Exploration and Characterization of Novel Glycoside Hydrolases from the Whole Genome of Lactobacillus ginsenosidimutans and Enriched Production of Minor Ginsenoside Rg3(S) by a Recombinant Enzymatic Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Strain Isolation and Fermentation of PPD-Mix-Type Major Ginsenosides
2.3. Exploration of Glycoside Hydrolase Gene Through Complete Genome Sequencing
2.3.1. Identification of Target Functional Genes
2.3.2. NCBI Accession Number
2.4. Multilocus Sequence Typing for Identification of Species-Specific Genes
2.5. Target Gene Identification, Cloning, and Expression and Biotransformation of Ginsenosides
Phylogenetic Analysis of BglL.gin-952
2.6. Characterization of Novel Recombinant Bgll.Gin-952 and Purification
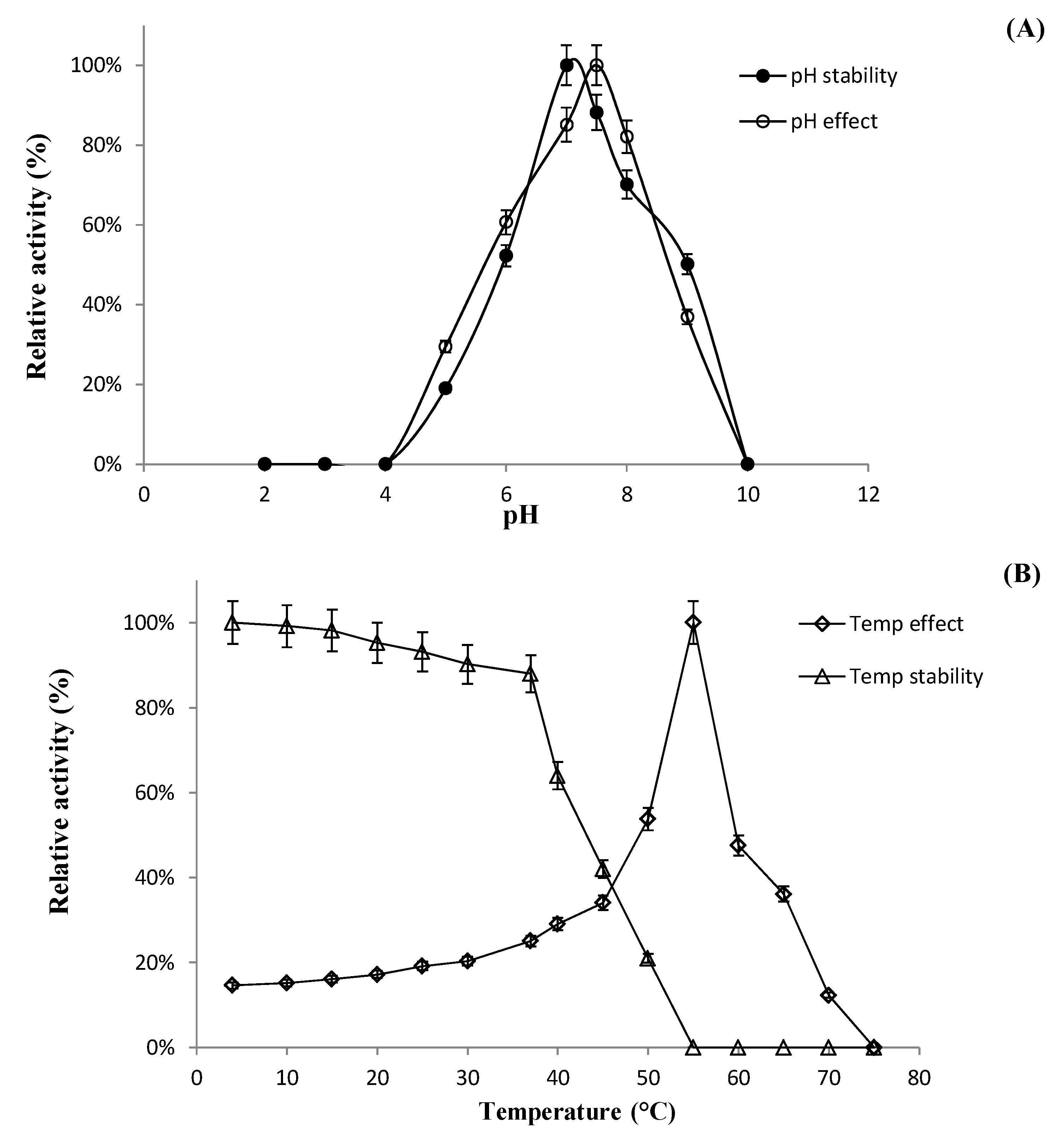
2.6.1. Effect of pH, Temperature, and Metal Ions on Recombinant Enzyme Activity
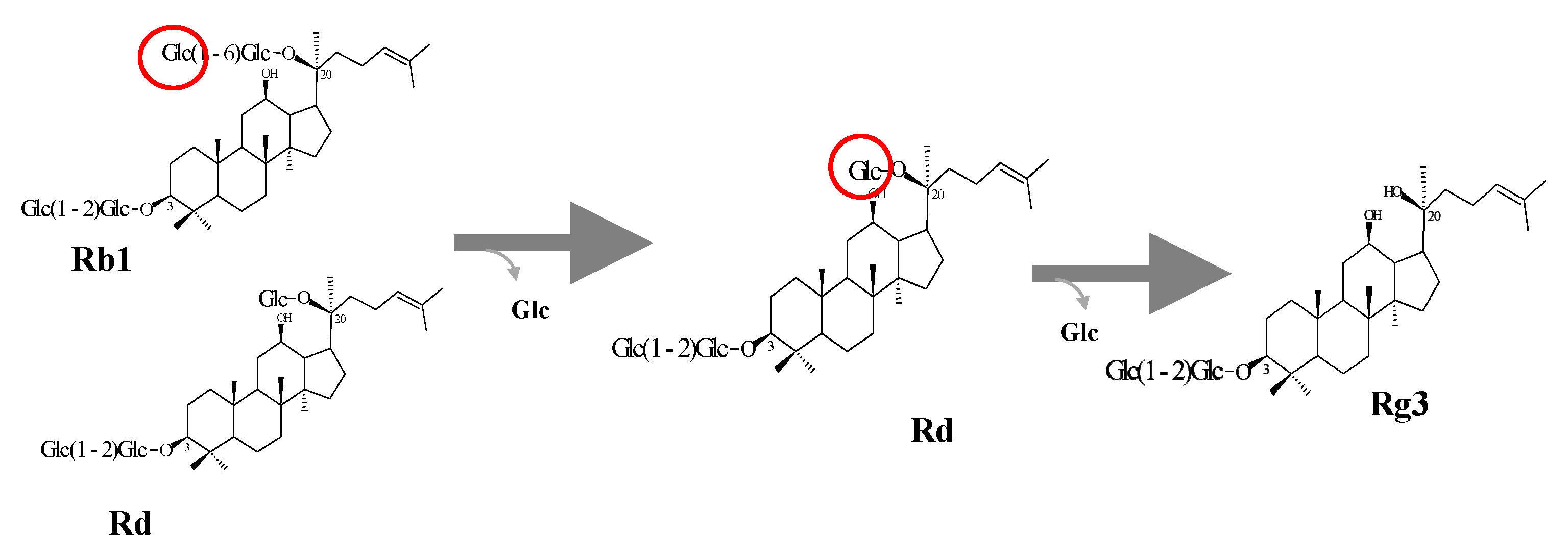
2.6.2. Enzymatic Transformation of Major Ginsenosides by Bgll.Gin-952
2.7. Chromatography
2.7.1. TLC analysis
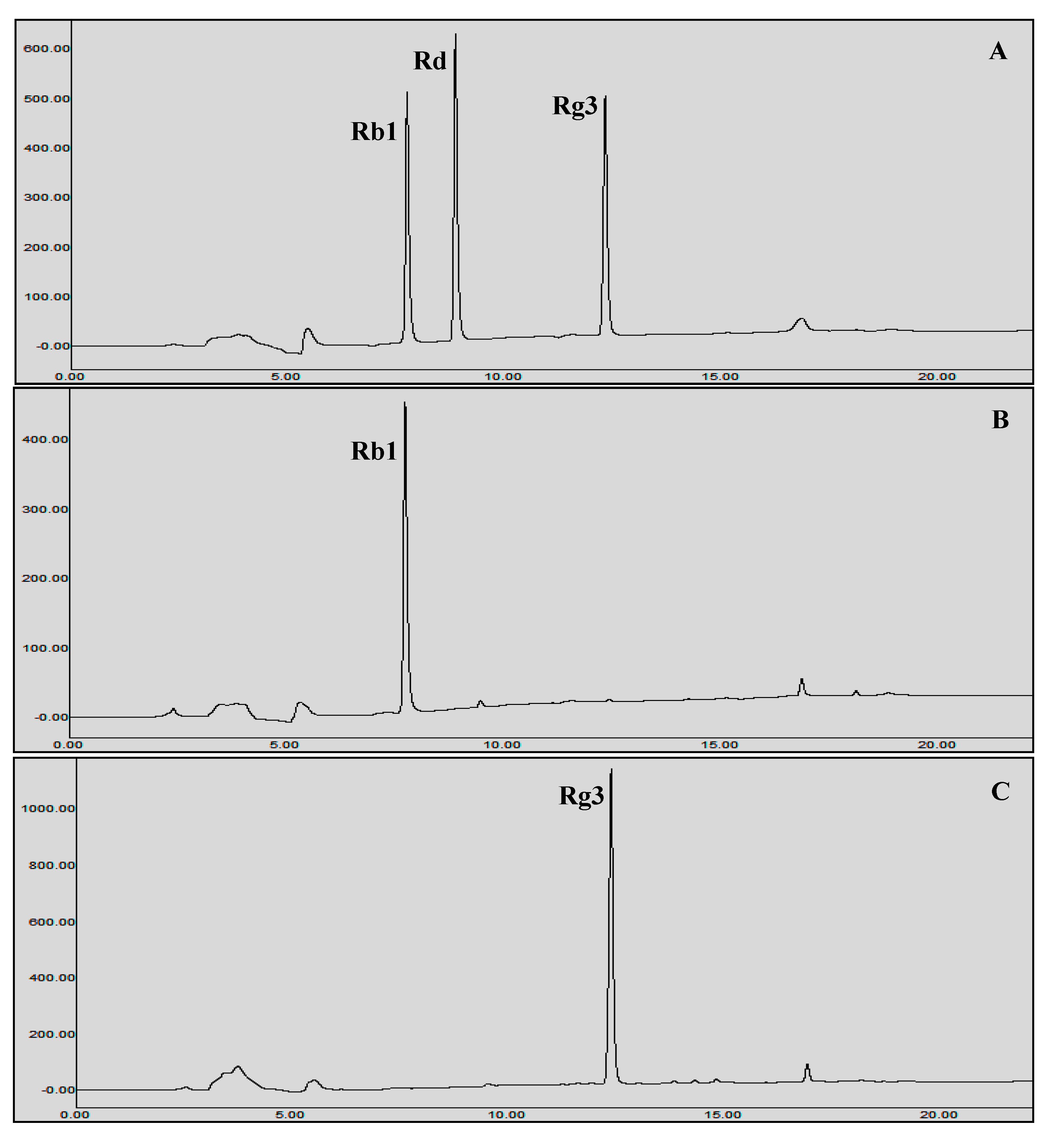
2.7.2. HPLC analysis
3. Results
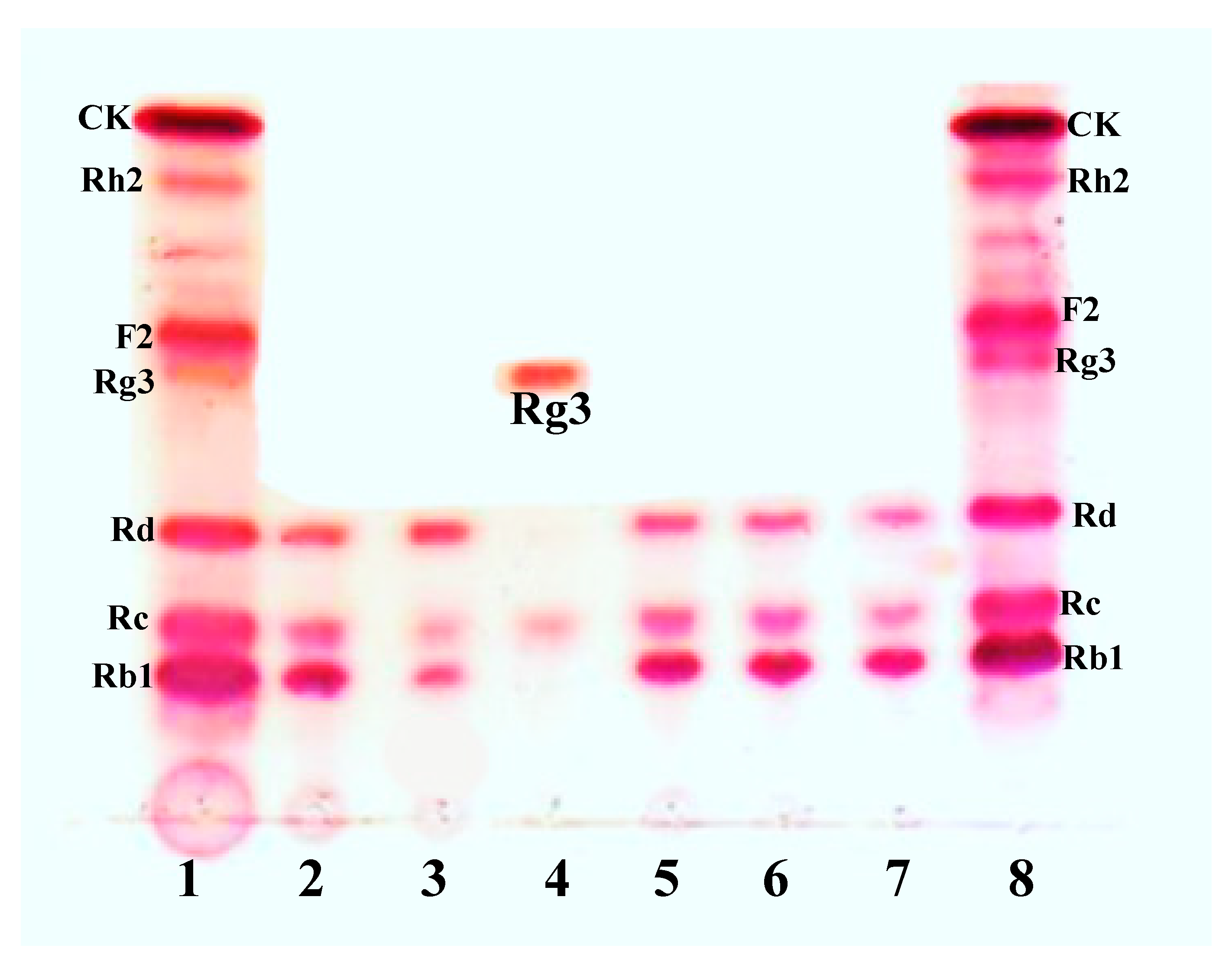
3.1. Bioconversion of PPD Mixtures by L. Ginsenosidimutans EMML 3041T
3.2. Genome Properties
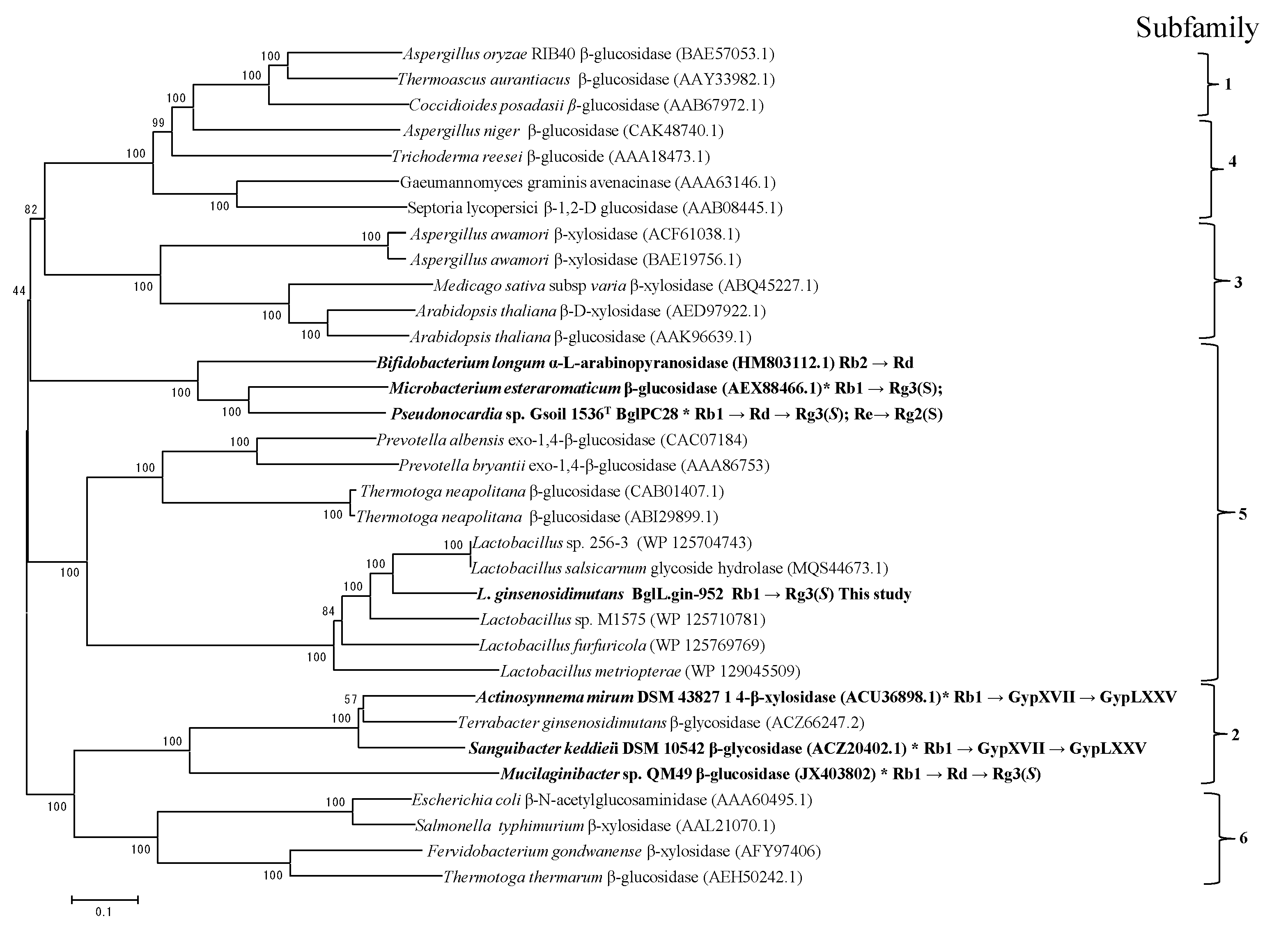
3.3. Phylogenetic Analysis of Sequence of BglL.Gin-952
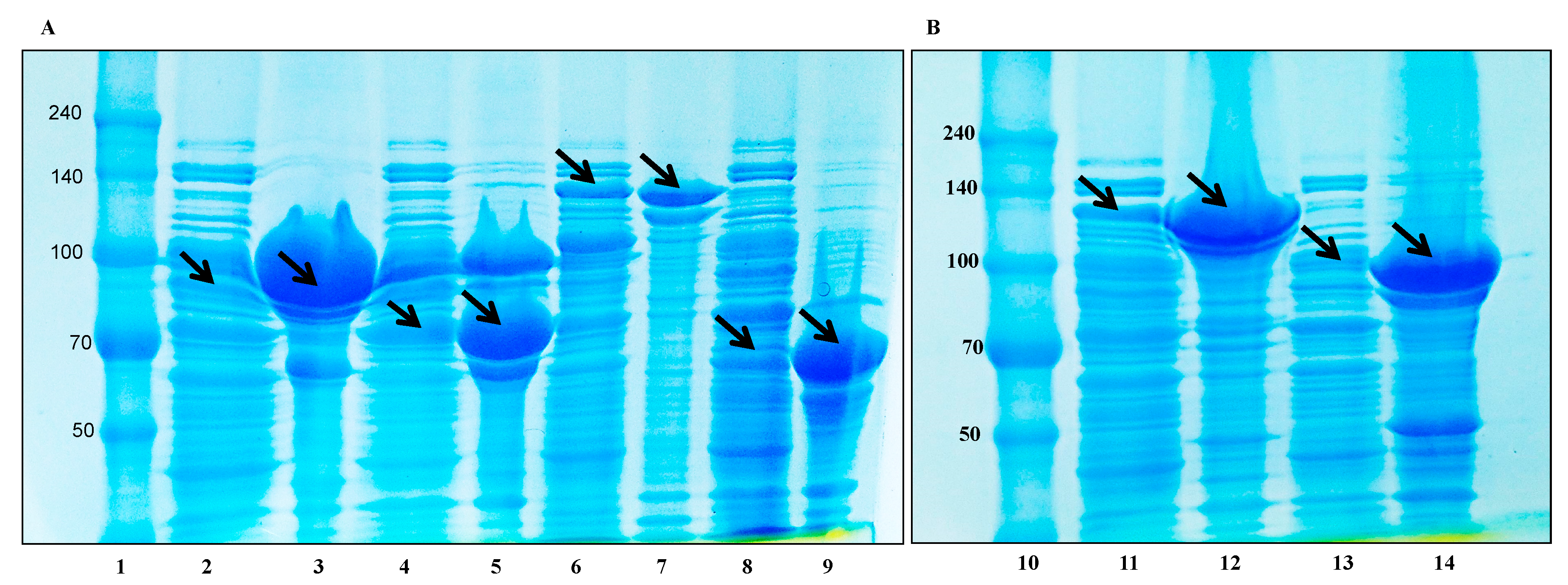
3.4. Molecular Cloning
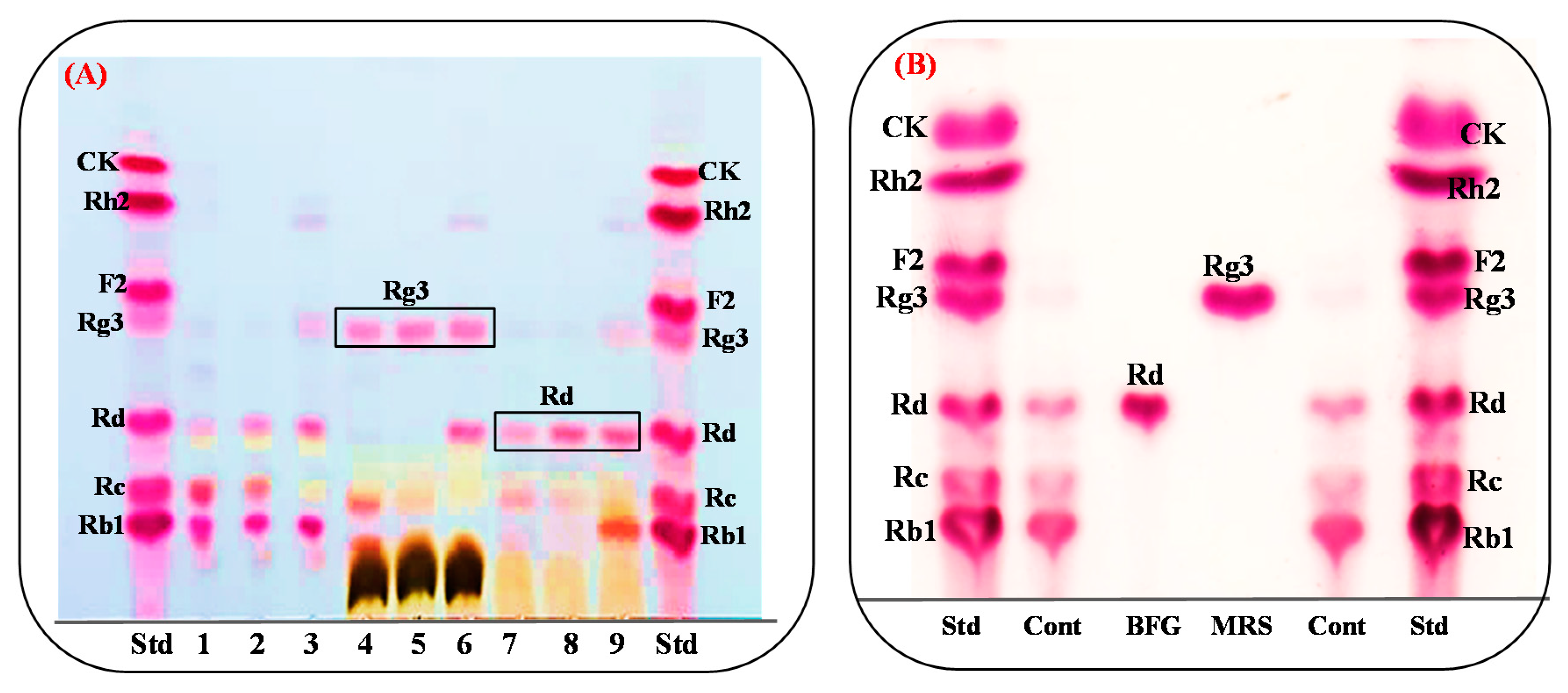
3.5. Protein Expression and Biotransformation of Ginsenosides by Bgll.Gin-952
Characterization of BglL.Gin952
3.6. Scaled-Up Biotransformation of Rg3(S)
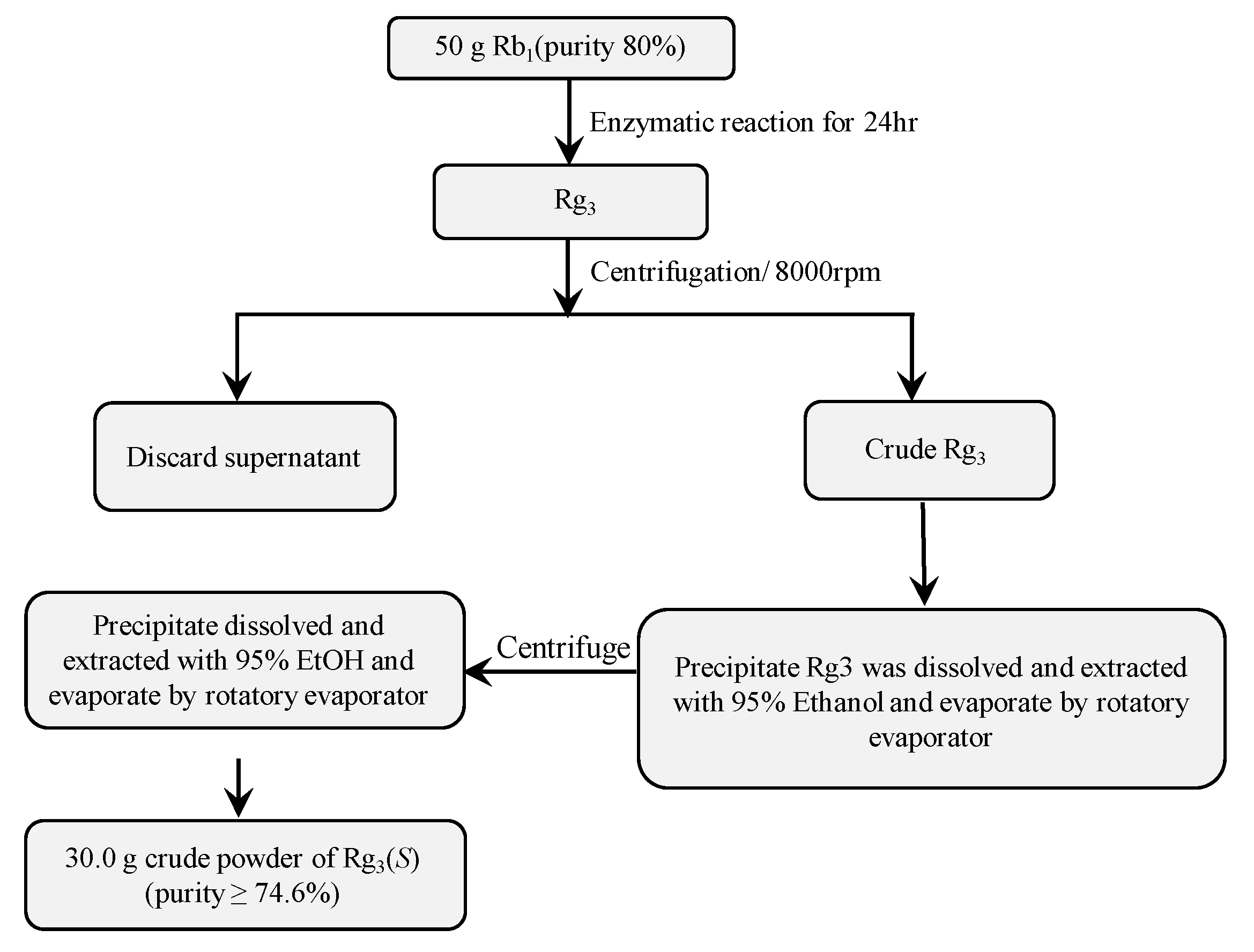
3.6.1. Preparation of High-Cell Density Culture of Recombinant Bgll.Gin-952
3.6.2. Gram Unit Production of Rg3(S)
3.7. Comparision of Rg3(S) Production between Bacterial Cell Fermentation and the Recombinat Enzyme Bgll.Gin952
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jia, L.; Zhao, Y. Current evaluation of the millennium phytomedicine-ginseng (I): Etymology, pharmacognosy, phytochemistry, market and regulations. Curr. Med. Chem. 2009, 16, 2475–2484. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, M.; Gu, J.; Meng, Z.-J.; Zhao, L.-C.; Zheng, Y.-N.; Chen, L.; Yang, G.-L. Hypoglycemic effect of protopanaxadiol-type ginsenosides and compound K on Type 2 Diabetes mice induced by High-Fat Diet combining with Streptozotocin via suppression of hepatic gluconeogenesis. Fitoterapia 2012, 83, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.H.; Siddiqi, M.Z.; Ahn, S.; Kim, Y.-J.; Yang, D.C. Ginsenoside Rh1 induces mouse osteoblast growth and differentiation through the bone morphogenetic protein 2/runt-related gene 2 signalling pathway. J. Pharm. Pharmacol. 2014, 66, 1763–1773. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Siddiqi, M.H.; Kim, Y.-J.; Jin, Y.; Huq, A.; Yang, D.-C. Effect of Fermented Red Ginseng Extract Enriched in Ginsenoside Rg3 on the Differentiation and Mineralization of Preosteoblastic MC3T3-E1 Cells. J. Med. Food 2015, 18, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.S.T.; Che, C.-M.; Leung, K.-W. Recent advances in ginseng as cancer therapeutics: A functional and mechanistic overview. Nat. Prod. Rep. 2015, 32, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Baatar, D.; Siddiqi, M.Z.; Im, W.T.; Khaliq, N.U.; Hwang, S.G. Anti-Inflammatory Effect of Ginsenoside Rh2-Mix on Lipopolysaccharide-Stimulated RAW 264.7 Murine Macrophage Cells. J. Med. Food 2018, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.Z.; Shafi, S.M.; Choi, K.D.; Im, W.-T.; Aslam, Z. Sphingobacterium jejuense sp. nov., with ginsenoside-converting activity, isolated from compost. Int. J. Syst. Evol. Microbiol. 2016, 66, 4433–4439. [Google Scholar] [CrossRef]
- An, D.S.; Cui, C.H.; Siddiqi, M.Z.; Yu, H.S.; Jin, F.X.; Kim, S.G.; Im, W.T. Gram-Scale Production of Ginsenoside F1 Using a Recombinant Bacterial β-Glucosidase. J. Microbiol. Biotechnol. 2017, 27, 1559–1565. [Google Scholar] [CrossRef]
- Christensen, L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv. Food Nutr. Res. 2009, 55, 1–99. [Google Scholar]
- Jia, L.; Zhao, Y.; Liang, X.-J. Current evaluation of the millennium phytomedicine- ginseng (II): Collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Curr. Med. Chem. 2009, 16, 2924–2942. [Google Scholar] [CrossRef]
- Mochizuki, M.; Yoo, Y.; Matsuzawa, K.; Sato, K.; Saiki, I.; Tonooka, S.; Samukawa, K.; Azuma, I. Inhibitory Effect of Tumor Metastasis in Mice by Saponins, Ginsenoside-Rb2, 20(R)- and 20(S)-Ginsenoside-Rg3, of Red ginseng. Boil. Pharm. Bull. 1995, 18, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Keum, Y.-S.; Han, S.S.; Chun, K.-S.; Park, K.-K.; Park, J.-H.; Lee, S.K.; Surh, Y.-J. Inhibitory effects of the ginsenoside Rg3 on phorbol ester-induced cyclooxygenase-2 expression, NF-kappaB activation and tumor promotion. Mutat. Res. Mol. Mech. Mutagen. 2003, 523–524, 75–85. [Google Scholar] [CrossRef]
- Han, B.; Park, M.; Han, Y.; Woo, L.; Sankawa, U.; Yahara, S.; Tanaka, O. Degradation of Ginseng Saponins under Mild Acidic Conditions. Planta Medica 1982, 44, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Iishi, H.; Tatsuta, M.; Baba, M.; Uehara, H.; Nakaizumi, A.; Shinkai, K.; Akedo, H.; Funai, H.; Ishiguro, S.; Kitagawa, I. Inhibition by ginsenoside Rg3 of bombesin-enhanced peritoneal metastasis of intestinal adenocarcinomas induced by azoxymethane in Wistar rats. Clin. Exp. Metastasis 1997, 15, 603–611. [Google Scholar] [CrossRef]
- Kim, D.G.; Jung, K.H.; Lee, D.G.; Yoon, J.H.; Choi, K.S.; Kwon, H.M.; Morgan, M.J.; Howang, S.S.; Kim, Y.S. 20(S)-Ginsenoside Rg3 is a novel inhibitor of autophagy and sensitizes hepatocellular carcinoma to doxorubicin. Oncotarget 2014, 5, 4438–4451. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lee, E.-H.; Ko, S.-R.; Choi, K.-J.; Park, J.-H.; Im, N.-S. Effects of ginsenosides Rg3 and Rh2 on the proliferation of prostate cancer cells. Arch. Pharmacal Res. 2004, 27, 429–435. [Google Scholar] [CrossRef]
- Xu, T.-M.; Xin, Y.; Cui, M.-H.; Jiang, X.; Gu, L.-P. Inhibitory effect of ginsenoside Rg3 combined with cyclophosphamide on growth and angiogenesis of ovarian cancer. Chin. Med. J. 2007, 120, 584–588. [Google Scholar] [CrossRef]
- Tian, J.; Zhang, S.; Li, G.; Liu, Z.; Xu, B. 20(S)-ginsenoside Rg3, a neuroprotective agent, inhibits mitochondrial permeability transition pores in rat brain. Phytotherapy Res. 2009, 23, 486–491. [Google Scholar] [CrossRef]
- In, J.G.; Kim, E.J.; Lee, B.S.; Park, M.H.; Yang, D.C. Saponin analysis and red ginseng production using the simplified method of Korean ginseng (Panax ginseng C.A. Meyer). Korean J. Plant Res. 2006, 19, 133–138. [Google Scholar]
- Jung, H.M.; Liu, Q.M.; Kim, J.W.; Lee, S.T.; Kim, S.C.; Im, W.T. Lactobacillus ginsenosidimutans sp. nov., isolated from kimchi with the ability to transform ginsenosides. Antonie van Leeuwenhoek 2013, 103, 867–876. [Google Scholar] [CrossRef]
- Sawatari, Y.; Hirano, T.; Yokota, A. Development of food grade media for the preparation of Lactobacillus plantarum starter culture. J. Gen. Appl. Microbiol. 2006, 52, 349–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Siddiqi, M.Z.; Muhammad Shafi, S.; Im, W.T. Complete genome sequencing of Arachidicoccus ginsenosidimutans sp. nov., and its application for production of minor ginsenosides by finding a novel ginsenoside-transforming b-glucosidase. RSC Adv. 2017, 7, 46745. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Boil. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Ruan, C.-C.; Zhang, H.; Zhang, L.-X.; Liu, Z.; Sun, G.-Z.; Lei, J.; Qin, Y.-X.; Zheng, Y.-N.; Li, X.; Pan, H.-Y. Biotransformation of Ginsenoside Rf to Rh1 by Recombinant β-Glucosidase. Molecules 2009, 14, 2043–2048. [Google Scholar] [CrossRef]
- Choi, S.-H.; Shin, T.-J.; Hwang, S.-H.; Lee, B.-H.; Kang, J.-Y.; Kim, H.-J.; Oh, J.-W.; Bae, C.-S.; Lee, S.-H.; Nah, S.-Y. Differential Effects of Ginsenoside Metabolites on HERG K+ Channel Currents. J. Ginseng Res. 2011, 35, 191–199. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Cui, C.H.; Park, S.K.; Han, N.S.; Kim, S.C.; Im, W.T. Comparative analysis of the expression level of recombinant ginsenoside-transforming β-glucosidase in GRAS hosts and mass production of the ginsenoside Rh2-Mix. PLoS ONE 2017, 12, e0176098. [Google Scholar] [CrossRef]
- Li, L.; Lee, S.J.; Yuan, Q.P.; Im, W.T.; Kim, S.C.; Han, N.N. Production of bioactive ginsenoside Rg3(S) and compound K using recombinant Lactococcus lactis. J. Ginseng Res. 2018, 24, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.-Q.; Na, J.R.; Bang, M.H.; Kim, M.K.; Yang, D.-C. Conversion of major ginsenoside Rb1 to 20(S)-ginsenoside Rg3 by Microbacterium sp. GS514. Phytochemistry 2008, 69, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.H.; Min, J.W.; Yang, D.U.; Kim, Y.J.; Yang, D.C. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant bglucosidase from Microbacterium esteraromaticum. Appl. Micro Biotech. 2012, 94, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Opassiri, R.; Pomthong, B.; Onkoksoong, T.; Akiyama, T.; Esen, A.; Cairns, J.R.K. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Boil. 2006, 6, 33. [Google Scholar]
- An, D.-S.; Cui, C.-H.; Lee, H.-G.; Wang, L.; Kim, S.C.; Lee, S.-T.; Jin, F.; Yu, H.; Chin, Y.-W.; Lee, H.-K.; et al. Identification and Characterization of a Novel Terrabacter ginsenosidimutans sp. nov. β-Glucosidase That Transforms Ginsenoside Rb1 into the Rare Gypenosides XVII and LXXV. Appl. Environ. Microbiol. 2010, 76, 5827–5836. [Google Scholar]
| Sets of Total Clones | Names of Clone Gene | Total Amino Acids | Common Name | Ginsenoside Conversion Activity | Primers Sequences (F, R) | Vector |
|---|---|---|---|---|---|---|
| 1 | α-glucosidase-556 | 556 | (AglL.gin-556) | - | GGTTCCGCGTGGATCCACAAAAAATTGGTGGCAAGGTT_F AACCTTGCCACCAATTTTTTGTGGATCCACGCGGAACC _R | pGEX 4T-1 |
| 2 | β-glucosidase-484 | 484 | (BglL.gin-484) | - | GGTTCCGCGTGGATCCCGCAAAAATGAGTATCCTTATT_F AATAAGGATACTCATTTTTGCGGGATCCACGCGGAACC_R | |
| 3 | β-glucosidase-902 | 902 | (BglL.gin-902) | - | GGTTCCGCGTGGATCCGAAAGCGAAATGTCACAACGAG_F CTCGTTGTGACATTTCGCTTTCGGATCCACGCGGAACC_R | |
| 4 | β-glucosidase-952 | 952 | (BglL.gin-952) | + | GGTTCCGCGTGGATCCGATAAAAAACAGAAGAGGATTG_F CAATCCTCTTCTGTTTTTTATCGGATCCACGCGGAACC_R | |
| 5 | α-galactosidase-556 | 556 | (AgalL.gin-556) | - | GGTTCCGCGTGGATCCTCTGTTAATGTTAATCAAAGTA_F TACTTTGATTAACATTAACAGAGGATCCACGCGGAACC_R | |
| 6 | α-galactosidase-319 | 319 | (AgalL.gin-319) | - | GGTTCCGCGTGGATCCGATTACACAAATAATAAGTTGC_F GCAACTTATTATTTGTGTAATCGGATCCACGCGGAACC_R | |
| 7 | β-galactosidase-629 | 629 | (BgalL.gin-629) | - | GGTTCCGCGTGGATCCCAAGCTGATATTAACTGGCTCG_F CGAGCCAGTTAATATCAGCTTGGGATCCACGCGGAACC_R |
| Features | Chromosome |
|---|---|
| Length (bp) | 2,590,556 |
| DNA coding region (bp) | 2,271,180 |
| G + C content (%) | 36.71 |
| Genes | 2574 |
| Pseudogenes | 26 |
| CDS | 2486 |
| rRNA gene | 9 |
| tRNA genes | 53 |
| Substrates | Relative Activity ± SD (%) |
|---|---|
| pNP-β-d-glucopyranoside | 100.0 ± 3.3 |
| pNP-β-d-galactopyranoside | 17.0 ± 1.0 |
| pNP-β-d-fucopyranoside | 5.3 ±0.3 |
| pNP-β-n-glucosaminide | ND |
| pNP-β-l-arabinopyranoside | ND |
| pNP-β-d-mannopyranoside | ND |
| pNP-β-d-xylopyranoside | ND |
| pNP-α-d-glucopyranoside | ND |
| pNP-α-d-arabinofuranoside | ND |
| pNP-α-d-arabinopyranoside | 14.5 ± 0.67 |
| pNP-α-d-mannopyranoside | ND |
| pNP-α-d-mannopyranoside | ND |
| oNP-β-d-glucopyranoside | ND |
| oNP-β-d-fucopyranoside | ND |
| oNP-α-d-galactopyranoside | ND |
| Metal Ions or Reagents | Relative Activity ± SD (%) at: | |
|---|---|---|
| 1 mM | 10 mM | |
| NaCl | 80.0 ± 6.7 | 85.6 ± 1.3 |
| KCl | 87.0 ± 0.7 | 85.3 ± 3.7 |
| CaCl2 | 81.3 ± 6.1 | 122.1 ± 0.9 |
| MgCl2 | 82.9 ± 0.2 | 71.6 ± 3.3 |
| CoCl2 | 84.2 ± 0.4 | 132.0 ± 3.9 |
| MnSO4 | 88.3 ± 4.0 | 155.5 ± 5.8 |
| MgSO4 | 91.6 ± 2.4 | 79.8 ± 0.3 |
| β-Mercaptoethanol | 88.6 ± 3.0 | 93.9 ± 0.3 |
| EDTA | 84.4 ± 3.8 | 78.7± 3.7 |
| Control | 100.0 ± 2.1 | 100.0 ± 5.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqi, M.Z.; Srinivasan, S.; Park, H.Y.; Im, W.-T. Exploration and Characterization of Novel Glycoside Hydrolases from the Whole Genome of Lactobacillus ginsenosidimutans and Enriched Production of Minor Ginsenoside Rg3(S) by a Recombinant Enzymatic Process. Biomolecules 2020, 10, 288. https://doi.org/10.3390/biom10020288
Siddiqi MZ, Srinivasan S, Park HY, Im W-T. Exploration and Characterization of Novel Glycoside Hydrolases from the Whole Genome of Lactobacillus ginsenosidimutans and Enriched Production of Minor Ginsenoside Rg3(S) by a Recombinant Enzymatic Process. Biomolecules. 2020; 10(2):288. https://doi.org/10.3390/biom10020288
Chicago/Turabian StyleSiddiqi, Muhammad Zubair, Sathiyaraj Srinivasan, Hye Yoon Park, and Wan-Taek Im. 2020. "Exploration and Characterization of Novel Glycoside Hydrolases from the Whole Genome of Lactobacillus ginsenosidimutans and Enriched Production of Minor Ginsenoside Rg3(S) by a Recombinant Enzymatic Process" Biomolecules 10, no. 2: 288. https://doi.org/10.3390/biom10020288
APA StyleSiddiqi, M. Z., Srinivasan, S., Park, H. Y., & Im, W.-T. (2020). Exploration and Characterization of Novel Glycoside Hydrolases from the Whole Genome of Lactobacillus ginsenosidimutans and Enriched Production of Minor Ginsenoside Rg3(S) by a Recombinant Enzymatic Process. Biomolecules, 10(2), 288. https://doi.org/10.3390/biom10020288






