GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans
Abstract
1. Introduction
2. The GAG-DB Database Construction and Utilization
2.1. Database Construction
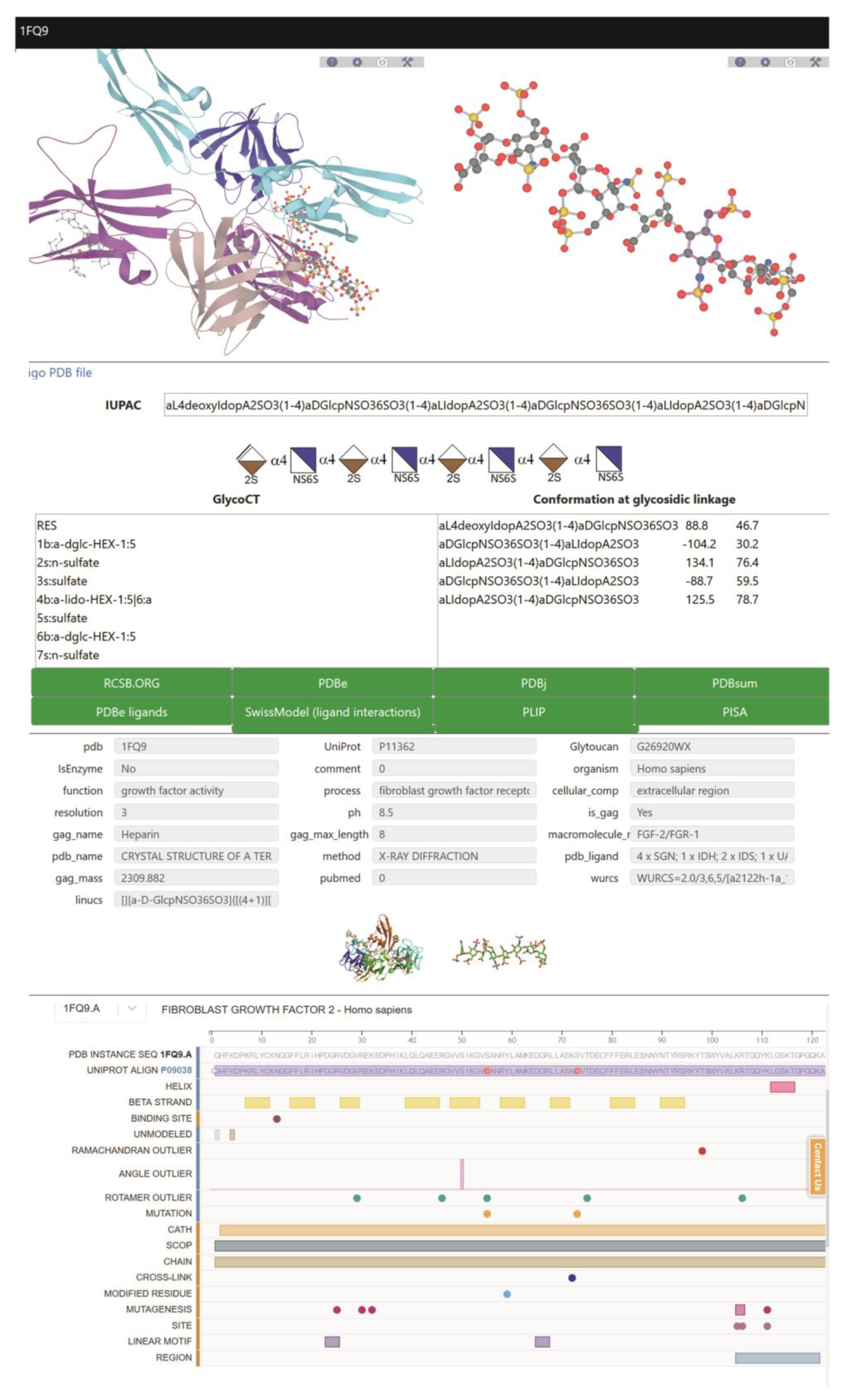
2.2. Description of the Search Interface
- The name of the polysaccharide gag_name, or its protein ligand, macromolecule_name.
- Cross-entries with external databases, such as pdb, UniProt, and repository GlytouCan.
- The biological role, such as function, process, or cellular compartment (compliant with Gene Ontology terms).
- The origin such as organism.
- The experimental condition(s) used to solve the structure: method and resolution.
- Characteristics of the GAG such as nature, (is_gag differentiates GAG and mimetics) and size (gag_max, gag-length, and gag-mass).
- Codes used for ligands in the Protein DataBase, pdb_ligand (nomenclature of the Chemical Component Dictionary. www.ebi.ac.uk/pdbe-srv/pdbechem/), or as encoded in LINUCS (Linear Notation for Unique description of Carbohydrate Sequences [58]), which provides access to WURCS (Web3 Unique Representation of Carbohydrate Structures [59]).
2.3. Curated Information for Each GAG Entry
3. Utilization of GAG-DB for Analysis of GAGs Structure and Conformation
3.1. Monosaccharides
3.2. Disaccharides
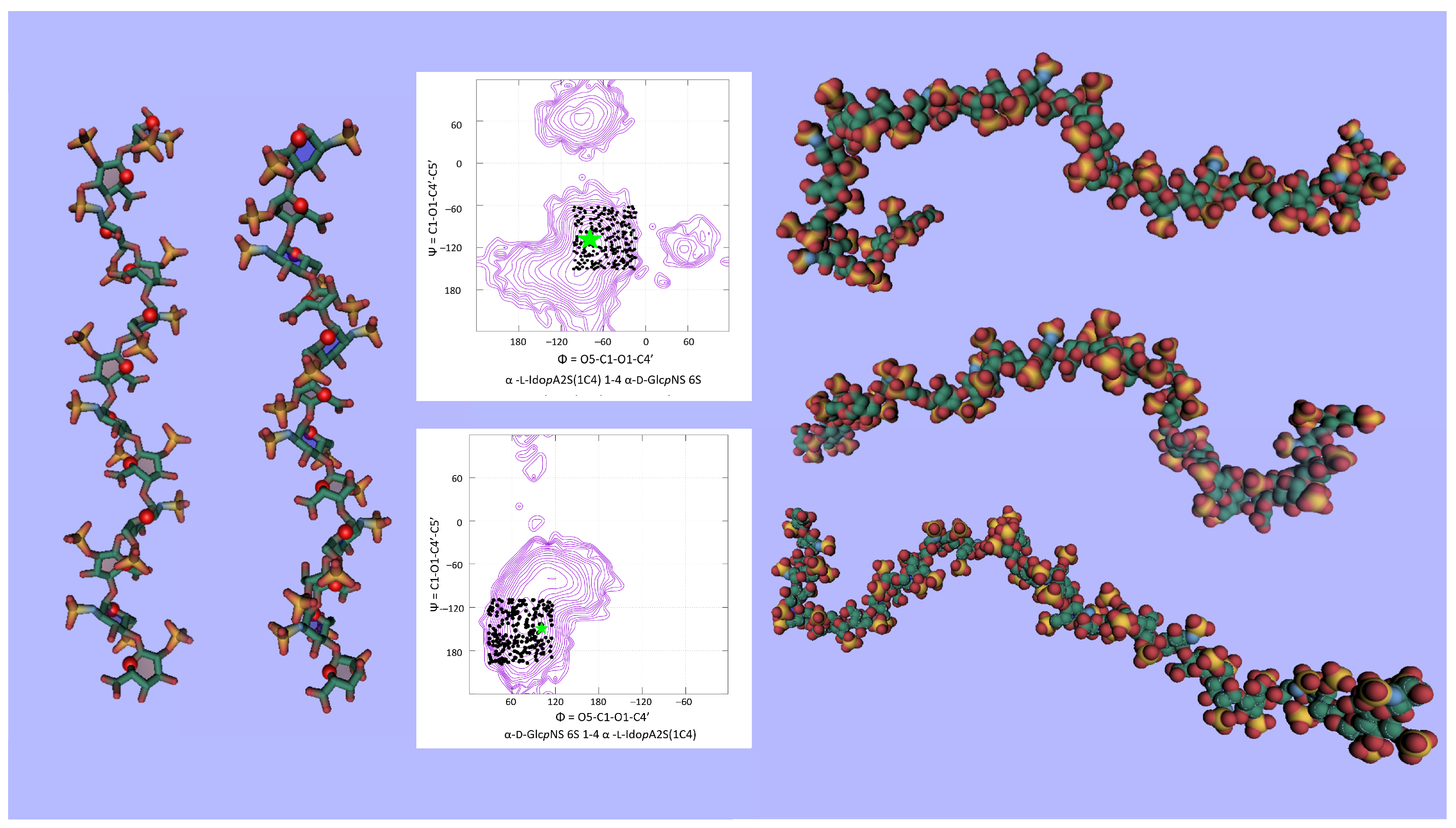
3.3. GAG Structures in the Solid-State
3.4. GAG Structures in Solution
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.K.; Piperigkou, Z.; Theocharis, A.D.; Watanabe, H.; Franchi, M.; Baud, S.; Brezillon, S.; Gotte, M.; Passi, A.; Vigetti, D.; et al. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem. Rev. 2018, 118, 9152–9232. [Google Scholar] [CrossRef] [PubMed]
- Mikami, T.; Kitagawa, H. Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 2013, 1830, 4719–4733. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J. Fell-Muir Lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. Int. J. Exp. Pathol. 2015, 96, 203–231. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Kusche-Gullberg, M. Heparan sulfate: Biosynthesis, structure, and function. Int. Rev. Cell. Mol. Biol. 2016, 325, 215–273. [Google Scholar] [CrossRef] [PubMed]
- Garantziotis, S.; Savani, R.C. Hyaluronan biology: A complex balancing act of structure, function, location and context. Matrix Biol. 2019, 78–79, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Caterson, B.; Melrose, J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 2018, 28, 182–206. [Google Scholar] [CrossRef]
- Pomin, V.H. Keratan sulfate: An up-to-date review. Int. J. Biol. Macromol. 2015, 72, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Clerc, O.; Mariethoz, J.; Rivet, A.; Lisacek, F.; Perez, S.; Ricard-Blum, S. A pipeline to translate glycosaminoglycan sequences into 3D models. Application to the exploration of glycosaminoglycan conformational space. Glycobiology 2019, 29, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Vallet, S.D.; Clerc, O.; Ricard-Blum, S. Glycosaminoglycan-protein Interactions: The first draft of the glycosaminoglycan interactome. J. Histochem. Cytochem. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Peysselon, F.; Ricard-Blum, S. Heparin-protein interactions: From affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. 2014, 35, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. Glycosaminoglycans: Major biological players. Glycoconj. J. 2017, 34, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 2011, 286, 19892–19904. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S.; Lisacek, F. Glycosaminoglycanomics: Where we are. Glycoconj. J. 2017, 34, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Aquino, R.S.; Park, P.W. Glycosaminoglycans and infection. Front. Biosci. 2016, 21, 1260–1277. [Google Scholar] [CrossRef]
- Burns, J.M.; Lewis, G.K.; DeVico, A.L. Soluble complexes of regulated upon activation, normal T cells expressed and secreted (RANTES) and glycosaminoglycans suppress HIV-1 infection but do not induce Ca(2+) signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 14499–14504. [Google Scholar] [CrossRef] [PubMed]
- Fatoux-Ardore, M.; Peysselon, F.; Weiss, A.; Bastien, P.; Pratlong, F.; Ricard-Blum, S. Large-scale investigation of Leishmania interaction networks with host extracellular matrix by surface plasmon resonance imaging. Infect. Immun. 2014, 82, 594–606. [Google Scholar] [CrossRef]
- Hsiao, F.S.; Sutandy, F.R.; Syu, G.D.; Chen, Y.W.; Lin, J.M.; Chen, C.S. Systematic protein interactome analysis of glycosaminoglycans revealed YcbS as a novel bacterial virulence factor. Sci. Rep. 2016, 6, 28425. [Google Scholar] [CrossRef]
- Jinno, A.; Park, P.W. Role of glycosaminoglycans in infectious disease. Methods Mol. Biol. 2015, 1229, 567–585. [Google Scholar] [CrossRef]
- Merida-de-Barros, D.A.; Chaves, S.P.; Belmiro, C.L.R.; Wanderley, J.L.M. Leishmaniasis and glycosaminoglycans: A future therapeutic strategy? Parasit. Vect. 2018, 11, 536. [Google Scholar] [CrossRef]
- Casu, B.; Naggi, A.; Torri, G. Re-visiting the structure of heparin. Carbohydr. Res. 2015, 403, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol nomenclature for graphical representations of glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.C.; Carvalho, M.S.; Han, X.; Xia, K.; Mikael, P.E.; Cabral, J.M.S.; Ferreira, F.C.; Linhardt, R.J. Compositional and structural analysis of glycosaminoglycans in cell-derived extracellular matrices. Glycoconj. J. 2019, 36, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N.; Linhardt, R.J. High-performance liquid chromatography-mass spectrometry for mapping and sequencing glycosaminoglycan-derived oligosaccharides. Nat. Protoc. 2010, 5, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wei, J.; Chopra, P.; Boons, G.J.; Lin, C.; Zaia, J. Sequencing heparan sulfate using HILIC LC-NETD-MS/MS. Anal. Chem. 2019, 91, 11738–11746. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Duan, J.; Leach, F.E., III; Toida, T.; Higashi, K.; Zhang, H.; Zhang, F.; Amster, I.J.; Linhardt, R.J. Sequencing the Dermatan Sulfate Chain of Decorin. J. Am. Chem. Soc. 2017, 139, 16986–16995. [Google Scholar] [CrossRef]
- Zaia, J. Glycosaminoglycan glycomics using mass spectrometry. Mol. Cell Proteom. 2013, 12, 885–892. [Google Scholar] [CrossRef]
- Langeslay, D.J.; Beni, S.; Larive, C.K. Detection of the 1H and 15N NMR resonances of sulfamate groups in aqueous solution: A new tool for heparin and heparan sulfate characterization. Anal. Chem. 2011, 83, 8006–8010. [Google Scholar] [CrossRef]
- Pomin, V.H. (1)H and (15)N NMR analyses on heparin, heparan sulfates and related monosaccharides concerning the chemical exchange regime of the N-sulfo-glucosamine sulfamate proton. Pharmaceuticals 2016, 9, 58. [Google Scholar] [CrossRef]
- Khan, S.; Fung, K.W.; Rodriguez, E.; Patel, R.; Gor, J.; Mulloy, B.; Perkins, S.J. The solution structure of heparan sulfate differs from that of heparin: Implications for function. J. Biol. Chem. 2013, 288, 27737–27751. [Google Scholar] [CrossRef]
- Jasnin, M. Use of neutrons reveals the dynamics of cell surface glycosaminoglycans. Methods Mol. Biol. 2012, 836, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Rubinson, K.A.; Chen, Y.; Cress, B.F.; Zhang, F.; Linhardt, R.J. Heparin’s solution structure determined by small-angle neutron scattering. Biopolymers 2016, 105, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Scherbinina, S.I.; Toukach, P.V. Three-dimensional structures of carbohydrates and where to find them. Intern. J. Mol. Sci. 2020, 21, 7702. [Google Scholar] [CrossRef] [PubMed]
- Samsonov, S.A.; Pisabarro, M.T. Computational analysis of interactions in structurally available protein-glycosaminoglycan complexes. Glycobiology 2016, 26, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, N.V.; Nagarajan, B.; Desai, U.R. So you think computational approaches to understanding glycosaminoglycan-protein interactions are too dry and too rigid? Think again! Curr. Opin. Struct. Biol. 2018, 50, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Sattelle, B.M.; Almond, A. Microsecond kinetics in model single- and double-stranded amylose polymers. Phys. Chem. Chem. Phys. 2014, 16, 8119–8126. [Google Scholar] [CrossRef] [PubMed]
- Sattelle, B.M.; Shakeri, J.; Cliff, M.J.; Almond, A. Proteoglycans and their heterogeneous glycosaminoglycans at the atomic scale. Biomacromolecules 2015, 16, 951–961. [Google Scholar] [CrossRef]
- Almond, A. Multiscale modeling of glycosaminoglycan structure and dynamics: Current methods and challenges. Curr. Opin. Struct. Biol. 2018, 50, 58–64. [Google Scholar] [CrossRef]
- Kolesnikov, A.L.; Budkov, Y.A.; Nogovitsyn, E.A. Coarse-grained model of glycosaminoglycans in aqueous salt solutions. A field-theoretical approach. J. Phys. Chem. B 2014, 118, 13037–13049. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Bichmann, L.; Pisabarro, M.T. Coarse-grained model of glycosaminoglycans. J. Chem. Inf. Model. 2015, 55, 114–124. [Google Scholar] [CrossRef]
- Whitmore, E.K.; Martin, D.; Guvench, O. Constructing 3-dimensional atomic-resolution models of nonsulfated glycosaminoglycans with arbitrary lengths using conformations from molecular dynamics. Intern. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019, 47, D376–D381. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bagdonas, H.; Ungar, D.; Agirre, J. Leveraging glycomics data in glycoprotein 3D structure validation with Privateer. Beilstein J. Org. Chem. 2020, 16, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Lutteke, T.; von der Lieth, C.W. pdb-care (PDB carbohydrate residue check): A program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinform. 2004, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Svobodova Varekova, R.; Pravda, L.; Ionescu, C.M.; Geidl, S.; Horsky, V.; Jaiswal, D.; Wimmerova, M.; Koca, J. ValidatorDB: Database of up-to-date validation results for ligands and non-standard residues from the Protein Data Bank. Nucleic Acids Res. 2015, 43, D369–D375. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Sarkar, A.; Rivet, A.; Breton, C.; Imberty, A. Glyco3D: A portal for structural glycosciences. Methods Mol. Biol. 2015, 1273, 241–258. [Google Scholar] [CrossRef]
- Bonnardel, F.; Mariethoz, J.; Salentin, S.; Robin, X.; Schroeder, M.; Perez, S.; Lisacek, F.; Imberty, A. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 2019, 47, D1236–D1244. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Mir, S.; Alhroub, Y.; Anyango, S.; Armstrong, D.R.; Berrisford, J.M.; Clark, A.R.; Conroy, M.J.; Dana, J.M.; Deshpande, M.; Gupta, D.; et al. PDBe: Towards reusable data delivery infrastructure at protein data bank in Europe. Nucleic Acids Res. 2018, 46, D486–D492. [Google Scholar] [CrossRef] [PubMed]
- Gutmanas, A.; Alhroub, Y.; Battle, G.M.; Berrisford, J.M.; Bochet, E.; Conroy, M.J.; Dana, J.M.; Fernandez Montecelo, M.A.; van Ginkel, G.; Gore, S.P.; et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2014, 42, 285–291. [Google Scholar] [CrossRef] [PubMed]
- PDBe-KB-Consortium. PDBe-KB: A community-driven resource for structural and functional annotations. Nucleic Acids Res. 2020, 48, D344–D353. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Vargas, A.; Delbianco, M.; Seeberger, P.H. Automated glycan assembly as an enabling technology. Curr. Opin. Chem. Biol. 2018, 46, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H.; Wang, X. Synthetic oligosaccharide libraries and microarray technology: A powerful combination for the success of current glycosaminoglycan interactomics. ChemMedChem 2018, 13, 648–661. [Google Scholar] [CrossRef] [PubMed]
- McNaught, A.D. Nomenclature of carbohydrates (recommendations 1996). Adv. Carbohydr. Chem. Biochem. 1997, 52, 43–177. [Google Scholar] [CrossRef] [PubMed]
- Herget, S.; Ranzinger, R.; Maass, K.; Lieth, C.W. GlycoCT-a unifying sequence format for carbohydrates. Carbohydr. Res. 2008, 343, 2162–2171. [Google Scholar] [CrossRef]
- Lutteke, T.; Bohne-Lang, A.; Loss, A.; Goetz, T.; Frank, M.; von der Lieth, C.W. GLYCOSCIENCES.de: An Internet portal to support glycomics and glycobiology research. Glycobiology 2006, 16, 71R–81R. [Google Scholar] [CrossRef]
- Tanaka, K.; Aoki-Kinoshita, K.F.; Kotera, M.; Sawaki, H.; Tsuchiya, S.; Fujita, N.; Shikanai, T.; Kato, M.; Kawano, S.; Yamada, I.; et al. WURCS: The Web3 unique representation of carbohydrate structures. J. Chem. Inf. Model. 2014, 54, 1558–1566. [Google Scholar] [CrossRef]
- Tiemeyer, M.; Aoki, K.; Paulson, J.; Cummings, R.D.; York, W.S.; Karlsson, N.G.; Lisacek, F.; Packer, N.H.; Campbell, M.P.; Aoki, N.P.; et al. GlyTouCan: An accessible glycan structure repository. Glycobiology 2017, 27, 915–919. [Google Scholar] [CrossRef]
- Sehnal, D.; Deshpande, M.; Varekova, R.S.; Mir, S.; Berka, K.; Midlik, A.; Pravda, L.; Velankar, S.; Koca, J. LiteMol suite: Interactive web-based visualization of large-scale macromolecular structure data. Nat. Methods 2017, 14, 1121–1122. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.S.; Bradley, A.R.; Valasatava, Y.; Duarte, J.M.; Prlic, A.; Rose, P.W. NGL viewer: Web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755–3758. [Google Scholar] [CrossRef] [PubMed]
- Kinjo, A.R.; Bekker, G.J.; Wako, H.; Endo, S.; Tsuchiya, Y.; Sato, H.; Nishi, H.; Kinoshita, K.; Suzuki, H.; Kawabata, T.; et al. New tools and functions in data-out activities at Protein Data Bank Japan (PDBj). Protein Sci. 2018, 27, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Jablonska, J.; Pravda, L.; Varekova, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Sillitoe, I.; Dawson, N.; Lewis, T.E.; Das, S.; Lees, J.G.; Ashford, P.; Tolulope, A.; Scholes, H.M.; Senatorov, I.; Bujan, A.; et al. CATH: Expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res. 2019, 47, D280–D284. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010, 31, 133–143. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Dimitropoulos, D.; Ionides, J.; Henrick, K. Using MSDchem to search the PDB ligand dictionary. Curr. Protoc. Bioinform. 2006. [Google Scholar] [CrossRef]
- Frank, M.; Lutteke, T.; von der Lieth, C.W. GlycoMapsDB: A database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res. 2007, 35, 287–290. [Google Scholar] [CrossRef]
- Smith, P.J.C.; Arnott, S. LALS, a linked-atom least-squares reciprocal-space refinement system incorporating stereochemical restraints to supplement sparse diffraction data. Acta Crystallogr. Sect. A Found. Crystallogr. 1978, 34, 3–11. [Google Scholar] [CrossRef]
- Guss, J.M.; Hukins, D.W.; Smith, P.J.; Winter, W.T.; Arnott, S. Hyaluronic acid: Molecular conformations and interactions in two sodium salts. J. Mol. Biol. 1975, 95, 359–384. [Google Scholar] [CrossRef]
- Mitra, A.K.; Arnott, S.; Sheehan, J.K. Hyaluronic acid: Molecular conformation and interactions in the tetragonal form of the potassium salt containing extended chains. J. Mol. Biol. 1983, 169, 813–827. [Google Scholar] [CrossRef]
- Winter, W.T.; Arnott, S. Hyaluronic acid: The role of divalent cations in conformation and packing. J. Mol. Biol. 1977, 117, 761–784. [Google Scholar] [CrossRef]
- Cael, J.J.; Winter, W.T.; Arnott, S. Calcium chondroitin 4-sulfate: Molecular conformation and organization of polysaccharide chains in a proteoglycan. J. Mol. Biol. 1978, 125, 21–42. [Google Scholar] [CrossRef]
- Millane, R.P.; Mitra, A.K.; Arnott, S. Chondroitin 4-sulfate: Comparison of the structures of the potassium and sodium salts. J. Mol. Biol. 1983, 169, 903–920. [Google Scholar] [CrossRef]
- Winter, W.T.; Arnott, S.; Isaac, D.H.; Atkins, E.D. Chondroitin 4-sulfate: The structure of a sulfated glycosaminoglycan. J. Mol. Biol. 1978, 125, 1–19. [Google Scholar] [CrossRef]
- Mitra, A.K.; Arnott, S.; Atkins, E.D.; Isaac, D.H. Dermatan sulfate: Molecular conformations and interactions in the condensed state. J. Mol. Biol. 1983, 169, 873–901. [Google Scholar] [CrossRef]
- Lee, S.C.; Guan, H.H.; Wang, C.H.; Huang, W.N.; Tjong, S.C.; Chen, C.J.; Wu, W.G. Structural basis of citrate-dependent and heparan sulfate-mediated cell surface retention of cobra cardiotoxin A3. J. Biol. Chem. 2005, 280, 9567–9577. [Google Scholar] [CrossRef]
- Arnott, S.; Gus, J.M.; Hukins, D.W.; Dea, I.C.; Rees, D.A. Conformation of keratan sulphate. J. Mol. Biol. 1974, 88, 175–184. [Google Scholar] [CrossRef]
- Haxaire, K.; Braccini, I.; Milas, M.; Rinaudo, M.; Perez, S. Conformational behavior of hyaluronan in relation to its physical properties as probed by molecular modeling. Glycobiology 2000, 10, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Tubiana, T.; Imberty, A.; Baaden, M. Three-dimensional representations of complex carbohydrates and polysaccharides—SweetUnityMol: A video game-based computer graphic software. Glycobiology 2015, 25, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Gor, J.; Mulloy, B.; Perkins, S.J. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: New insight into heparin-protein complexes. J. Mol. Biol. 2010, 395, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Carvajal, M.A.; Imberty, A.; Perez, S. Conformational behavior of chondroitin and chondroitin sulfate in relation to their physical properties as inferred by molecular modeling. Biopolymers 2003, 69, 15–28. [Google Scholar] [CrossRef] [PubMed]

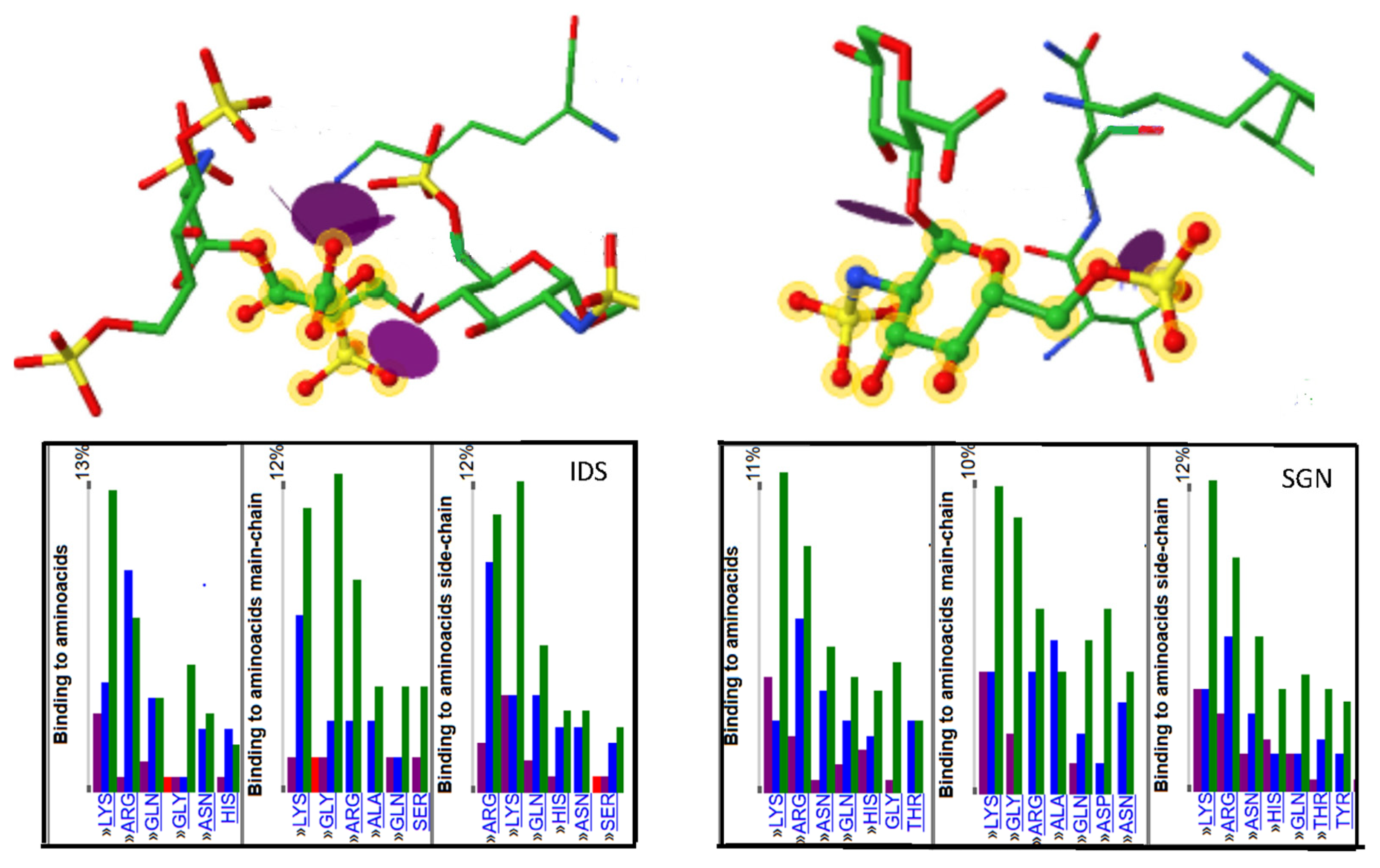
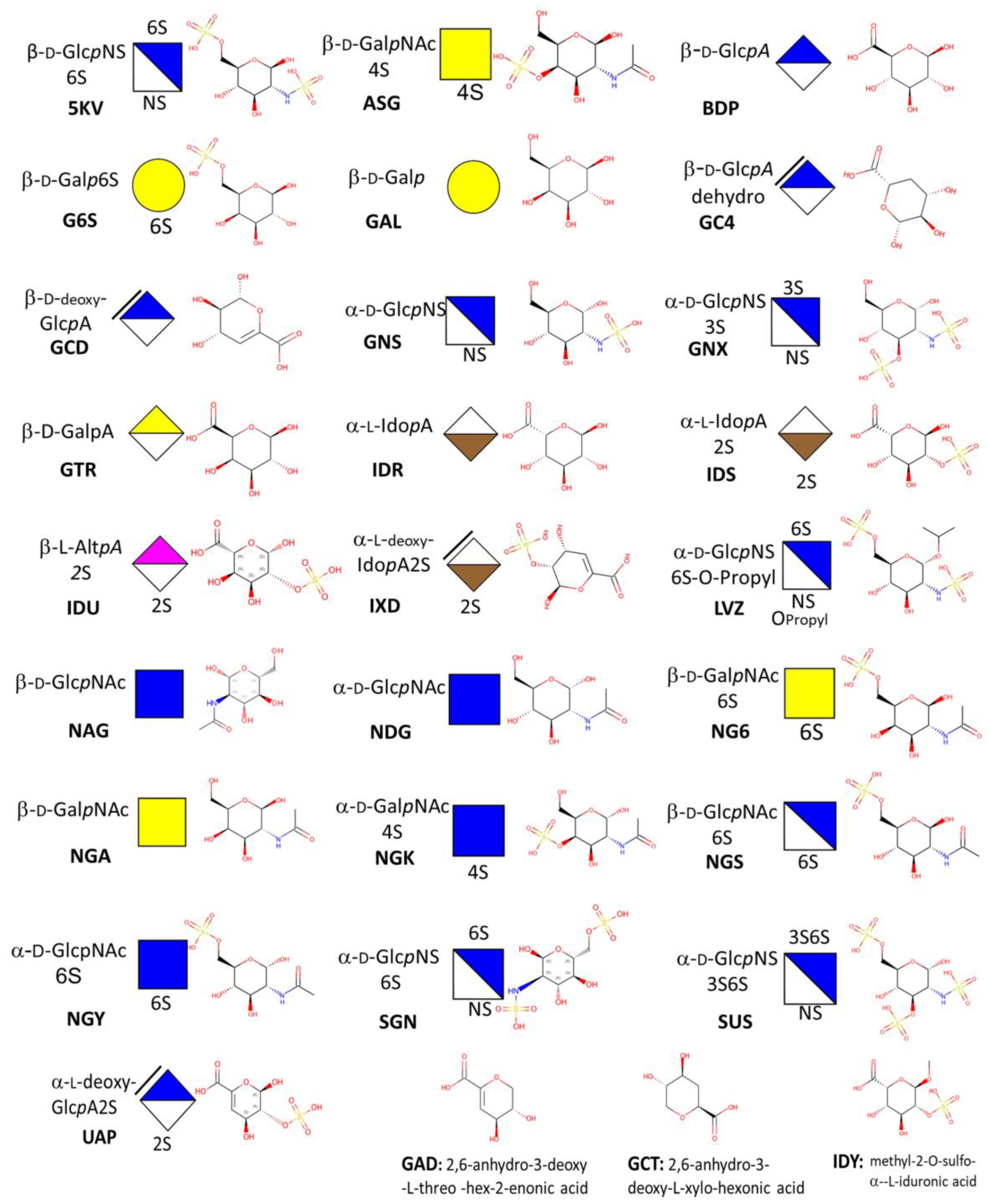
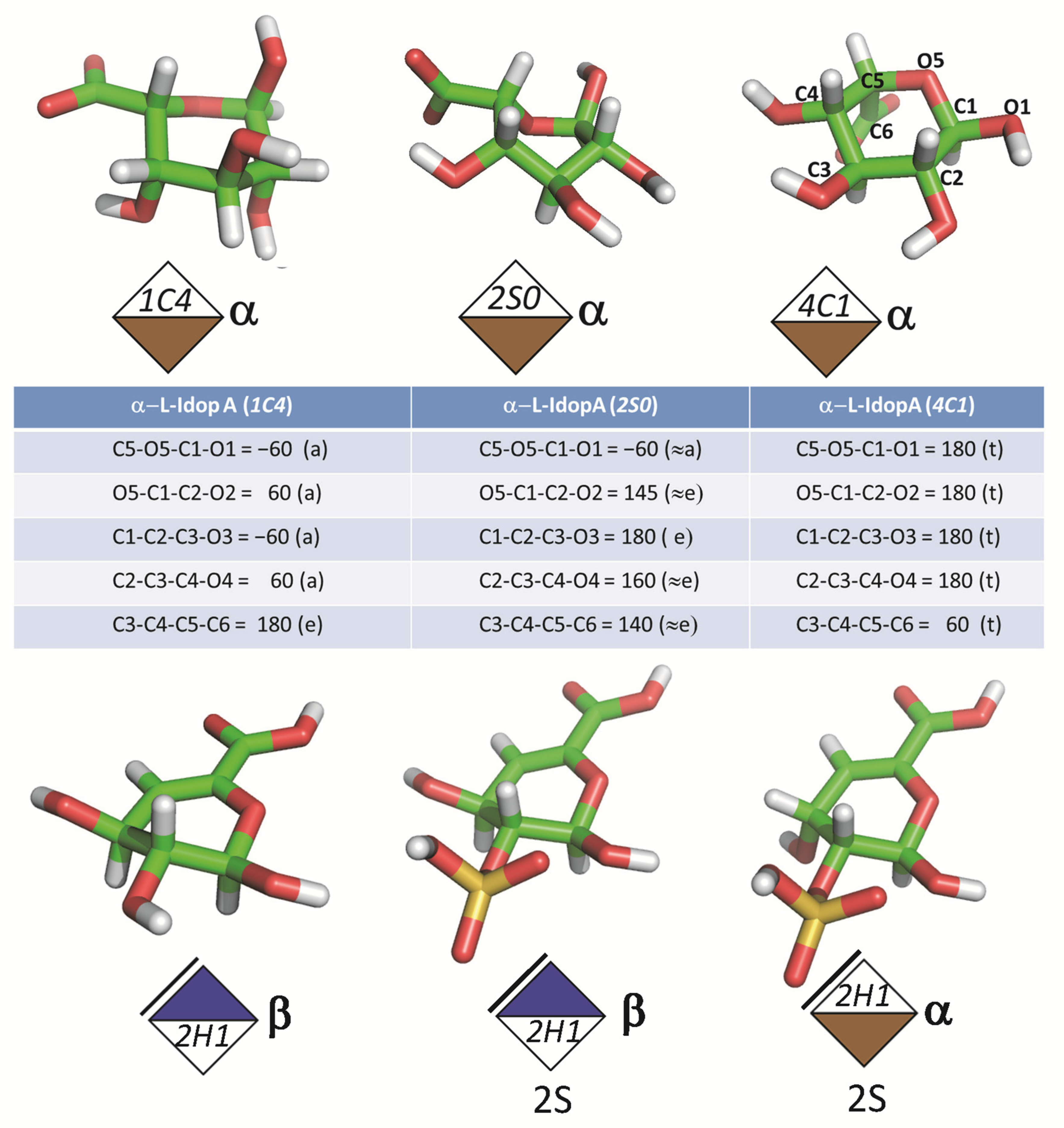
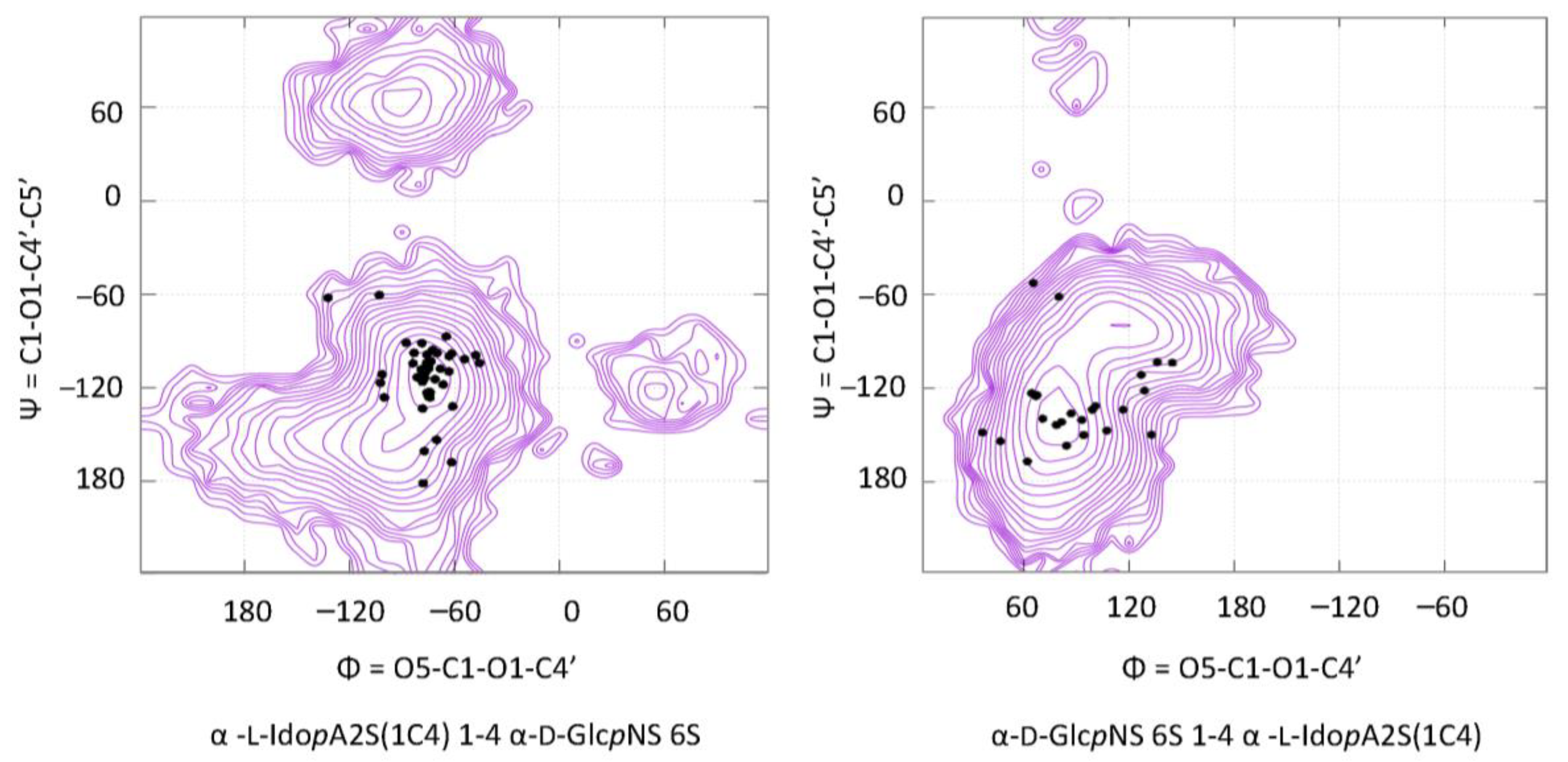
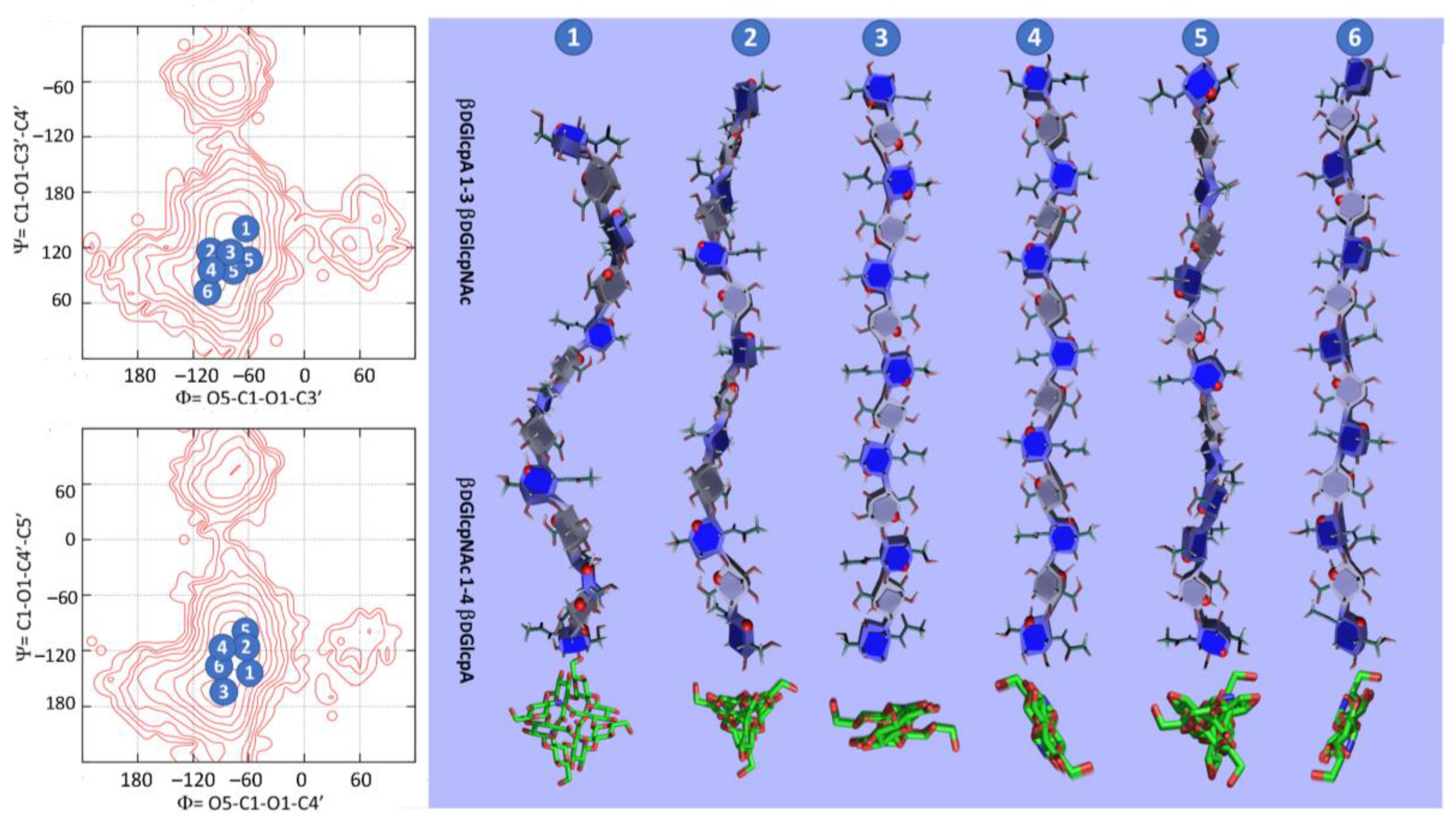
| Major Disaccharides Found in the GAG-Protein Complexes Extracted from the PDB | Number |
|---|---|
| α-d-GlcpNS (1-4) β-d-GlcpA | 21 |
| α-d-GlcpNS(6S) (1-4) α-l-IdopA(2S) [1C4] | 42 |
| α-d-GlcpNS(6S) (1-4) α-l-IdopA(2S) [2S0] | 15 |
| α-l-IdopA(2S) [1C4] (1-4) α-d-GlcpNS(6S) | 40 |
| α-l-IdopA(2S) [2S0] (1-4) α-d-GlcpNS(6S) | 16 |
| β-d-GlcpA (1-3) β-d-GlcpNAc | 13 |
| β-d-GlcpA (1-4) α-d-GlcpNS | 15 |
| β-d-GlcpNAc (1-4) β-d-GlcpA | 12 |
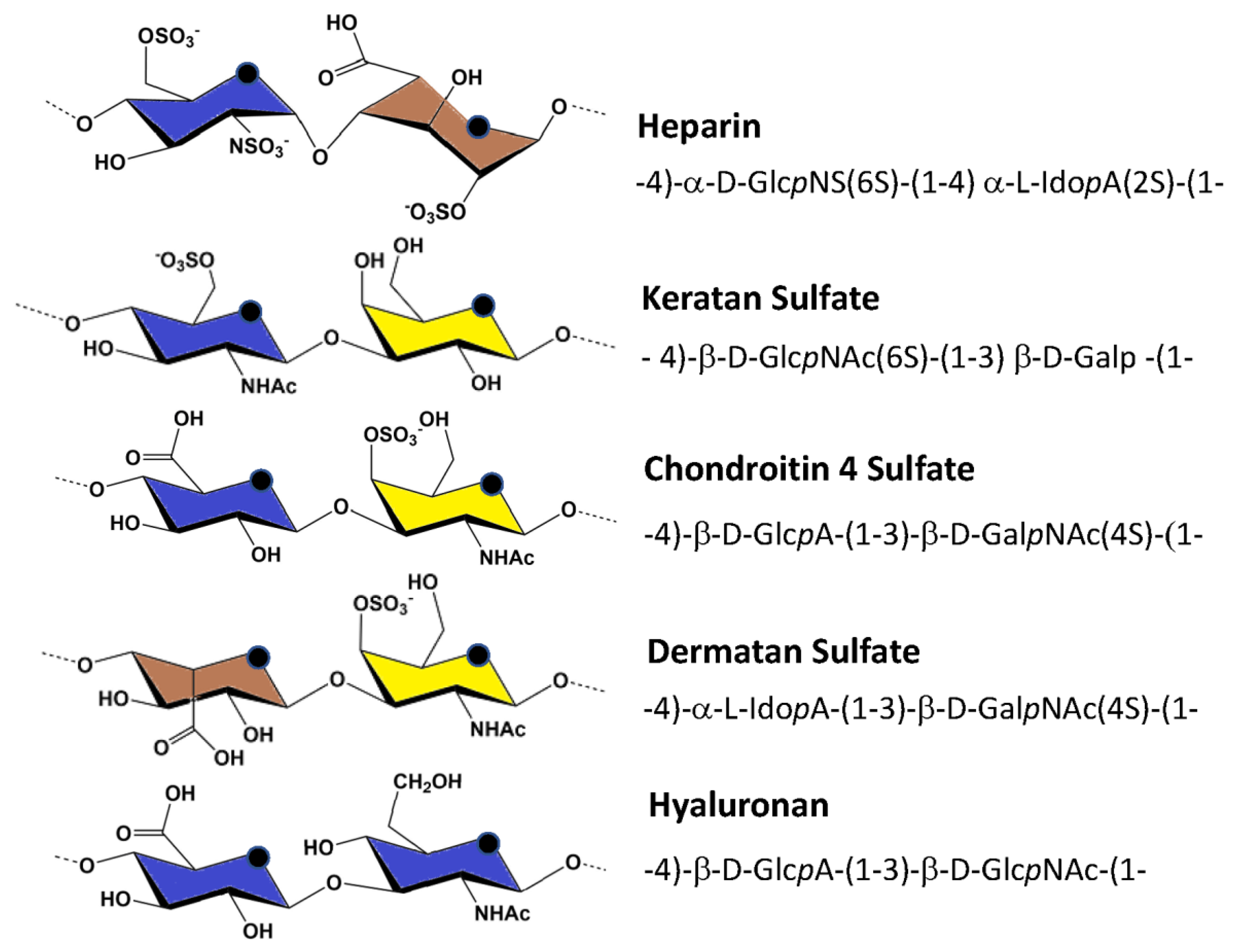
| Glycosaminoglycans | Structure of the Main Repeating Disaccharides | Helix Symmetry | Ref. |
|---|---|---|---|
| Hyaluronan | -4)-β-d-GlcpA-(1-3)-β-d-GlcpNAc-(1- | 21, 32, 43 | [72,73,74] |
| Chondroitin-4-sulfate | -4)-β-d-GlcpA-(1-3)-β-d-GalpNAc(4S)-(1- | 21, 32 | [75,76,77] |
| Chondroitin-6-sulfate | -4)-β-d-GlcpA-(1-3)-β-d-GalpNAc(6S)-(1- | 21, 32, 83 | |
| Dermatan sulfate | -4)-α-l-IdopA-(1-3)-β-d-GalpNAc(4S)-(1- | 21, 32, 83 | [78] |
| Heparin | -4)-α-l-IdopA(2S)-(1-4)-α-d-GlcpNS(6S)-(1 | (Na+) 21 | |
| Heparan sulfate | -4)-β-d-GlcpA-(1-4)-α-d-GlcpNAc-(1- | 21 | [79] |
| Keratan sulfate | -3)-β-d-Galp-(1-4)-β-d-GlcpNAc(6S)-(1- | 21 | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez, S.; Bonnardel, F.; Lisacek, F.; Imberty, A.; Ricard Blum, S.; Makshakova, O. GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans. Biomolecules 2020, 10, 1660. https://doi.org/10.3390/biom10121660
Pérez S, Bonnardel F, Lisacek F, Imberty A, Ricard Blum S, Makshakova O. GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans. Biomolecules. 2020; 10(12):1660. https://doi.org/10.3390/biom10121660
Chicago/Turabian StylePérez, Serge, François Bonnardel, Frédérique Lisacek, Anne Imberty, Sylvie Ricard Blum, and Olga Makshakova. 2020. "GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans" Biomolecules 10, no. 12: 1660. https://doi.org/10.3390/biom10121660
APA StylePérez, S., Bonnardel, F., Lisacek, F., Imberty, A., Ricard Blum, S., & Makshakova, O. (2020). GAG-DB, the New Interface of the Three-Dimensional Landscape of Glycosaminoglycans. Biomolecules, 10(12), 1660. https://doi.org/10.3390/biom10121660