Integrated Analysis of Transcriptomic and Metabolomic Data Reveals the Mechanism by Which LED Light Irradiation Extends the Postharvest Quality of Pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee)
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material and LED Treatment
2.2. RNA-Seq Library Preparation and Sequencing
2.3. Transcriptome Data Analysis
2.3.1. Quality Control
2.3.2. Comparative Analysis
2.3.3. Functional Annotation of Genes
2.3.4. Quantification of Gene Expression Levels
2.3.5. Differential Expression Analysis
2.4. Sample Preparation and Metabolomic Analysis
2.5. Transcriptome and Metabolome Coalition Analysis
2.5.1. Correlation Analysis
2.5.2. Correlation Network Visualization
2.5.3. Canonical Correlation Analysis
2.6. Glucosinolates
2.7. Chlorophyll
2.8. Chlorophyllide a
2.9. Statistical Analyses
3. Results and Discussion
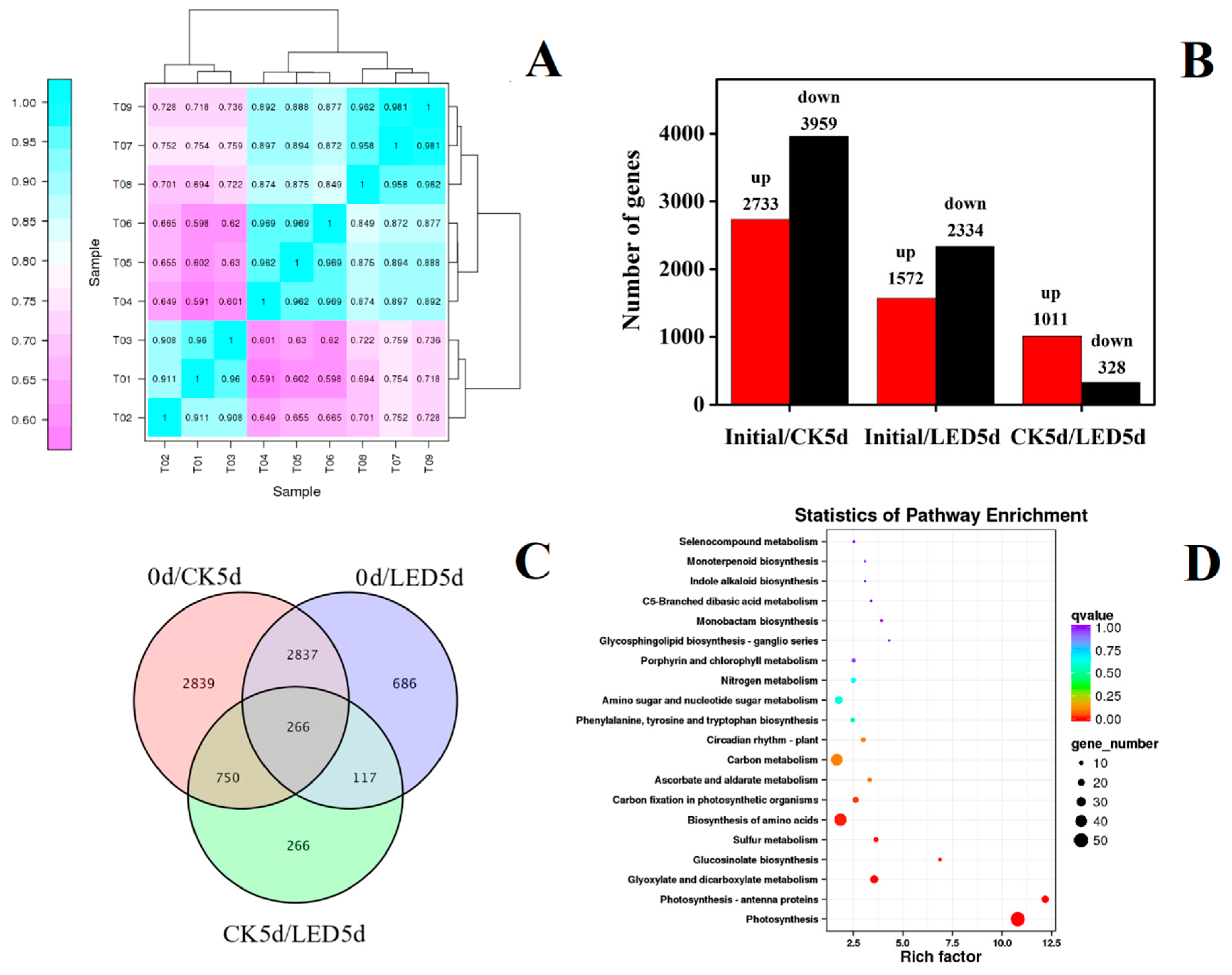
3.1. Transcriptome Response and Joint Analysis
3.2. Photosynthesis
3.3. Chlorophyll Synthesis
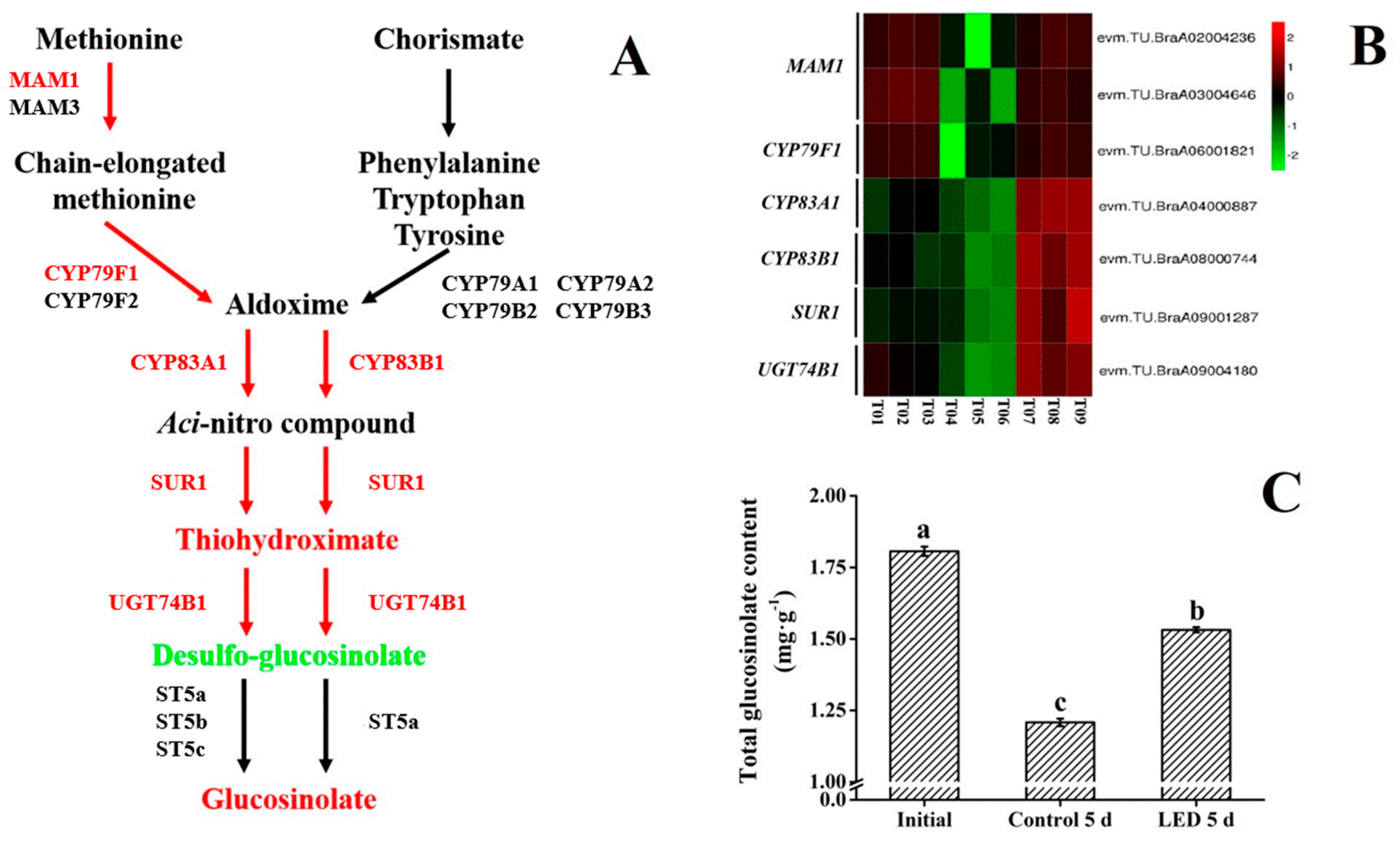
3.4. Glucosinolate Biosynthesis
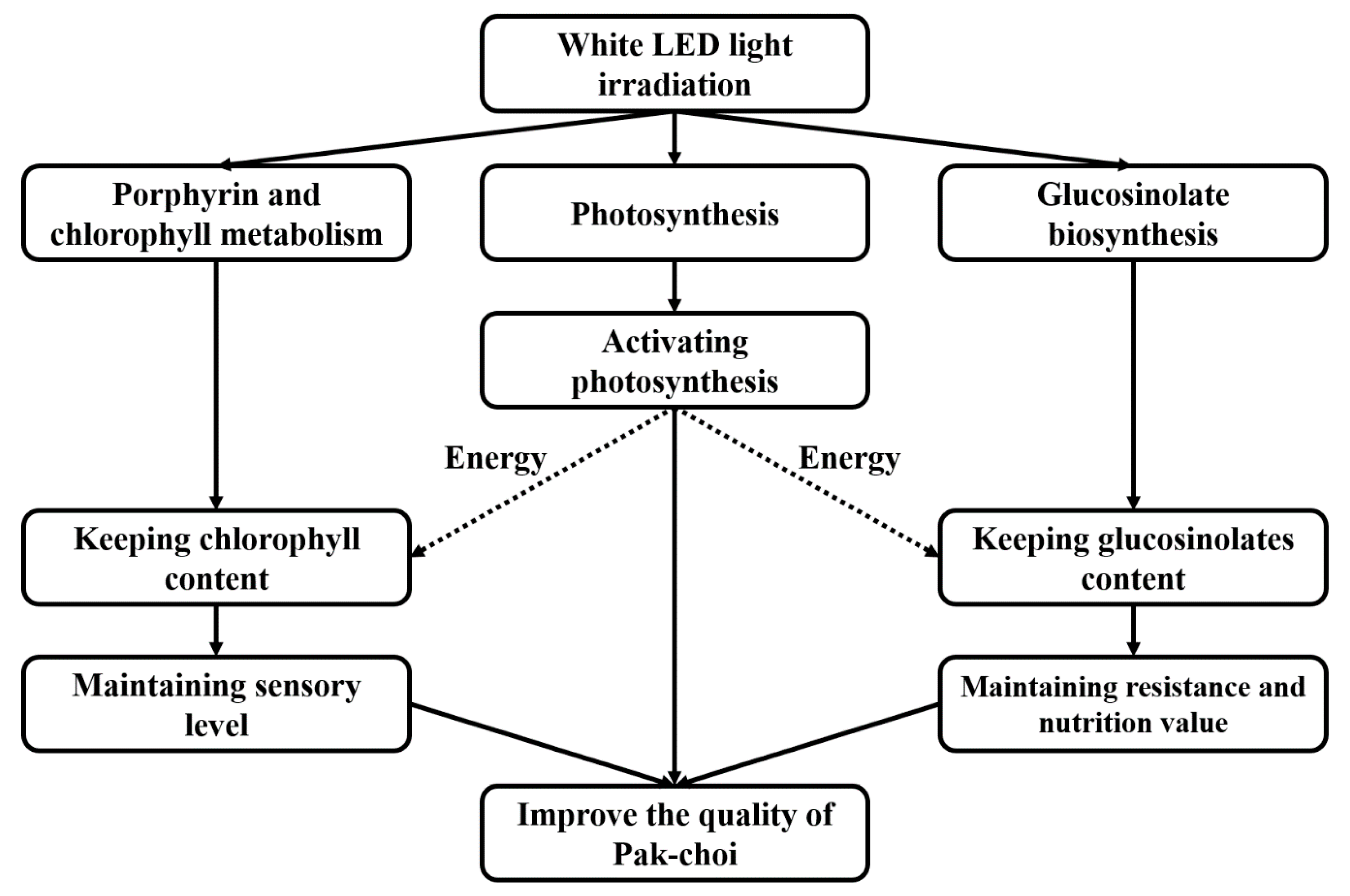
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Qiu, Y.; Li, X.X.; Zhi, H.Y.; Shen, D.; Lu, P. Differential expression of salt tolerance related genes in brassica campestris L. ssp. chinensis (L.) makino var. communis tsen et lee. J. Zhejiang Univ. Sci. B 2009, 10, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.; Fenwick, G.R. Glucosinolate content of brassica, vegetables: Analysis of twenty-four cultivars of calabrese (green sprouting broccoli, brassica oleracea, L. var. botrytis subvar. cymosa lam.). Food Chem. 1987, 25, 259–268. [Google Scholar] [CrossRef]
- Rybarczyk-Plonska, A.; Hansen, M.K.; Wold, A.B.; Hagen, S.F.; Borge, G.I.A.; Bengtsson, G.B. Vitamin c in broccoli (brassica oleracea L. var. italica) flower buds as affected by postharvest light, UV-B irradiation and temperature. Postharvest Biol. Technol. 2014, 98, 82–89. [Google Scholar] [CrossRef]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Keck, A.S.; Finley, J.W. Cruciferous vegetables: Cancer protective mechanisms of glucosinolate hydrolysis products and selenium. Integr. Cancer Ther. 2004, 3, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Song, H.J.; Song, K.B. Effect of combined treatment of aqueous chlorine dioxide with ultraviolet-C on the quality of red chicory and pak choi during storage. J. Korean Soc. Food Sci. Nutr. 2011, 40, 245–252. [Google Scholar] [CrossRef]
- Kramchote, S.; Srilaong, V.; Wongs-Aree, C.; Kanlayanarat, S. Low temperature storage maintains postharvest quality of cabbage (Brassica oleraceae var. capitata L.) in supply chain. Int. Food Res. J. 2012, 19, 759–763. [Google Scholar]
- Samuolienė, G.; Sirtautas, R.; Brazaitytė, A.; Duchovskis, P. Led lighting and seasonality effects antioxidant properties of baby leaf lettuce. Food Chem. 2012, 134, 1494–1499. [Google Scholar] [CrossRef]
- Morrow, R.C. LED lighting in horticulture. HortScience 2008, 43, 1947–1950. [Google Scholar] [CrossRef]
- Bourget, C.M. An introduction to light-emitting diodes. Hortscience 2008, 43, 1944–1946. [Google Scholar] [CrossRef]
- Hasperué, J.H.; Guardianelli, L.; Rodoni, L.M.; Chaves, A.R.; Martínez, G.A. Continuous white–blue led light exposition delays postharvest senescence of broccoli. LWT-Food Sci. Technol. 2015, 65, 495–502. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Setiawan, C.K.; Yamawaki, K.; Asai, T.; Nishikawa, F.; Nishikawab, F.; Maezawac, S.; Satod, H.; Kanemitsue, N.; et al. Effect of red and blue led light irradiation on ascorbate content and expression of genes related to ascorbate metabolism in postharvest broccoli. Postharvest Biol. Technol. 2014, 94, 97–103. [Google Scholar] [CrossRef]
- Liu, S.; Hu, L.; Jiang, D.; Xi, W. Effect of Post-Harvest LED and UV Light Irradiation on the Accumulation of Flavonoids and Limonoids in the Segments of Newhall Navel Oranges (Citrus sinensis Osbeck). Molecules 2019, 24, 1755. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Xu, F.; Zhou, W. Effect of LED irradiation on the ripening and nutritional quality of postharvest banana fruit. J. Sci. Food Agric. 2018, 98, 5486–5493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.H.; Zuo, J.H.; Xu, D.Y.; Gao, L.P.; Wang, Q.; Jiang, A.L. Low intensity white light-emitting diodes (LED) application to delay senescence and maintain quality of postharvest pakchoi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Sci. Hortic. 2020, 262, 109060. [Google Scholar] [CrossRef]
- Zhou, F.H.; Zuo, J.H.; Gao, L.P.; Sui, Y.; Wang, Q.; Jiang, A.L.; Shi, J.Y. An untargeted metabolomic approach reveals significant postharvest alterations in vitamin metabolism in response to LED irradiation in pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Metabolomics 2019, 15, 155. [Google Scholar] [CrossRef]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Roessner, U.; Luedemann, A.; Brust, D.; Fiehn, O.; Linke, T.; Willmitzer, L.; Fernie, A. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 2001, 13, 11–29. [Google Scholar] [CrossRef]
- Goossens, A.; Hãkkinen, S.T.; Laakso, I.; Seppãnen-Laakso, T.; Biondi, S.; Sutter, V.; Lammertyn, F.; Nuutila, A.; Söderlund, H.; Zabeau, M.; et al. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA 2003, 100, 8595–8600. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Yano, M.; Goodenowe, D.B.; Kanaya, S.; Kimura, T.; Awazuhara, M.; Arita, M.; Fujiwara, T.; Saito, K. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10205–10210. [Google Scholar] [CrossRef]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chang, J.; Ma, Q.; Chen, L.; Liu, S.; Jin, S.; Han, J.; Xu, R.; Zhu, A.; Gao, J.; et al. Network analysis of postharvest senescence process in citrus fruits revealed by transcriptomic and metabolomic profiling. Plant Physiol. 2015, 168, 357–376. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. Analysis of the coding and non-coding RNA transcriptomes in response to bell pepper chilling. Int. J. Mol. Sci. 2018, 19, 2001. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc, Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2015, 88, 524–545. [Google Scholar] [CrossRef]
- Hsu, F.C.; Wirtz, M.; Heppel, S.C.; Bogs, J.; Krämer, U.; Khan, M.S.; Bub, A.; Hell, R.; Rausch, T. Generation of Se-fortified broccoli as functional food: Impact of Se fertilization on S metabolism. Plant Cell Environ. 2011, 34, 192–207. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, S.; Chen, Y.J.; Xu, H.; LI, Y.G.; Zhang, Y.W.; Luan, F.S. Ozone sensitivity of four pakchoi cultivars with different leaf colors: Physiological and biochemical mechanisms. Photosynthetica 2016, 55, 478–490. [Google Scholar] [CrossRef]
- Hasperué, J.H.; Rodoni, L.M.; Guardianelli, L.M.; Chaves, A.R.; Martínez, G.A. Use of LED light for Brussels sprouts postharvest conservation. Sci. Hortic. 2016, 213, 281–286. [Google Scholar]
- Tanai, C.; Shengxi, S.; Peter, J.N. Enhancing photosynthesis in plants: The light reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar]
- Favre, N.; Bárcena, A.; Bahima, J.V.; Martínez, G.; Costa, L. Pulses of low intensity light as promising technology to delay postharvest senescence of broccoli. Postharvest Biol. Technol. 2018, 142, 107–114. [Google Scholar] [CrossRef]
- Braidot, E.; Petrussa, E.; Peresson, C.; Patui, S.; Bertolini, A.; Tubarob, F.; Wählbyc, U.; Coanc, M.; Vianelloa, A.; Zancani, M. Low-intensity light cycles improve the quality of lamb’s lettuce (Valerianella olitoria [L.] Pollich) during storage at low temperature. Postharvest Biol. Technol. 2014, 90, 15–23. [Google Scholar] [CrossRef]
- Wang, H.; Qian, Z.; Ma, S.; Zhou, Y.; Patrick, J.W.; Duan, X.; Qu, H. Energy status of ripening and postharvest senescent fruit of litchi (Litchi chinensis Sonn.). BMC Plant Biol. 2013, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Adyanthaya, I.; KWON, Y.I.; Apostolidis, E.; Shetty, K. Apple postharvest preservation is linked to phenolic content and superoxide dismutase activity. J. Food Biochem. 2009, 33, 535–556. [Google Scholar] [CrossRef]
- Büchert, A.M.; Civello, P.M.; Martínez, G.A. Effect of hot air, UV-C, white light and modified atmosphere treatments on expression of chlorophyll degrading genes in postharvest broccoli (Brassica oleracea L.) florets. Sci. Hortic. 2011, 127, 214–219. [Google Scholar] [CrossRef]
- Jin, P.; Yao, D.; Xu, F.; Wang, H.; Zheng, Y. Effect of light on quality and bioactive compounds in postharvest broccoli florets. Food Chem. 2015, 172, 705–709. [Google Scholar] [CrossRef]
- Zhang, S.; Zuo, L.; Zhang, J.; Chen, P.; Wang, J.; Yang, M. Transcriptome analysis of ulmus pumila ‘jinye’ responses to different shading involved in chlorophyll metabolism. Tree Genet. Genomes 2017, 13, 64. [Google Scholar] [CrossRef]
- Kannangara, C.G.; Gough, S.P. Biosynthesis of Δ-aminolevulinate in greening barley leaves: Glutamate 1-semialdehyde aminotransferase. Carlsberg Res. Commun. 1978, 43, 185–194. [Google Scholar] [CrossRef][Green Version]
- Tottey, S.; Block, M.A.; Allen, M.; Westergren, T.; Albrieux, C.; Scheller, H.; Merchant, S.; Jensen, P. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 2003, 100, 16119–16124. [Google Scholar] [CrossRef]
- Nagata, N.; Tanaka, R.; Tanaka, A. The major route for chlorophyll synthesis includes [3, 8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 1803–1808. [Google Scholar] [CrossRef]
- Watson, G.W.; Beaver, L.M.; Williams, D.E.; Dashwood, R.H.; Ho, E. Phytochemicals from cruciferous vegetables, epigenetics, and prostate cancer prevention. AAPS J. 2013, 15, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.-I. The R2R3-MYB transcription factor HAGL/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Ménard, R.; Larue, J.P.; Silué, D.; Thouvenot, D. Glucosinolates in cauliflower as biochemical markers for resistance against downy mildew. Phytochemistry 1999, 52, 29–35. [Google Scholar] [CrossRef]
- Mithen, R.F.; Lewis, B.G.; Fenwick, G.R. In vitro activity of glucosinolates and their products against leptosphaeria maculans. Fungal Biol. 1986, 87, 433–440. [Google Scholar] [CrossRef]
- Mari, M.; Iori, R.; Leoni, O.; Marchi, A. In vitro activity of glucosinolate-derived isothiocyanates against postharvest fruit pathogens. Ann. Appl. Biol. 1993, 123, 155–164. [Google Scholar] [CrossRef]
- Hanson, P.; Yang, R.Y.; Chang, L.C.; Ledesma, L.; Ledesma, D. Contents of carotenoids, ascorbic acid, minerals and total glucosinolates in leafy brassica pakchoi (Brassica rapa L. chinensis) as affected by season and variety. J. Sci. Food Agric. 2009, 89, 906–914. [Google Scholar] [CrossRef]
- Steinbrecher, A.; Rohrmann, S.; Timofeeva, M.; Risch, A.; Jansen, E.; Linseisen, J. Dietary glucosinolate intake, polymorphisms in selected biotransformation enzymes, and risk of prostate cancer. Cancer Epidemiol. Biomark. Prev. 2010, 19, 135–143. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Kopsell, D.A.; Sams, C.E.; Morrow, R.C. Blue wavelengths from LED lighting increase nutritionally important metabolites in specialty crops. HortScience 2015, 50, 1285–1288. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Zuo, J.; Zhou, F.; Shi, J.; Xu, D.; Hu, W.; Jiang, A.; Liu, Y.; Wang, Q. Integrated Analysis of Transcriptomic and Metabolomic Data Reveals the Mechanism by Which LED Light Irradiation Extends the Postharvest Quality of Pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Biomolecules 2020, 10, 252. https://doi.org/10.3390/biom10020252
Yan Z, Zuo J, Zhou F, Shi J, Xu D, Hu W, Jiang A, Liu Y, Wang Q. Integrated Analysis of Transcriptomic and Metabolomic Data Reveals the Mechanism by Which LED Light Irradiation Extends the Postharvest Quality of Pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Biomolecules. 2020; 10(2):252. https://doi.org/10.3390/biom10020252
Chicago/Turabian StyleYan, Zhicheng, Jinhua Zuo, Fuhui Zhou, Junyan Shi, Dongying Xu, Wenzhong Hu, Aili Jiang, Yao Liu, and Qing Wang. 2020. "Integrated Analysis of Transcriptomic and Metabolomic Data Reveals the Mechanism by Which LED Light Irradiation Extends the Postharvest Quality of Pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee)" Biomolecules 10, no. 2: 252. https://doi.org/10.3390/biom10020252
APA StyleYan, Z., Zuo, J., Zhou, F., Shi, J., Xu, D., Hu, W., Jiang, A., Liu, Y., & Wang, Q. (2020). Integrated Analysis of Transcriptomic and Metabolomic Data Reveals the Mechanism by Which LED Light Irradiation Extends the Postharvest Quality of Pak-choi (Brassica campestris L. ssp. chinensis (L.) Makino var. communis Tsen et Lee). Biomolecules, 10(2), 252. https://doi.org/10.3390/biom10020252





