2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA †
Abstract
1. Introduction
2. Materials and Methods
2.1. General Procedure for the Synthesis of 1,4-Disubstituted Thiosemicarbazide Derivatives (1,2)
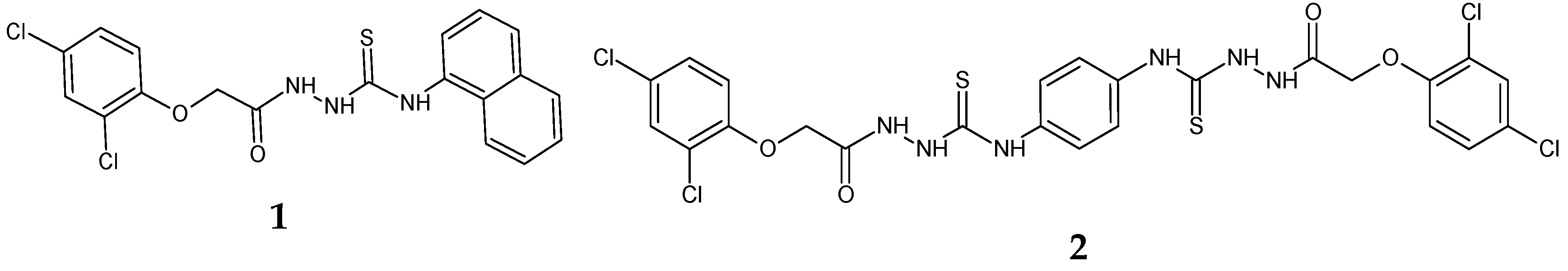
2.1.1. 1-(2,4-Dichlorophenoxy)acetyl-4-(1-naphtyl)thiosemicarbazide (1) [21]
2.1.2. 1-[[2-(2,4-Dichlorophenoxy)acetyl]amino]-3-[4-[[[2-(2,4-dichlorophenoxy)acetyl]amino] carbamoylamino]phenyl]thiourea (2)
2.2. Cell Culturing
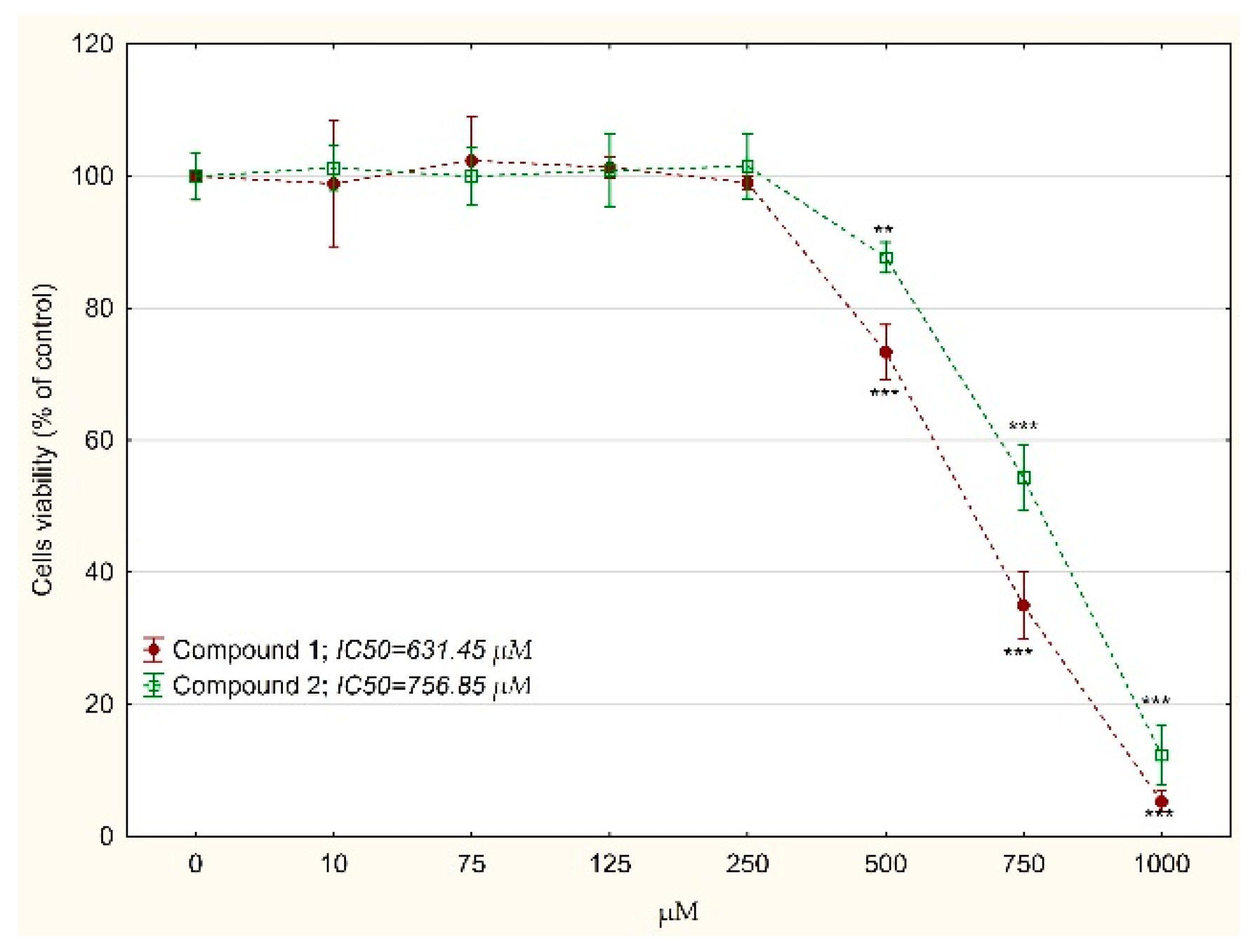
2.3. The Cytotoxicity Analysis
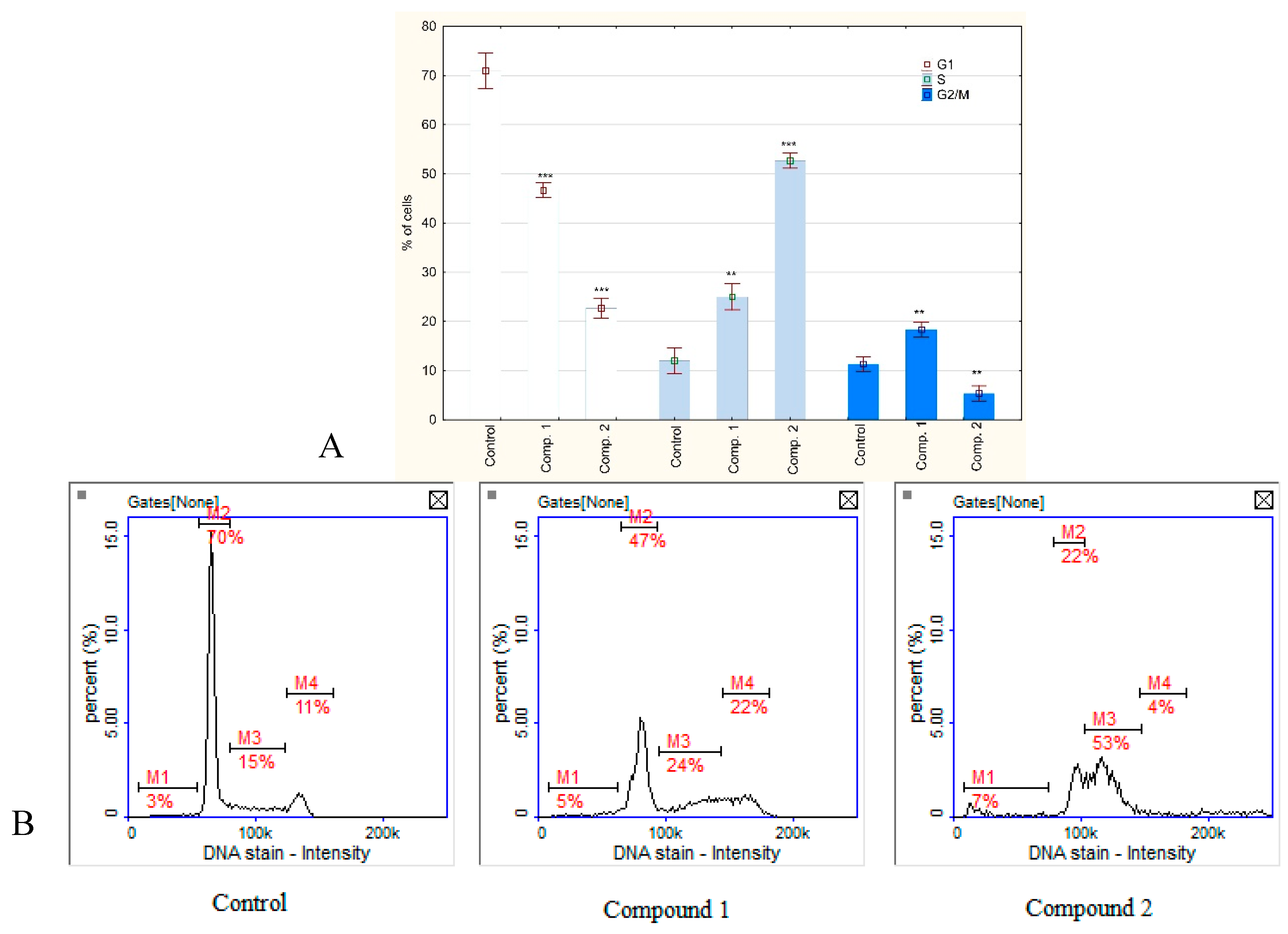
2.4. Cell Cycle Analysis
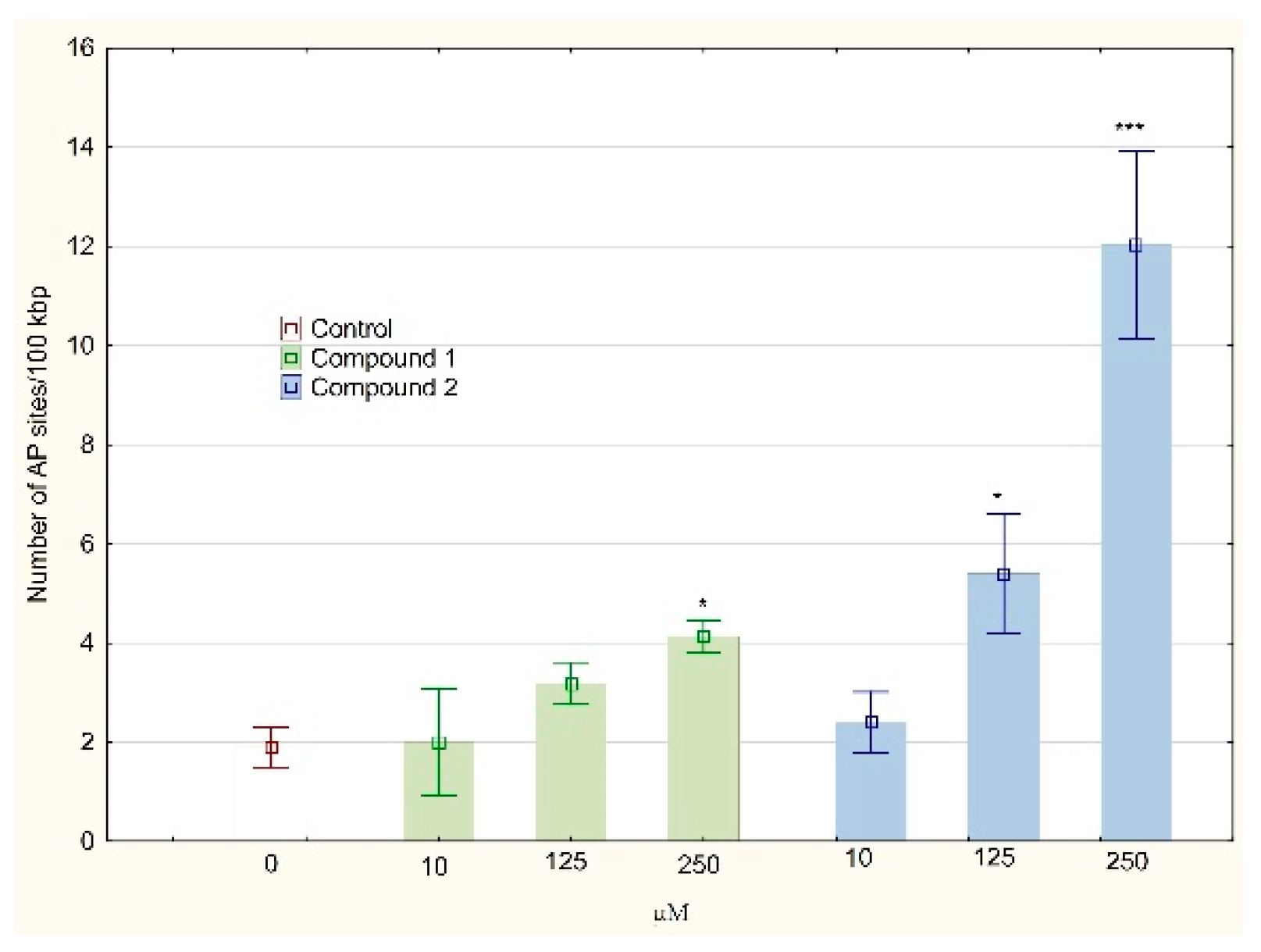
2.5. Determination of DNA Oxidative Damage
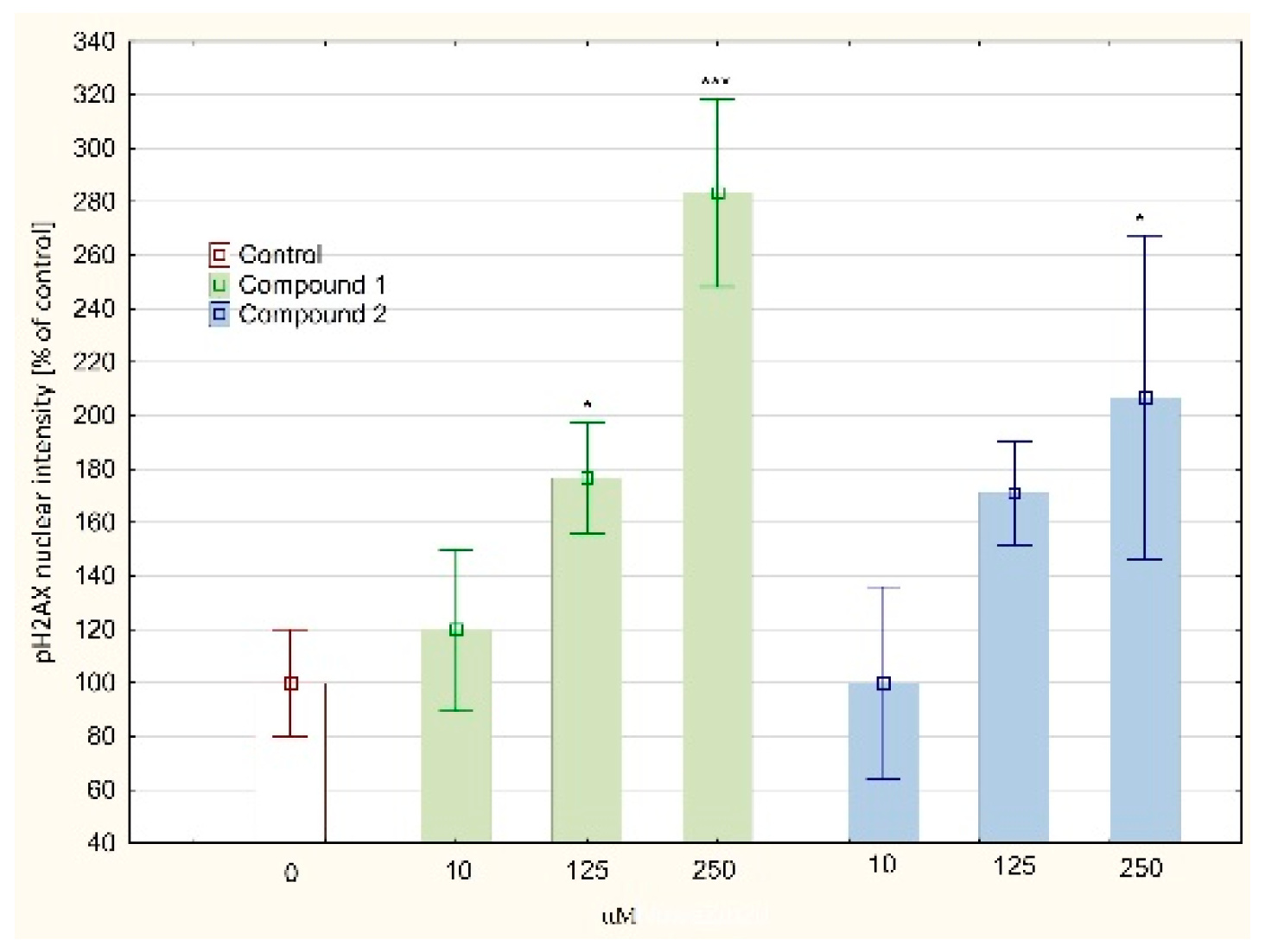
2.6. DNA Damage–Double Strand Breaks (DSB)
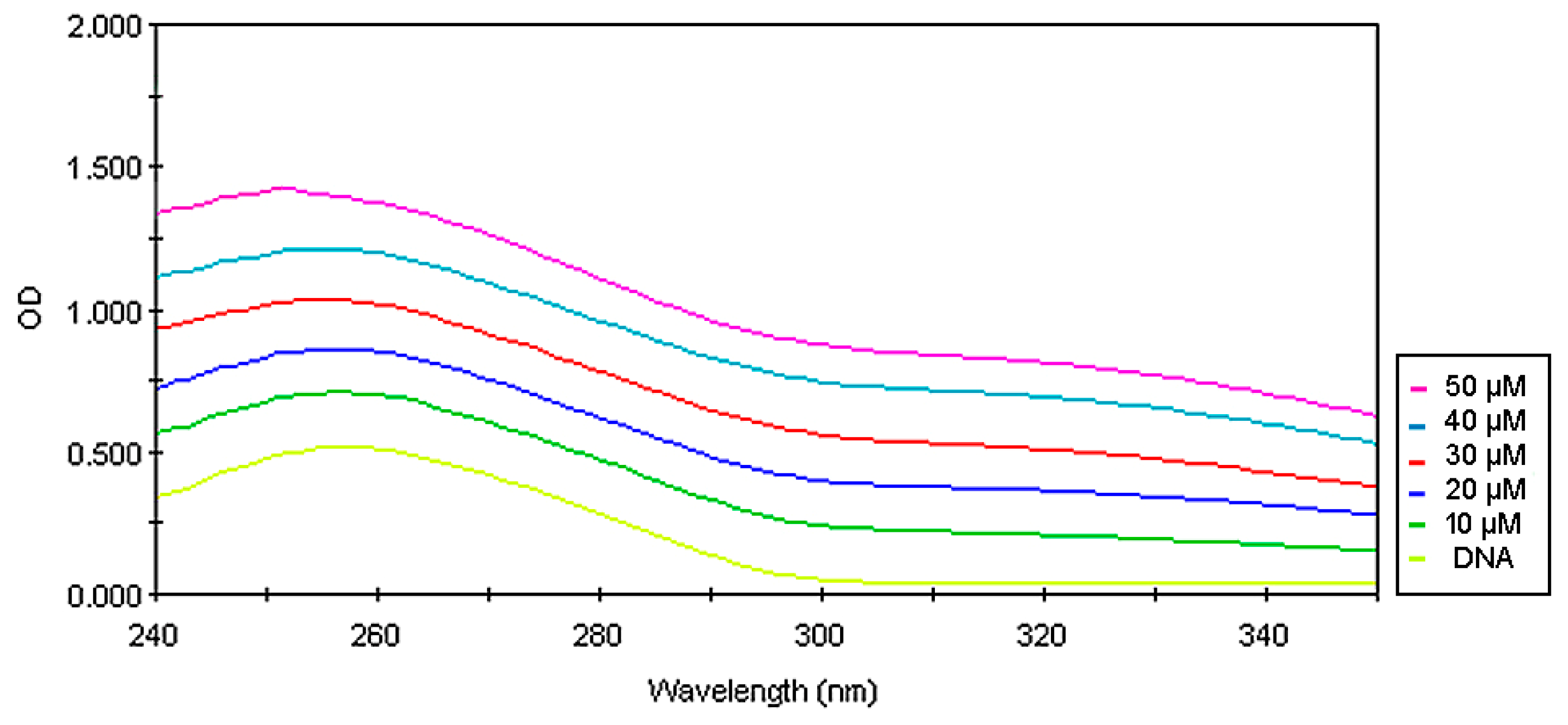
2.7. Spectrophotometry Study on the Interaction of the Compounds with ds-DNA
2.8. Statistical Analysis
2.9. Molecular Modeling
2.9.1. Compound Preparation
2.9.2. Molecular Target
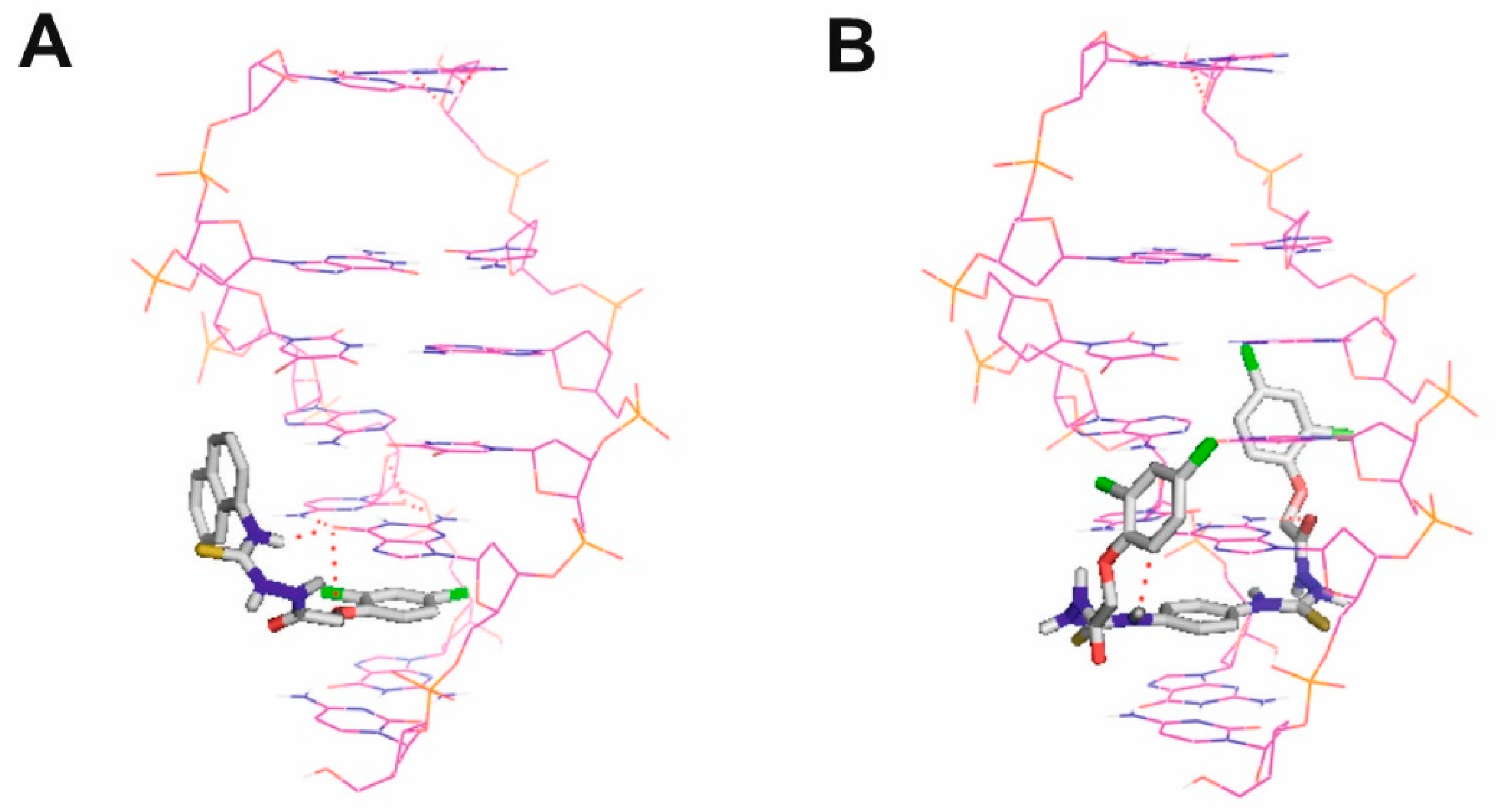
2.9.3. Molecular Docking
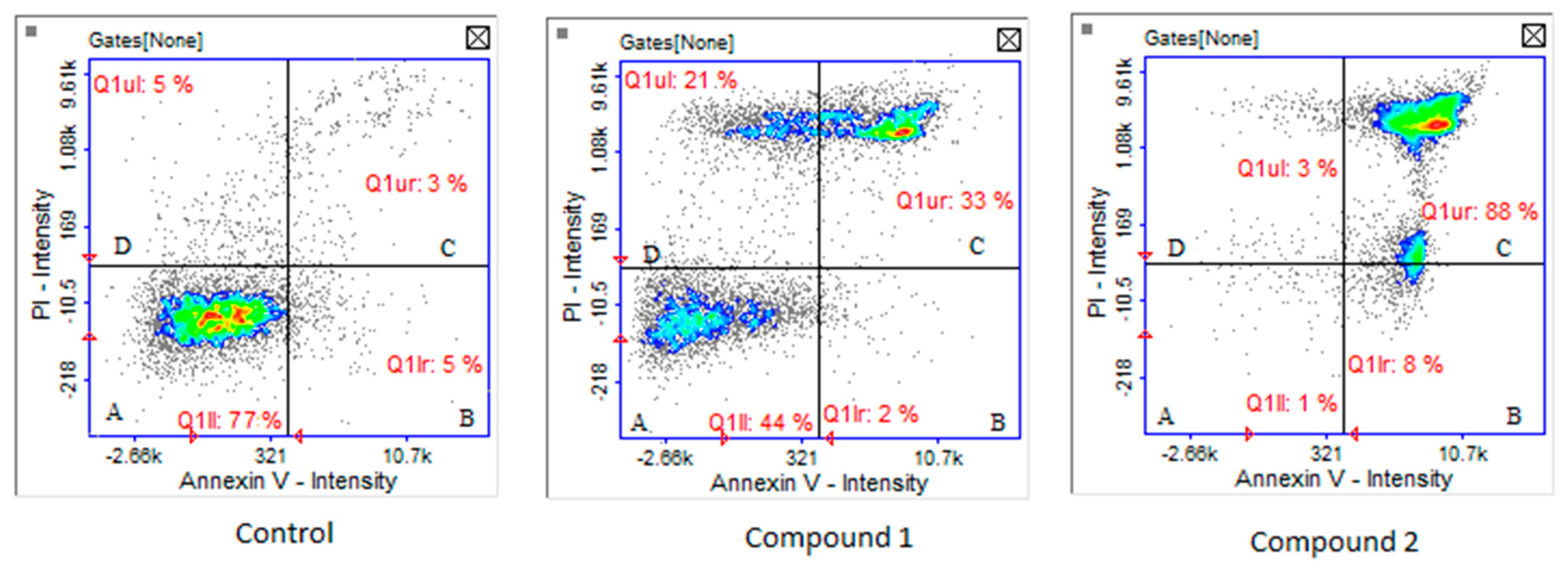
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | abasic sites |
| DAPI | 4′,6-Diamidine-2′-phenylindole dihydrochloride |
| DSB | double strands breaks |
| MTT | Methylthiazolyldiphenyl-tetrazolium bromide |
| ROS | Reactive Oxygen Species |
| SP | Standard Precision |
References
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends-An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef]
- Available online: https://www.who.int/health-topics/cancer (accessed on 14 December 2019).
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, G.J.A.; Maciejewski, R.; Polkowski, W.P. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef]
- Pérez-Herrero, E.; Fernandez-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef]
- Sun, J.; Wei, Q.; Zhou, Y.; Wang, J.; Liu, Q.; Xu, H. A systematic analysis of FDA-approved anticancer drugs. BMC Syst. Boil. 2017, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wu, K.-J.; Liu, J.-B.; Zhou, X.-M.; Leung, C.-H.; Ma, D.-L. A dual-functional molecular strategy for in situ suppressing and visualizing of neuraminidase in aqueous solution using iridium(iii) complexes. Chem. Commun. 2019, 55, 6353–6356. [Google Scholar] [CrossRef]
- Yang, G.-J.; Ko, C.-N.; Zhong, H.-J.; Leung, C.-H.; Ma, D.-L. Structure-Based Discovery of a Selective KDM5A Inhibitor that Exhibits Anti-Cancer Activity via Inducing Cell Cycle Arrest and Senescence in Breast Cancer Cell Lines. Cancers 2019, 11, 92. [Google Scholar] [CrossRef]
- Xiao, M.; Lai, W.; Wang, F.; Li, L.; Fan, C.; Pei, H. Programming Drug Delivery Kinetics for Active Burst Release with DNA Toehold Switches. J. Am. Chem. Soc. 2019, 141, 20354–20364. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Park, O.K.; Kim, J.; Shin, K.; Pack, C.G.; Kim, K.; Ko, G.; Lee, N.; Kwon, S.-H.; Hyeon, T. Deep Tumor Penetration of Drug-Loaded Nanoparticles by Click Reaction-Assisted Immune Cell Targeting Strategy. J. Am. Chem. Soc. 2019, 141, 13829–13840. [Google Scholar] [CrossRef]
- Pitucha, M.; Rzymowska, J.; Olender, A.; Grzybowska-Szatkowska, L. Synthesis of 1,6-hexanediyl-bis(semicarbazides) and 1,6-hexanediyl-bis(1,2,4-triazol-5-ones) and their antiproliferative and antimicrobial activity. J. Serb. Chem. Soc. 2012, 77, 1–8. [Google Scholar] [CrossRef]
- Kaczor, A.A.; Pitucha, M.; Karczmarzyk, Z.; Wysocki, W.; Rzymowska, J.; Matosiuk, D. Structural and molecular docking studies of 4-benzyl-3-[(1-methylpyrrol-2-yl)methyl]-4,5-dihydro-1H-1,2,4-triazol-5-one with anticancer activity. Med. Chem. 2013, 9, 313–328. [Google Scholar] [CrossRef]
- Pitucha, M.; Woś, M.; Miazga-Karska, M.; Klimek, K.; Mirosław, B.; Pachuta-Stec, A.; Gładysz, A.; Ginalska, G. Synthesis, antibacterial and antiproliferative potential of some new 1-pyridinecarbonyl-4-substituted thiosemicarbazide derivatives. Med. Chem. Res. 2016, 25, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Wos, M.; Miazga-Karska, M.; Kaczor, A.A.; Klimek, K.; Karczmarzyk, Z.; Kowalczuk, D.; Wysocki, W.; Ginalska, G.; Urbanczyk-Lipkowska, Z.; Morawiak, M.; et al. Novel thiosemicarbazide derivatives with 4-nitrophenyl group as multi-target drugs: α-glucosidase inhibitors with antibacterial and antiproliferative activity. Biomed. Pharmacother. 2017, 93, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Shefali, A.; Shilpi, A.; Shailey, S. Anticancer activity of thiosemicarbazides/thiosemicarbazones: A review. Int. J. Pharm. Pharm. Sci. 2014, 6, 34–41. [Google Scholar]
- Singhal, S.; Arora, S.; Agarwal, S.; Sharma, R.; Singhal, N. A review on potential biological activities of thiosemicarbazides. WJPPS 2013, 2, 4661–4681. [Google Scholar]
- Reshma, N.J.; Avinash, D.S. Antibacterial activity of thiosemicarbazide derivatives. Der Pharma Chemica 2013, 5, 45–49. [Google Scholar]
- Zhang, X.; Lei, P.; Sun, T.; Jin, X.; Yang, X.; Ling, Y. Design, Synthesis, and Fungicidal Activity of Novel Thiosemicarbazide Derivatives Containing Piperidine Fragments. Molecules 2017, 22, 2085. [Google Scholar] [CrossRef]
- Chen, R.; Huo, L.; Jaiswal, Y.; Huang, J.; Zhong, Z.; Zhong, J.; Williams, L.; Xia, X.; Liang, Y.; Yan, Z. Design, Synthesis, Antimicrobial, and Anticancer Activities of Acridine Thiosemicarbazides Derivatives. Molecules 2019, 24, 2065. [Google Scholar] [CrossRef]
- He, J.; Wang, X.; Zhao, X.; Liang, Y.; He, H.; Fu, L. Synthesis and antitumor activity of novel quinazoline derivatives containing thiosemicarbazide moiety. Eur. J. Med. Chem. 2012, 54, 925–930. [Google Scholar] [CrossRef]
- Pitucha, M.; Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Wos, M.; Chudzik, K.; Ginalska, G.; Fruzinski, A. Synthesis, In Vitro Screening and Docking Studies of New Thiosemicarbazide Derivatives as Antitubercular Agents. Molecules 2019, 24, 251. [Google Scholar] [CrossRef]
- Sen Gupta, A.K.; Misra, H.K. Studies on potential pesticides. Part XIII: Synthesis and evaluation of S-(3-substituted phenoxymethyl-4-aryl/ayclohexyl-4H-1,2,4-triazol-5-yl)-2-mercaptomethylbenzimidazo-les for antibacterial and insecticidal activities. J. Indian. Chem. Soc. 1981, 58, 508–511. [Google Scholar]
- AAT Bioquest, Inc. Quest Graph™ IC50 Calculator. Available online: www.aatbio.com/tools/ic50-calculator (accessed on 14 January 2020).
- Release, S. Schrödinger Release 2019-4: LigPrep; Schrödinger, LLC: New York, NY, USA, 2019. [Google Scholar]
- Kaczor, A.A.; Silva, A.G.; Loza, M.I.; Kolb, P.; Castro, M.; Poso, A. Structure-Based Virtual Screening for Dopamine D 2 Receptor Ligands as Potential Antipsychotics. ChemMedChem 2016, 11, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Kaczor, A.A.; Targowska-Duda, K.M.; Budzyńska, B.; Biała, G.; Silva, A.G.; Castro, M. In vitro, molecular modeling and behavioral studies of 3-{[4-(5-methoxy-1H-indol-3-yl)-1,2,3,6-tetrahydropyridin-1-yl]methyl}-1,2-dihydroquinolin-2-one (D2AAK1) as a potential antipsychotic. Neurochem. Int. 2016, 96, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Kaczor, A.A.; Jörg, M.; Capuano, B. The dopamine D2 receptor dimer and its interaction with homobivalent antagonists: Homology modeling, docking and molecular dynamics. J. Mol. Model. 2016, 22, 203. [Google Scholar] [CrossRef] [PubMed]
- Kondej, M.; Wróbel, T.M.; Silva, A.G.; Stępnicki, P.; Koszła, O.; Kędzierska, E.; Bartyzel, A.; Biała, G.; Matosiuk, D.; Loza, M.I.; et al. Synthesis, pharmacological and structural studies of 5-substituted-3-(1-arylmethyl-1,2,3,6-tetrahydropyridin-4-yl)-1H-indoles as multi-target ligands of aminergic GPCRs. Eur. J. Med. Chem. 2019, 180, 673–689. [Google Scholar] [CrossRef]
- Kondej, M.; Bartyzel, A.; Pitucha, M.; Wróbel, T.M.; Silva, A.G.; Matosiuk, D.; Castro, M.; Kaczor, A.A. Synthesis, Structural and Thermal Studies of 3-(1-Benzyl-1,2,3,6-tetrahydropyridin-4-yl)-5-ethoxy-1H-indole (D2AAK1_3) as Dopamine D2 Receptor Ligand. Molecules 2018, 23, 2249. [Google Scholar] [CrossRef]
- Release, S. Schrödinger Release 2019-4: Epik; Schrödinger, LLC: New York, NY, USA, 2019. [Google Scholar]
- Todd, A.K.; Adams, A.; Thorpe, J.H.; Denny, W.A.; Wakelin, L.P.G.; Cardin, C.J. Major Groove Binding and ‘DNA-Induced’ Fit in the Intercalation of a Derivative of the Mixed Topoisomerase I/II PoisonN-(2-(Dimethylamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray Structure Complexed to d(CG(5-BrU)ACG)2at 1.3-Å Resolution. J. Med. Chem. 1999, 42, 536–540. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Release, S. Schrödinger Release 2019-4: BioLuminate; Schrödinger, LLC: New York, NY, USA, 2019. [Google Scholar]
- The PyMOL Molecular Graphics System; Version 2.0; Schrödinger, LLC: New York, NY, USA, 2019.
- Xu, Z.-Y.; Tang, J.-N.; Xie, H.-X.; Du, Y.-A.; Huang, L.; Yu, P.-F.; Cheng, X.-D. 5-Fluorouracil Chemotherapy of Gastric Cancer Generates Residual Cells with Properties of Cancer Stem Cells. Int. J. Boil. Sci. 2015, 11, 284–294. [Google Scholar] [CrossRef]
- Bathaie, S.Z.; Nikfarjam, L.; Rahmanpour, R.; Moosavi-Movahedi, A. Spectroscopic studies of the interaction of aspirin and its important metabolite, salicylate ion, with DNA, A·T and G·C rich sequences. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 77, 1077–1083. [Google Scholar] [CrossRef]
- Dehghan, G.; Dolatabadi, J.E.N.; Jouyban, A.; Zeynali, K.A.; Ahmadi, S.M.; Kashanian, S. Spectroscopic Studies on the Interaction of Quercetin–Terbium(III) Complex with Calf Thymus DNA. DNA Cell Boil. 2011, 30, 195–201. [Google Scholar] [CrossRef]
- Maga, G.; van Loon, B.; Crespan, E.; Villani, G.; Hübscher, U. The block of DNA polymerase delta strand displacement activity by an abasic site can be rescued by the concerted action of DNA polymerase beta and Flap endonuclease 1. J. Biol. Chem. 2009, 284, 14267–14275. [Google Scholar] [CrossRef] [PubMed]
- Herrick, J.; Bensimon, A. Global regulation of genome duplication in eukaryotes: An overview from the epifluorescence microscope. Chromosoma 2008, 117, 243–260. [Google Scholar] [CrossRef][Green Version]
- Cann, K.L.; Hicks, G.G. Regulation of the cellular DNA double-strand break response. Biochem. Cell Boil. 2007, 85, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Chastain, P.D.; Nakamura, J.; Swenberg, J.; Kaufman, D. Nonrandom AP site distribution in highly proliferative cells. FASEB J. 2006, 20, 2612–2614. [Google Scholar] [CrossRef]
- Gilad, Y.; Senderowitz, H. Docking studies on DNA intercalators. J. Chem. Inf. Model. 2014, 54, 96–107. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitucha, M.; Korga-Plewko, A.; Kozyra, P.; Iwan, M.; Kaczor, A.A. 2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA. Biomolecules 2020, 10, 296. https://doi.org/10.3390/biom10020296
Pitucha M, Korga-Plewko A, Kozyra P, Iwan M, Kaczor AA. 2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA. Biomolecules. 2020; 10(2):296. https://doi.org/10.3390/biom10020296
Chicago/Turabian StylePitucha, Monika, Agnieszka Korga-Plewko, Pawel Kozyra, Magdalena Iwan, and Agnieszka A. Kaczor. 2020. "2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA" Biomolecules 10, no. 2: 296. https://doi.org/10.3390/biom10020296
APA StylePitucha, M., Korga-Plewko, A., Kozyra, P., Iwan, M., & Kaczor, A. A. (2020). 2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA. Biomolecules, 10(2), 296. https://doi.org/10.3390/biom10020296






