratase, mitochondrial | 0.5 | 0.03689 | M |
| Q6P1J1 | Crmp1 | Crmp1 protein | 0.5 | 0.00045 | CK, C, N |
| B2CY77 | Rpsa | Laminin receptor (Fragment) | 0.5 | 0.01872 | C, ERS, N, PM |
| Q922F4 | Tubb6 | Tubulin beta-6 chain | 0.5 | 0.00002 | CK |
| Q9ERD7 | Tubb3 | Tubulin beta-3 chain | 0.5 | 0.00003 | CK |
| Q60930 | Vdac2 | Voltage-dependent anion-selective channel protein 2 | 0.5 | 0.00247 | M |
| P14152 | Mdh1 | Malate dehydrogenase, cytoplasmic | 0.5 | 0.00092 | C, M |
| A6ZI44 | Aldoa | Fructose-bisphosphate aldolase | 0.5 | 0.00156 | CK, C, ERS, M, N, PM |
| Q80X68 | Csl | Citrate synthase | 0.5 | 0.00001 | N |
| A2AQ07 | Tubb1 | Tubulin beta-1 chain | 0.5 | 0.00009 | CK |
| P01831 | Thy1 | Thy-1 membrane glycoprotein | 0.6 | 0.00272 | C, ER, PM |
| A0A1S6GWG6 | Uncharacterized protein | 0,6 | 0.00144 | ||
| Q3UBZ3 | Capza2 | Putative uncharacterized protein | 0.6 | 0.03767 | CK |
| Q9D6M3 | Slc25a22 | Mitochondrial glutamate carrier 1 | 0.6 | 0.02851 | M |
| O08749 | Dld | Dihydrolipoyl dehydrogenase, mitochondrial | 0.6 | 0.00293 | M, N |
| Q91VA7 | Idh3b | Isocitrate dehydrogenase [NAD] subunit, mitochondrial | 0.6 | 0.0105 | M |
| O88712 | Ctbp1 | C-terminal-binding protein 1 | 0.6 | 0.00177 | N |
| Q8C3L6 | Atp6v1b1 | Putative uncharacterized protein | 0.6 | 0.04137 | PM |
| P70333 | Hnrnph2 | Heterogeneous nuclear ribonucleoprotein H2 | 0.6 | 0.00158 | N |
| P14094 | Atp1b1 | Sodium/potassium-transporting ATPase subunit beta-1 | 0.7 | 0.00001 | PM |
| O55100 | Syngr1 | Synaptogyrin-1 | 0.7 | 0.03643 | PM |
| Q9R0P9 | Uchl1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 0.7 | 0.00142 | C, ER, PM |
| P05063 | Aldoc | Fructose-bisphosphate aldolase C | 0.7 | 0.00002 | C, M |
| Q61990 | Pcbp2 | Poly(rC)-binding protein 2 | 0.8 | 0.03331 | C, N |
| Q8R464 | Cadm4 | Cell adhesion molecule 4 | 0.8 | 0.01837 | PM |
| P21279 | Gnaq | Guanine nucleotide-binding protein G(q) subunit alpha | 0.9 | 0.04159 | C, N, PM |
| Q00898 | Serpina1 | Alpha-1-antitrypsin 1-5 | 1.2 | 0.04259 | ER, ERS, GA |
| P63330 | Ppp2ca | Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | 1.4 | 0.00163 | CK, C, N, PM |
| P03995 | Gfap | Glial fibrillary acidic protein | 1.4 | 0.00001 | CK, Ly |
| REACTOME Pathways | Mus musculus REFLIST | Client Input | Client Input (Raw p-Value) |
|---|---|---|---|
| Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane (R-MMU-190840) | 14 | 7 | 1.45 × 10−11 |
| Citric acid cycle (TCA cycle) (R-MMU-71403) | 22 | 8 | 3.68 × 10−12 |
| RHO GTPases activate IQGAPs (R-MMU-5626467) | 25 | 8 | 8.61 × 10−12 |
| Carboxyterminal post-translational modifications of tubulin (R-MMU-8955332) | 25 | 7 | 4.00 × 10−10 |
| Recycling pathway of L1 (R-MMU-437239) | 34 | 9 | 1.74 × 10−12 |
| Lysine catabolism (R-MMU-71064) | 12 | 3 | 7.16 × 10−05 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) (R-MMU-3371497) | 51 | 12 | 1.00 × 10−15 |
| COPI-independent Golgi-to-ER retrograde traffic (R-MMU-6811436) | 47 | 10 | 6.42 × 10−13 |
| Gluconeogenesis (R-MMU-70263) | 35 | 7 | 3.07 × 10−09 |
| Serotonin Neurotransmitter Release Cycle (R-MMU-181429) | 17 | 3 | 1.76 × 10−04 |
| Pyruvate metabolism and Citric Acid (TCA) cycle (R-MMU-71406) | 51 | 9 | 4.21 × 10−11 |
| GABA synthesis, release, reuptake and degradation (R-MMU-888590) | 19 | 3 | 2.35 × 10−04 |
| Dopamine Neurotransmitter Release Cycle (R-MMU-212676) | 22 | 3 | 3.47 × 10−04 |
| Glyoxylate metabolism and glycine degradation (R-MMU-389661) | 30 | 4 | 3.64 × 10−05 |
| Kinesins (R-MMU-983189) | 54 | 7 | 4.54 × 10−08 |
| Intraflagellar transport (R-MMU-5620924) | 54 | 7 | 4.54 × 10−08 |
| Recruitment of NuMA to mitotic centrosomes (R-MMU-380320) | 86 | 11 | 6.28 × 10−12 |
| RAF activation (R-MMU-5673000) | 25 | 3 | 4.89 × 10−04 |
| COPI-mediated anterograde transport (R-MMU-6807878) | 98 | 11 | 2.29 × 10−11 |
| The role of GTSE1 in G2/M progression after G2 checkpoint (R-MMU-8852276) | 72 | 8 | 1.45 × 10−08 |
| Loss of Nlp from mitotic centrosomes (R-MMU-380259) | 67 | 7 | 1.77 × 10−07 |
| AURKA Activation by TPX2 (R-MMU-8854518) | 70 | 7 | 2.33 × 10−07 |
| MHC class II antigen presentation (R-MMU-2132295) | 120 | 12 | 9.50 × 10−12 |
| Recruitment of mitotic centrosome proteins and complexes (R-MMU-380270) | 76 | 7 | 3.91 × 10−07 |
| Regulation of PLK1 Activity at G2/M Transition (R-MMU-2565942) | 85 | 7 | 7.92 × 10−07 |
| Hedgehog “on” state (R-MMU-5632684) | 103 | 8 | 1.91 × 10−07 |
| ER to Golgi Anterograde Transport (R-MMU-199977) | 155 | 12 | 1.52 × 10−10 |
| COPI-dependent Golgi-to-ER retrograde traffic (R-MMU-6811434) | 92 | 7 | 1.30 × 10−06 |
| Anchoring of the basal body to the plasma membrane (R-MMU-5620912) | 93 | 7 | 1.40 × 10−06 |
| RHO GTPases Activate Formins (R-MMU-5663220) | 133 | 10 | 7.13 × 10−09 |
| Resolution of Sister Chromatid Cohesion (R-MMU-2500257) | 121 | 9 | 4.53 × 10−08 |
| Hedgehog “off” state (R-MMU-5610787) | 109 | 8 | 2.87 × 10−07 |
| Glycolysis (R-MMU-70171) | 63 | 4 | 5.22 × 10−04 |
| Separation of Sister Chromatids (R-MMU-2467813) | 185 | 10 | 1.36 × 10−07 |
| Mitotic Anaphase (R-MMU-68882) | 188 | 10 | 1.57 × 10−07 |
| Neutrophil degranulation (R-MMU-6798695) | 553 | 14 | 3.17 × 10−06 |
| Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane (R-MMU-190840) | 14 | 7 | 1.45 × 10−11 |
| Citric acid cycle (TCA cycle) (R-MMU-71403) | 22 | 8 | 3.68 × 10−12 |
| RHO GTPases activate IQGAPs (R-MMU-5626467) | 25 | 8 | 8.61 × 10−12 |
| Carboxyterminal post-translational modifications of tubulin (R-MMU-8955332) | 25 | 7 | 4.00 × 10−10 |
| Recycling pathway of L1 (R-MMU-437239) | 34 | 9 | 1.74 × 10−12 |
| Lysine catabolism (R-MMU-71064) | 12 | 3 | 7.16 × 10−05 |
| Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion (R-MMU-434316) | 8 | 2 | 1.33 × 10−03 |
| HSP90 chaperone cycle for steroid hormone receptors (SHR) (R-MMU-3371497) | 51 | 12 | 1.00 × 10−15 |
| COPI-independent Golgi-to-ER retrograde traffic (R-MMU-6811436) | 47 | 10 | 6.42 × 10−13 |
| Gluconeogenesis (R-MMU-70263) | 35 | 7 | 3.07 × 10−09 |
| Serotonin Neurotransmitter Release Cycle (R-MMU-181429) | 17 | 3 | 1.76 × 10−04 |
| Pyruvate metabolism and Citric Acid (TCA) cycle (R-MMU-71406) | 51 | 9 | 4.21 × 10−11 |
| GABA synthesis, release, reuptake and degradation (R-MMU-888590) | 19 | 3 | 2.35 × 10−04 |
| Dopamine Neurotransmitter Release Cycle (R-MMU-212676) | 22 | 3 | 3.47 × 10−04 |
| Glyoxylate metabolism and glycine degradation (R-MMU-389661) | 30 | 4 | 3.64 × 10−05 |
| Kinesins (R-MMU-983189) | 54 | 7 | 4.54 × 10−08 |
| Intraflagellar transport (R-MMU-5620924) | 54 | 7 | 4.54 × 10−08 |
| Recruitment of NuMA to mitotic centrosomes (R-MMU-380320) | 86 | 11 | 6.28 × 10−12 |
| RAF activation (R-MMU-5673000) | 25 | 3 | 4.89 × 10−04 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Pasquale, V.; Costanzo, M.; Siciliano, R.A.; Mazzeo, M.F.; Pistorio, V.; Bianchi, L.; Marchese, E.; Ruoppolo, M.; Pavone, L.M.; Caterino, M. Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain. Biomolecules 2020, 10, 355. https://doi.org/10.3390/biom10030355
De Pasquale V, Costanzo M, Siciliano RA, Mazzeo MF, Pistorio V, Bianchi L, Marchese E, Ruoppolo M, Pavone LM, Caterino M. Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain. Biomolecules. 2020; 10(3):355. https://doi.org/10.3390/biom10030355
Chicago/Turabian StyleDe Pasquale, Valeria, Michele Costanzo, Rosa Anna Siciliano, Maria Fiorella Mazzeo, Valeria Pistorio, Laura Bianchi, Emanuela Marchese, Margherita Ruoppolo, Luigi Michele Pavone, and Marianna Caterino. 2020. "Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain" Biomolecules 10, no. 3: 355. https://doi.org/10.3390/biom10030355
APA StyleDe Pasquale, V., Costanzo, M., Siciliano, R. A., Mazzeo, M. F., Pistorio, V., Bianchi, L., Marchese, E., Ruoppolo, M., Pavone, L. M., & Caterino, M. (2020). Proteomic Analysis of Mucopolysaccharidosis IIIB Mouse Brain. Biomolecules, 10(3), 355. https://doi.org/10.3390/biom10030355












