Comparing Interfacial Trp, Interfacial His and pH Dependence for the Anchoring of Tilted Transmembrane Helical Peptides
Abstract
1. Introduction
2. Materials and Methods
2.1. Synthesis and Purification of 2H-labeled Peptides
2.2. Circular Dichroism (CD) Spectroscopy
2.3. 2H and 31P NMR Spectroscopy Using Oriented Bilayer Samples
2.4. Analysis of Helix Orientation and Dynamics from 2H NMR Data
3. Results
3.1. Comparison of H with G, K, R and W at Positions 2 and 22
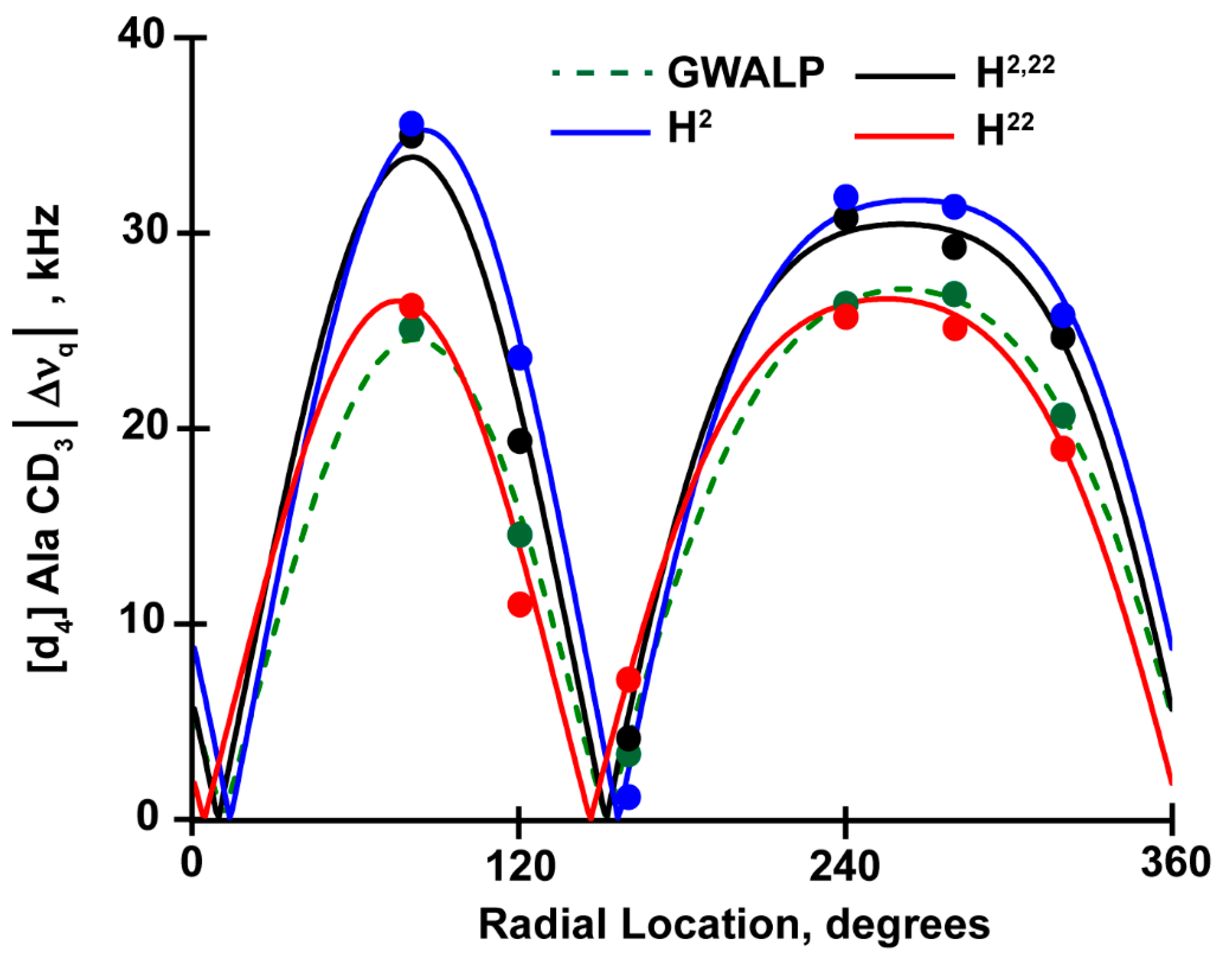
3.2. Comparison of H2,22 with H2 and H22
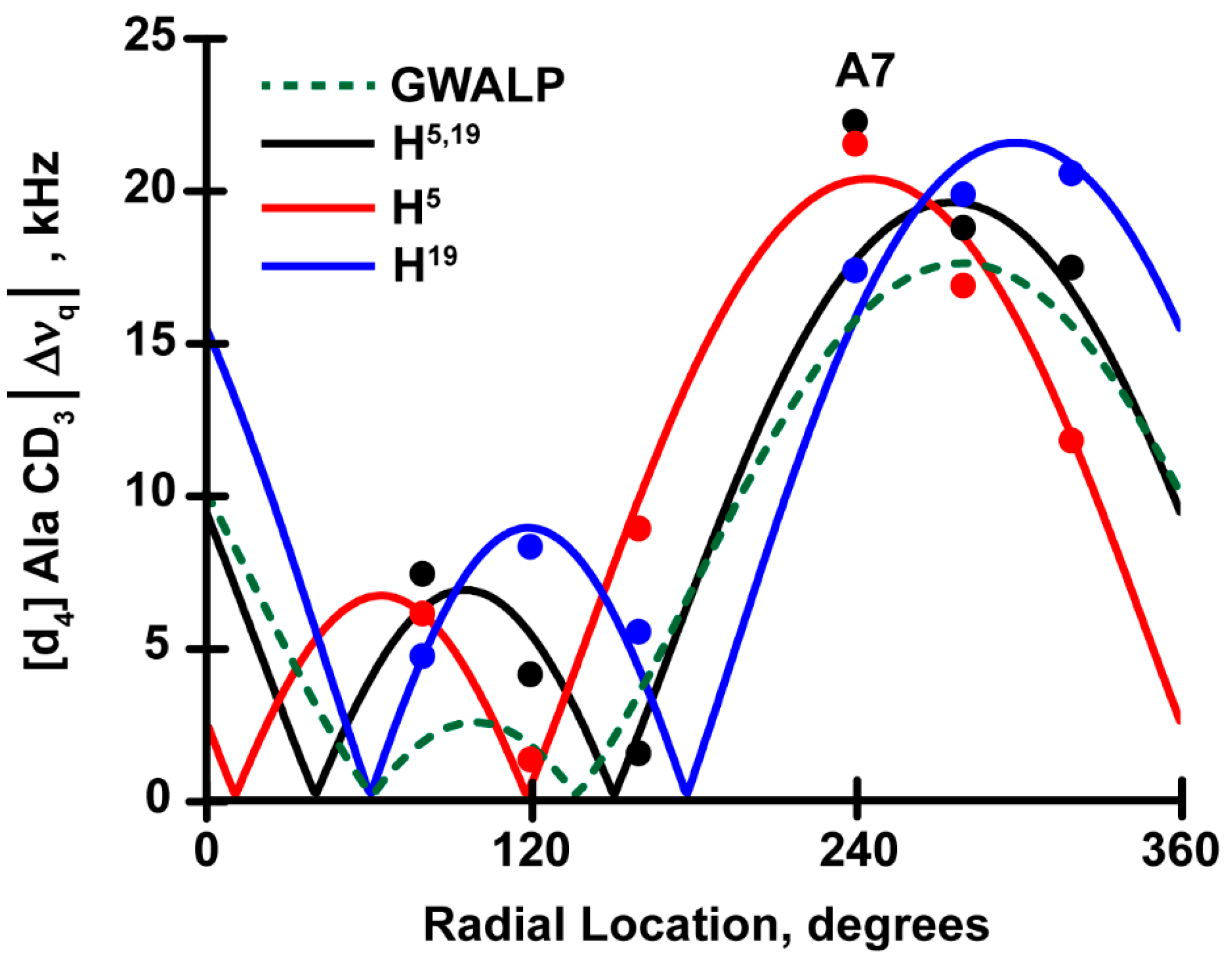
3.3. Comparison of H5,19 with W5,19 and mixed Interfacial Pairs, H5W19 and W5H19
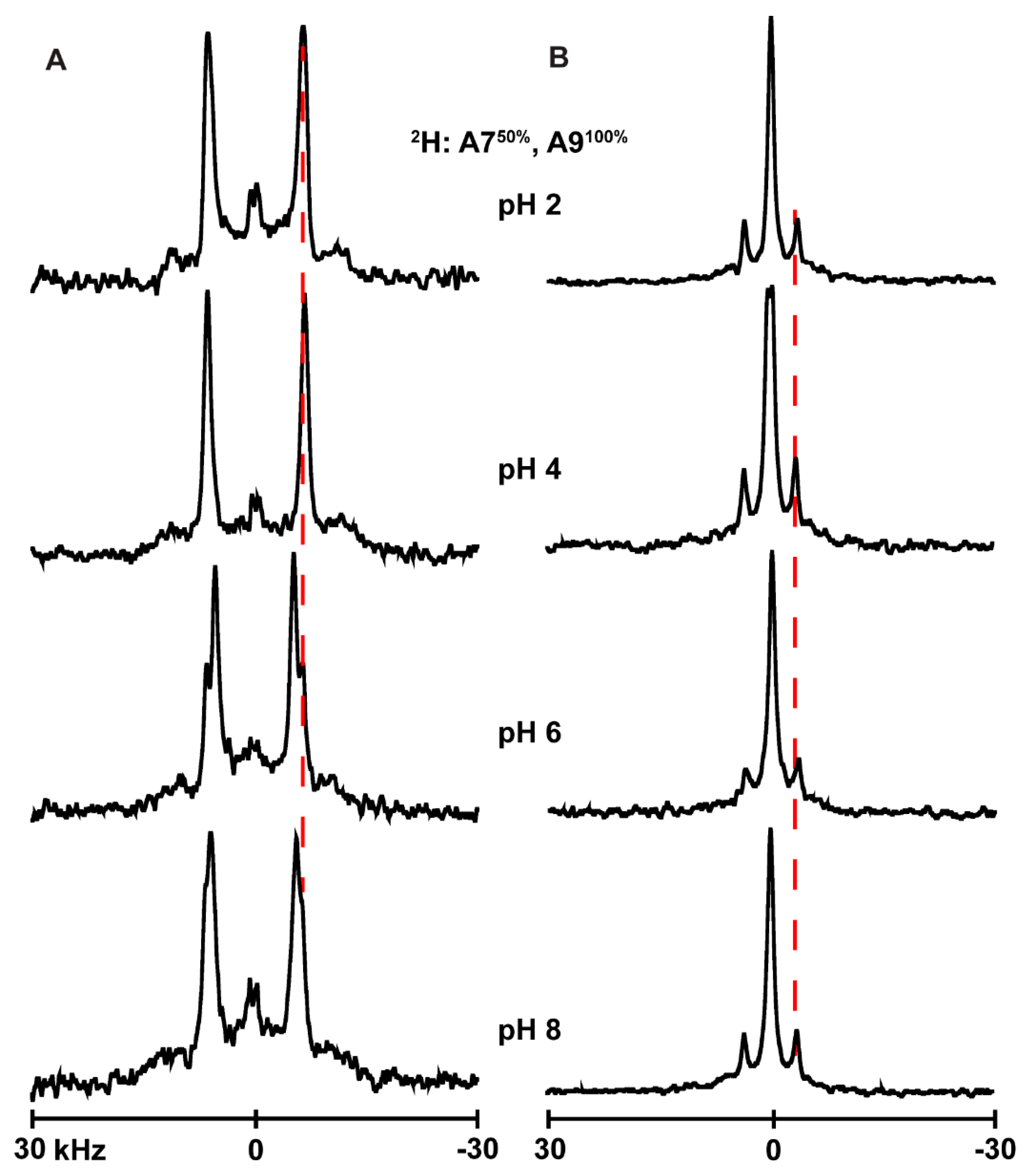
3.4. Ionization of Histidines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ulmschneider, M.B.; Sansom, M.S.P. Amino acid distributions in integral membrane protein structures. BBA-Biomembranes 2001, 1512, 1–14. [Google Scholar] [CrossRef]
- Wimley, W.C. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci. 2002, 11, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Granseth, E.; von Heijne, G.; Elofsson, A. A Study of the Membrane–Water Interface Region of Membrane Proteins. J. Mol. Biol. 2005, 346, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ulmschneider, M.B.; Sansom, M.S.P.; Di Nola, A. Properties of integral membrane protein structures: Derivation of an implicit membrane potential. Proteins: Struct. Func. Bioinformatics 2005, 59, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Adamian, L.; Nanda, V.; DeGrado, W.F.; Liang, J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins: Struct. Func. Bioinformatics 2005, 59, 496–509. [Google Scholar] [CrossRef]
- Senes, A.; Chadi, D.C.; Law, P.B.; Walters, R.F.S.; Nanda, V.; DeGrado, W.F. Ez, a Depth-dependent Potential for Assessing the Energies of Insertion of Amino Acid Side-chains into Membranes: Derivation and Applications to Determining the Orientation of Transmembrane and Interfacial Helices. J. Mol. Biol. 2007, 366, 436–448. [Google Scholar] [CrossRef]
- Landolt-Marticorena, C.; Williams, K.A.; Deber, C.M.; Reithmeier, R.A. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 1993, 229, 602–608. [Google Scholar] [CrossRef]
- Wallin, E.; von Heijne, G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998, 7, 1029–1038. [Google Scholar] [CrossRef]
- Wimley, W.C.; White, S.H. Membrane partitioning: Distinguishing bilayer effects from the hydrophobic effect. Biochemistry 1993, 32, 6307–6312. [Google Scholar] [CrossRef]
- Wimley, W.C.; Creamer, T.P.; White, S.H. Solvation Energies of Amino Acid Side Chains and Backbone in a Family of Host–Guest Pentapeptides. Biochemistry 1996, 35, 5109–5124. [Google Scholar] [CrossRef]
- Yau, W.-M.; Wimley, W.C.; Gawrisch, K.; White, S.H. The Preference of Tryptophan for Membrane Interfaces. Biochemistry 1998, 37, 14713–14718. [Google Scholar] [CrossRef] [PubMed]
- Vostrikov, V.V.; Grant, C.V.; Daily, A.E.; Opella, S.J.; Koeppe, R.E., II. Comparison of "Polarization inversion with spin exchange at magic angle" and "geometric analysis of labeled alanines" methods for transmembrane helix alignment. J. Am. Chem. Soc. 2008, 130, 12584–12585. [Google Scholar] [CrossRef] [PubMed]
- Vostrikov, V.V.; Daily, A.E.; Greathouse, D.V.; Koeppe, R.E., II. Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J. Biol. Chem. 2010, 285, 31723–31730. [Google Scholar] [CrossRef]
- Killian, J.A.; Salemink, I.; De Planque, M.R.; Lindblom, G.; Koeppe, R.E., II; Greathouse, D.V. Induction of non-bilayer structures in diacylphosphatidylcholine model membranes by transmembrane α-helical peptides. Importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry 1996, 35, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Strandberg, E.; Ozdirekcan, S.; Rijkers, D.T.S.; van der Wel, P.C.A.; Koeppe, R.E., II; Liskamp, R.M.J.; Killian, J.A. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by H-2 solid-state NMR. Biophys. J. 2004, 86, 3709–3721. [Google Scholar] [CrossRef]
- van der Wel, P.C.; Strandberg, E.; Killian, J.A.; Koeppe, R.E., II. Geometry and intrinsic tilt of a tryptophan-anchored transmembrane alpha-helix determined by 2H NMR. Biophys. J. 2002, 83, 1479–1488. [Google Scholar] [CrossRef]
- Vostrikov, V.V.; Grant, C.V.; Opella, S.J.; Koeppe, R.E., II. On the combined analysis of (2)H and (1)(5)N/(1)H solid-state NMR data for determination of transmembrane peptide orientation and dynamics. Biophys. J. 2011, 101, 2939–2947. [Google Scholar] [CrossRef]
- Strandberg, E.; Esteban-Martin, S.; Ulrich, A.S.; Salgado, J. Hydrophobic mismatch of mobile transmembrane helices: Merging theory and experiments. Biochim. Biophys. Acta 2012, 1818, 1242–1249. [Google Scholar] [CrossRef]
- Sparks, K.A.; Gleason, N.J.; Gist, R.; Langston, R.; Greathouse, D.V.; Koeppe, R.E., II. Comparisons of interfacial Phe, Tyr, and Trp residues as determinants of orientation and dynamics for GWALP transmembrane peptides. Biochemistry 2014, 53, 3637–3645. [Google Scholar] [CrossRef]
- Martfeld, A.N.; Greathouse, D.V.; Koeppe, R.E., II. Ionization Properties of Histidine Residues in the Lipid Bilayer Membrane Environment. J. Biol. Chem. 2016, 291, 19146–19156. [Google Scholar] [CrossRef]
- Lee, S.A.; Eyeson, R.; Cheever, M.L.; Geng, J.; Verkhusha, V.V.; Burd, C.; Overduin, M.; Kutateladze, T.G. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA 2005, 102, 13052. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, T.; Mueller, D.S.; Mark, A.E.; Young, P.R.; Kobe, B. The Role of Histidine Residues in Low-pH-Mediated Viral Membrane Fusion. Structure 2006, 14, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- Bustad, H.J.; Skjaerven, L.; Ying, M.; Halskau, Ø.; Baumann, A.; Rodriguez-Larrea, D.; Costas, M.; Underhaug, J.; Sanchez-Ruiz, J.M.; Martinez, A. The peripheral binding of 14–3-3γ to membranes involves isoform-specific histidine residues. PloS ONE 2012, 7, e49671. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, J.; Andersson, H.; Nilsson, I.; White, S.H.; von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.P.; Fleming, K.G. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc. Natl. Acad. Sci. USA 2011, 108, 10174. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Gessmann, D.; Naveed, H.; Liang, J. Outer Membrane Protein Folding and Topology from a Computational Transfer Free Energy Scale. J. Am. Chem. Soc. 2016, 138, 2592–2601. [Google Scholar] [CrossRef] [PubMed]
- Iyer, B.R.; Vetal, P.V.; Noordeen, H.; Zadafiya, P.; Mahalakshmi, R. Salvaging the Thermodynamic Destabilization of Interface Histidine in Transmembrane β-Barrels. Biochemistry 2018, 57, 6669–6678. [Google Scholar] [CrossRef]
- Gleason, N.J.; Greathouse, D.V.; Grant, C.V.; Opella, S.J.; Koeppe, R.E., II. Single Tryptophan and Tyrosine Comparisons in the N-terminal and C-terminal Interface Regions of Transmembrane GWALP Peptides. J. Phys. Chem. B 2013, 117, 13786–13794. [Google Scholar] [CrossRef][Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Kortenaar, P.B.W.; Dijk, B.G.; Peeters, J.M.; Raagen, B.J.; Adams, P.J.; Tesser, G.I. Rapid and efficient method for the preparation of Fmoc-amino acids starting from 9-fluorenylmethanol. Int. J. Pept. Prot. Res. 1986, 27, 398–400. [Google Scholar] [CrossRef]
- Greathouse, D.V.; Koeppe, R.E., II; Providence, L.L.; Shobana, S.; Andersen, O.S. Design and characterization of gramicidin channels. Methods Enzymol. 1999, 294, 525–550. [Google Scholar]
- Davis, J.H.; Jeffrey, K.R.; Valic, M.I.; Bloom, M.; Higgs, T.P. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett. 1976, 390–394. [Google Scholar] [CrossRef]
- Aisenbrey, C.; Bechinger, B. Investigations of Polypeptide Rotational Diffusion in Aligned Membranes by 2H and 15N Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2004, 126, 16676–16683. [Google Scholar] [CrossRef] [PubMed]
- Afrose, F.; McKay, M.J.; Mortazavi, A.; Suresh Kumar, V.; Greathouse, D.V.; Koeppe, R.E., II. Transmembrane Helix Integrity versus Fraying To Expose Hydrogen Bonds at a Membrane-Water Interface. Biochemistry 2019, 58, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Rajagopalan, V.; Sparks, K.A.; Greathouse, D.V.; Koeppe, R.E., II. Juxta-terminal Helix Unwinding as a Stabilizing Factor to Modulate the Dynamics of Transmembrane Helices. ChemBioChem 2016, 17, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Strandberg, E.; Grau-Campistany, A.; Wadhwani, P.; Burck, J.; Rabanal, F.; Ulrich, A.S. Helix Fraying and Lipid-Dependent Structure of a Short Amphipathic Membrane-Bound Peptide Revealed by Solid-State NMR. J. Phys. Chem. B 2018, 122, 6236–6250. [Google Scholar] [CrossRef] [PubMed]
- Gleason, N.J.; Vostrikov, V.V.; Greathouse, D.V.; Koeppe, R.E., II. Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc. Natl. Acad. Sci. USA 2013, 110, 1692–1695. [Google Scholar] [CrossRef]
- Gleason, N.J.; Vostrikov, V.V.; Greathouse, D.V.; Grant, C.V.; Opella, S.J.; Koeppe, R.E., II. Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry 2012, 51, 2044–2053. [Google Scholar] [CrossRef]
- Tang, Y.; Zaitseva, F.; Lamb, R.A.; Pinto, L.H. The Gate of the Influenza Virus M2 Proton Channel Is Formed by a Single Tryptophan Residue. J. Biol. Chem. 2002, 277, 39880–39886. [Google Scholar] [CrossRef]
- Venkataraman, P.; Lamb, R.A.; Pinto, L.H. Chemical Rescue of Histidine Selectivity Filter Mutants of the M2 Ion Channel of Influenza A Virus. J. Biol. Chem. 2005, 280, 21463–21472. [Google Scholar] [CrossRef]
- Okada, A.; Miura, T.; Takeuchi, H. Protonation of Histidine and Histidine–Tryptophan Interaction in the Activation of the M2 Ion Channel from Influenza A Virus. Biochemistry 2001, 40, 6053–6060. [Google Scholar] [CrossRef]
- Takeuchi, H.; Okada, A.; Miura, T. Roles of the histidine and tryptophan side chains in the M2 proton channel from influenza A virus. FEBS Lett. 2003, 552, 35–38. [Google Scholar] [CrossRef]
- Paterson, R.G.; Takeda, M.; Ohigashi, Y.; Pinto, L.H.; Lamb, R.A. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology 2003, 306, 7–17. [Google Scholar] [CrossRef]
- Mould, J.A.; Paterson, R.G.; Takeda, M.; Ohigashi, Y.; Venkataraman, P.; Lamb, R.A.; Pinto, L.H. Influenza B Virus BM2 Protein Has Ion Channel Activity that Conducts Protons across Membranes. Developmental Cell 2003, 5, 175–184. [Google Scholar] [CrossRef]
- Fernández-Recio, J.; Vázquez, A.; Civera, C.; Sevilla, P.; Sancho, J. The Tryptophan/Histidine interaction in α-helices. J. Mol. Biol. 1997, 267, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Otomo, K.; Toyama, A.; Miura, T.; Takeuchi, H. Interactions Between Histidine and Tryptophan Residues in the BM2 Proton Channel from Influenza B Virus. J. Biochem. 2009, 145, 543–554. [Google Scholar] [CrossRef]
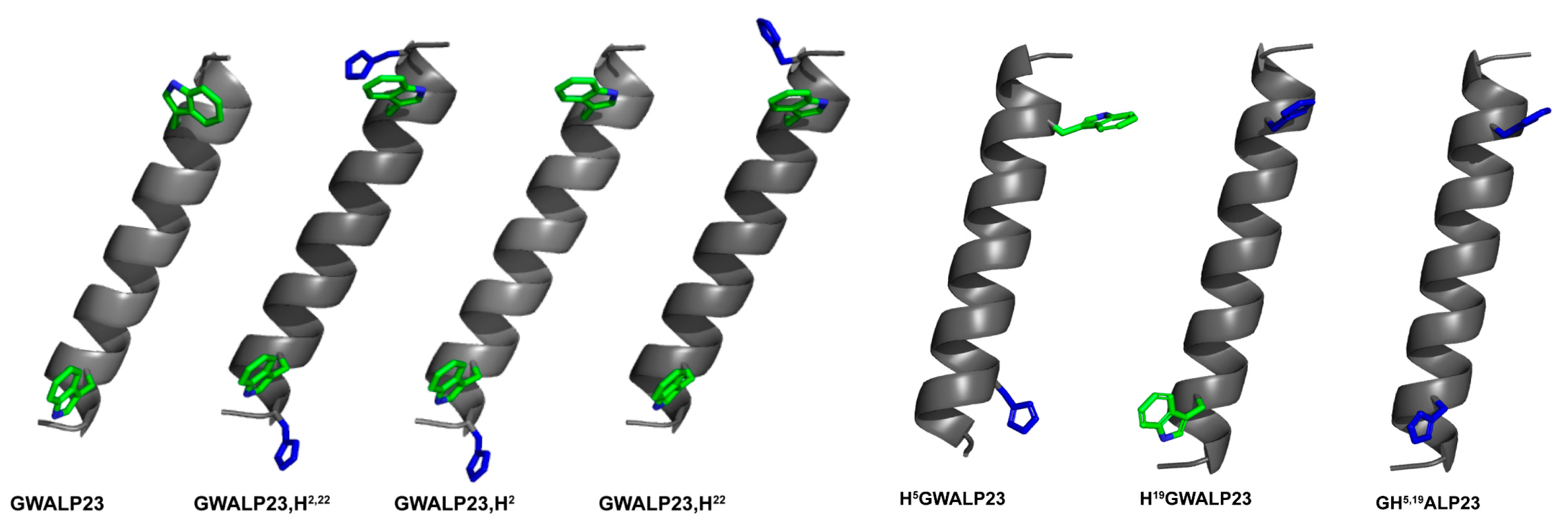





| Name of peptide | Sequence a | Reference |
|---|---|---|
| GWALP23 | acetyl-GGALWLALALALALALALWLAGA-amide | [12] |
| H2,22WALP23 | acetyl-GHALWLALALALALALALWLAHA-amide | This work |
| H2GWALP23 | acetyl-GHALWLALALALALALALWLAGA-amide | This work |
| H22GWALP23 | acetyl-GGALWLALALALALALALWLAHA-amide | This work |
| H5GWALP23 | acetyl-GGALHLALALALALALALWLAGA-amide | This work |
| H19GWALP23 | acetyl-GGALWLALALALALALALHLAGA-amide | This work |
| GH5,19ALP23 | acetyl-GGALHLALALALALALALHLAGA-amide | This work |
| Lipid | Peptide | [d4] Ala CD3 Quadrupolar Splittings in kHza | |||||
|---|---|---|---|---|---|---|---|
| 7 | 9 | 11 | 13 | 15 | 17 | ||
| DLPC | GWALP23 | 26.4 | 25.5 | 26.9 | 14.6 | 20.7 | 3.4 |
| H2,22 | 30.8 | 35 | 29.3 | 19.4 | 24.7 | 4.2 | |
| H2 | 31.9 | 35.6 | 31.4 | 23.7 | 25.8 | 1.2 | |
| H22 | 26.3 | 24.3 | 25.4 | 11.7 | 19.0 | 7.2 | |
| DOPC | GWALP23 | 16.6 | 1.7 | 16.7 | 1.5 | 15.4 | 2.6 |
| H2,22 | 15.4 | 1.2 | 17.0 | 1.6 | 16.0 | 2.1 | |
| H2 | 19.1 | 6.3 | 19.5 | 5.2 | 17.4 | 1.6 | |
| H22 | 14.0 | 1.4 | 14.1 | 1.1 | 12.5 | 4.1 | |
| H5,19 | 22.3 | 7.3 | 18.6 | 3.8 | 17.5 | 1.1 | |
| H5 | 22.1 | 6.0 | 16.8 | 1.2 | 11.7 | 8.5 | |
| H19 | 17.3 | 4.6 | 19.8 | 8.2 | 20.5 | 5.4 | |
| Peptide | Lipid | GALA Analysis | Modified Gaussian Analysisa | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| τ | ρ | Szz | RMSD (kHz) | τo | ρo | στ | σρ | RMSD (kHz) | ||
| GWALPb | DLPC | 20.7° | 307° | 0.71 | 0.66 | 23° | 304° | 15° | 33° | 0.7 |
| H2,22 | 26° | 304° | 0.74 | 1.05 | 24° | 303° | 10° | 16° | 1.43 | |
| H2 | 26° | 308° | 0.77 | 0.86 | 33° | 305° | 10° | 36° | 0.61 | |
| H22 | 22.7° | 297° | 0.67 | 0.66 | 17° | 299° | 10° | 8° | 1.22 | |
| GWALPb | DOPC | 6° | 323° | 0.87 | 0.61 | 9° | 321° | 9° | 48° | 0.7 |
| H2,22 | 6° | 329° | 0.86 | 0.33 | 10° | 326° | 10° | 56° | 0.56 | |
| H2 | 8.7° | 319° | 0.83 | 0.52 | 12° | 318° | 10° | 46° | 0.52 | |
| H22 | 6° | 315° | 0.73 | 0.35 | 9° | 319° | 10° | 70° | 0.45 | |
| cH5,19 | 8.7° | 319° | 0.80 | 0.97 | 10° | 319° | 10° | 34° | 1.1 | |
| cH5 | 8.7° | 292° | 0.76 | 0.37 | 9° | 290° | 10° | 32° | 0.6 | |
| H19 | 9.7° | 343° | 0.83 | 0.97 | 11° | 342° | 10° | 28° | 0.84 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afrose, F.; Koeppe II, R.E. Comparing Interfacial Trp, Interfacial His and pH Dependence for the Anchoring of Tilted Transmembrane Helical Peptides. Biomolecules 2020, 10, 273. https://doi.org/10.3390/biom10020273
Afrose F, Koeppe II RE. Comparing Interfacial Trp, Interfacial His and pH Dependence for the Anchoring of Tilted Transmembrane Helical Peptides. Biomolecules. 2020; 10(2):273. https://doi.org/10.3390/biom10020273
Chicago/Turabian StyleAfrose, Fahmida, and Roger E. Koeppe II. 2020. "Comparing Interfacial Trp, Interfacial His and pH Dependence for the Anchoring of Tilted Transmembrane Helical Peptides" Biomolecules 10, no. 2: 273. https://doi.org/10.3390/biom10020273
APA StyleAfrose, F., & Koeppe II, R. E. (2020). Comparing Interfacial Trp, Interfacial His and pH Dependence for the Anchoring of Tilted Transmembrane Helical Peptides. Biomolecules, 10(2), 273. https://doi.org/10.3390/biom10020273





