Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring—An Inactive Enzyme with Intact Substrate Binding
Abstract
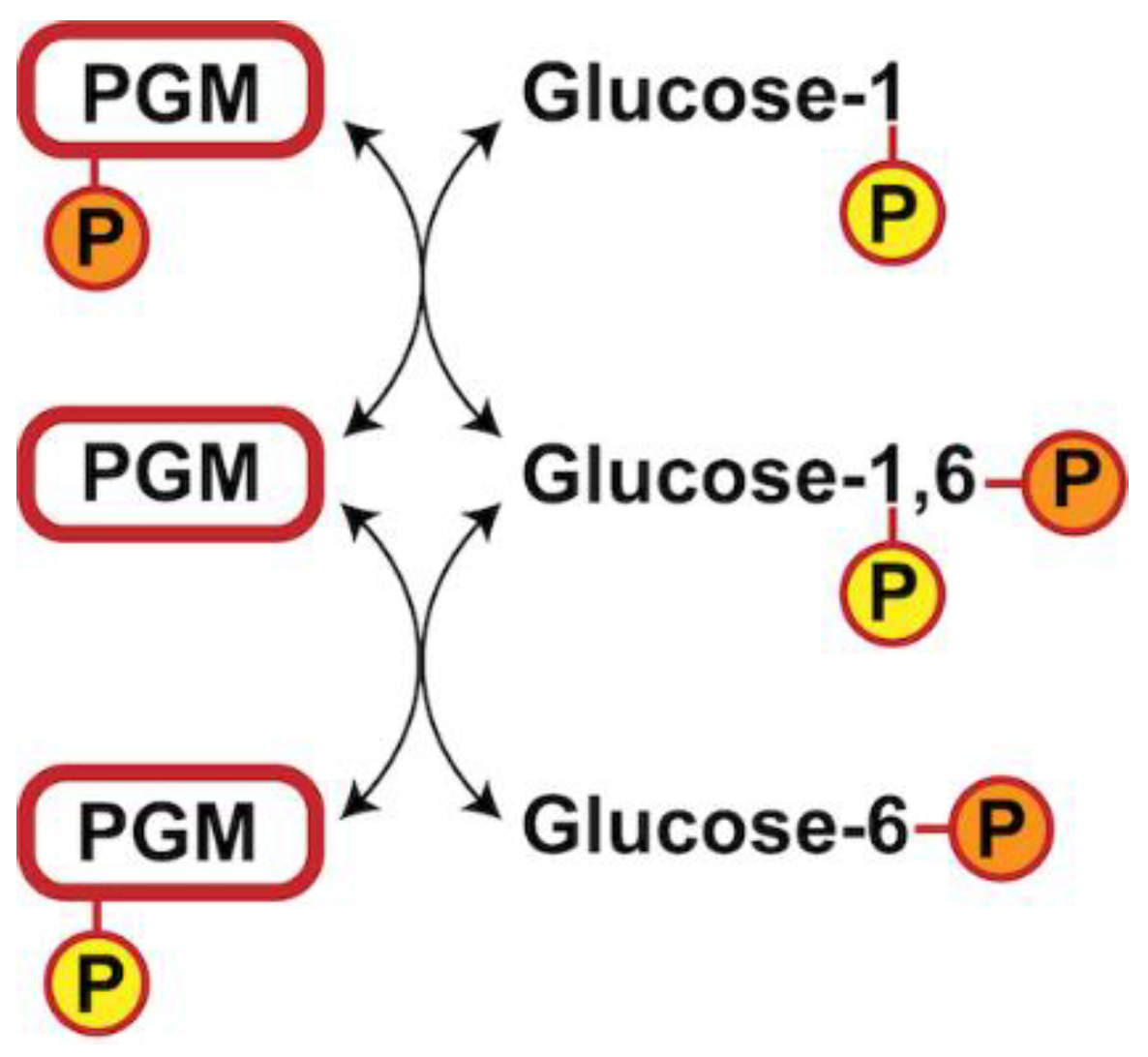
1. Introduction
2. Materials and Methods
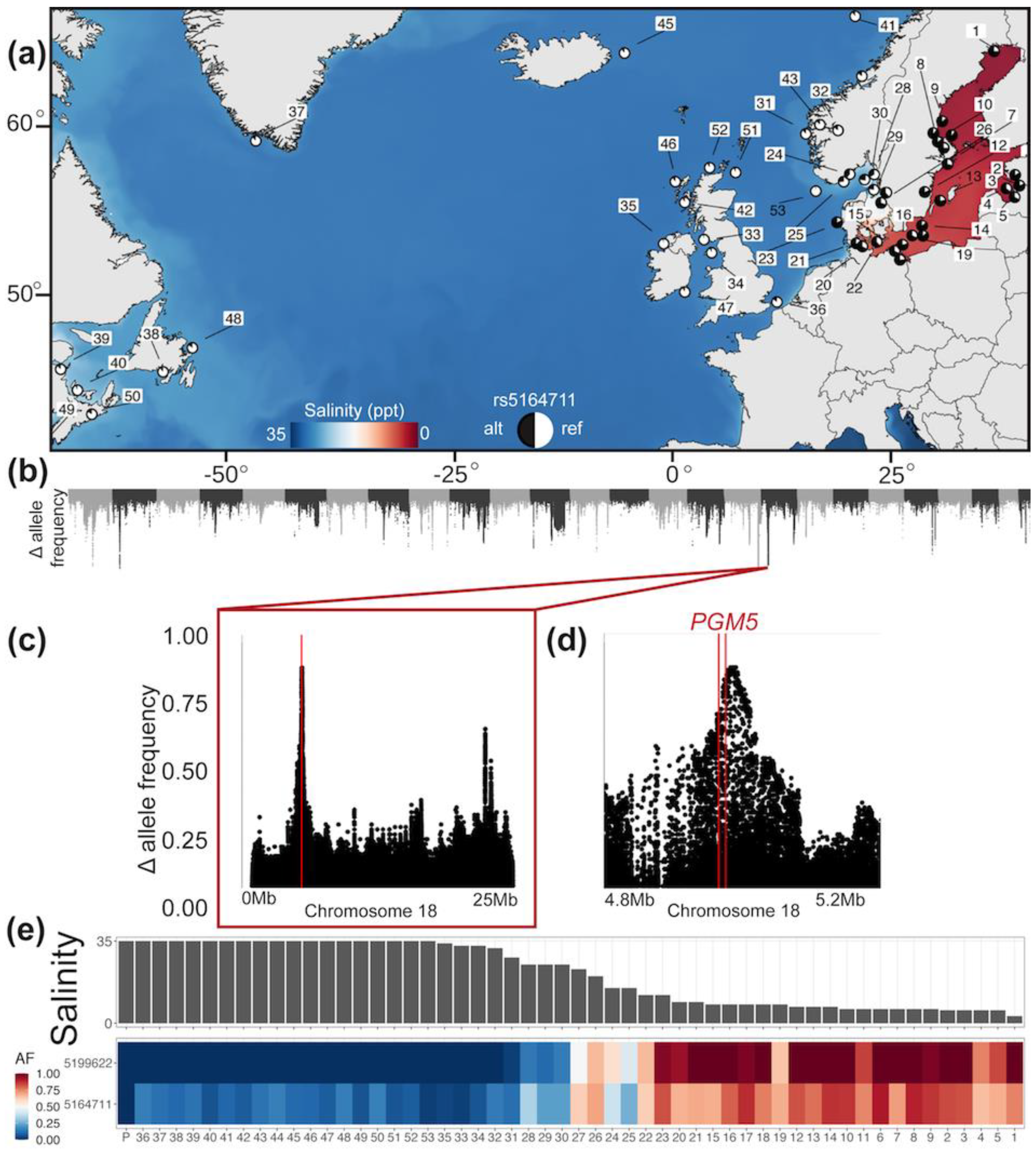
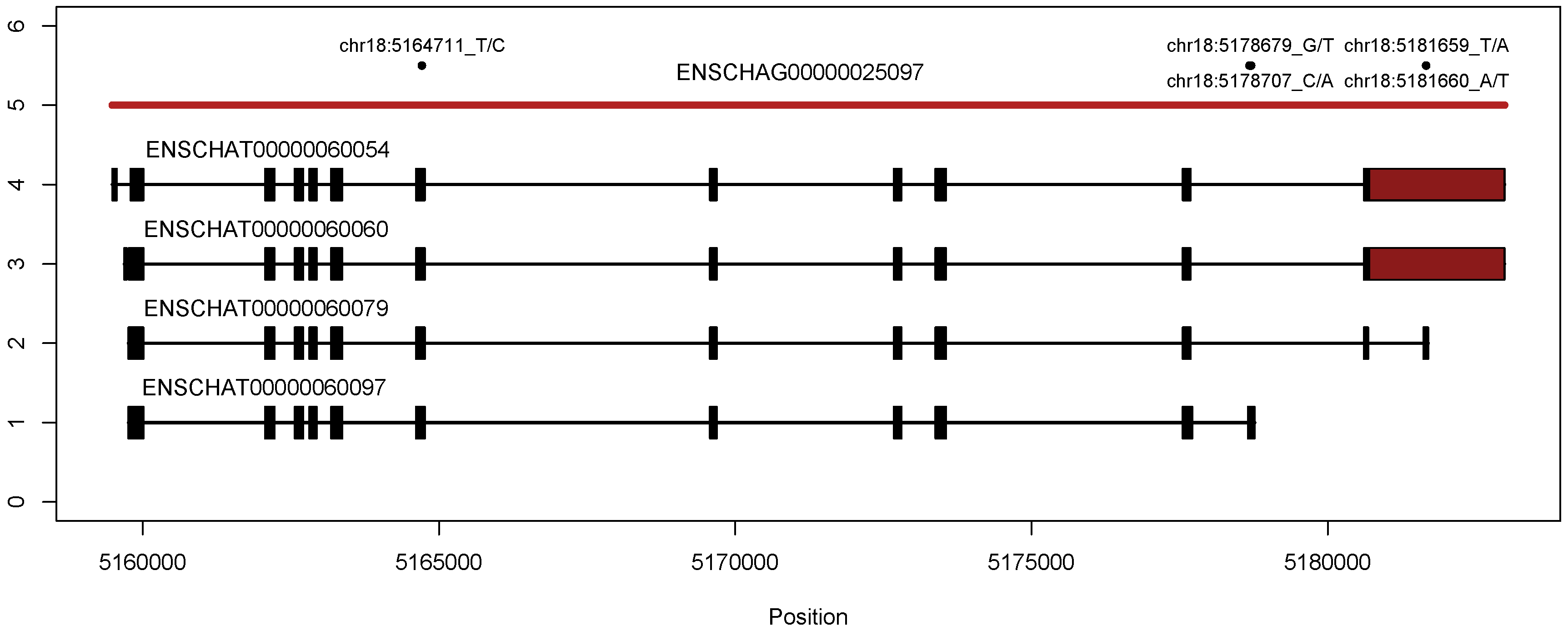
2.1. Genetic Analysis of PGM5 in the Atlantic Herring
2.2. Molecular Cloning
2.3. Protein Expression and Purification
2.4. Protein Crystallization and Structure Determination
2.5. Enzymatic Assays
2.6. Differential Scanning Fluorimetry
2.7. Circular Dichroism Spectroscopy
2.8. Structure Analysis
2.9. Multiple Sequence Alignment
3. Results
3.1. Genetic Analysis of PGM5 in the Atlantic Herring
3.2. Structure Determination of PGM5 from Herring
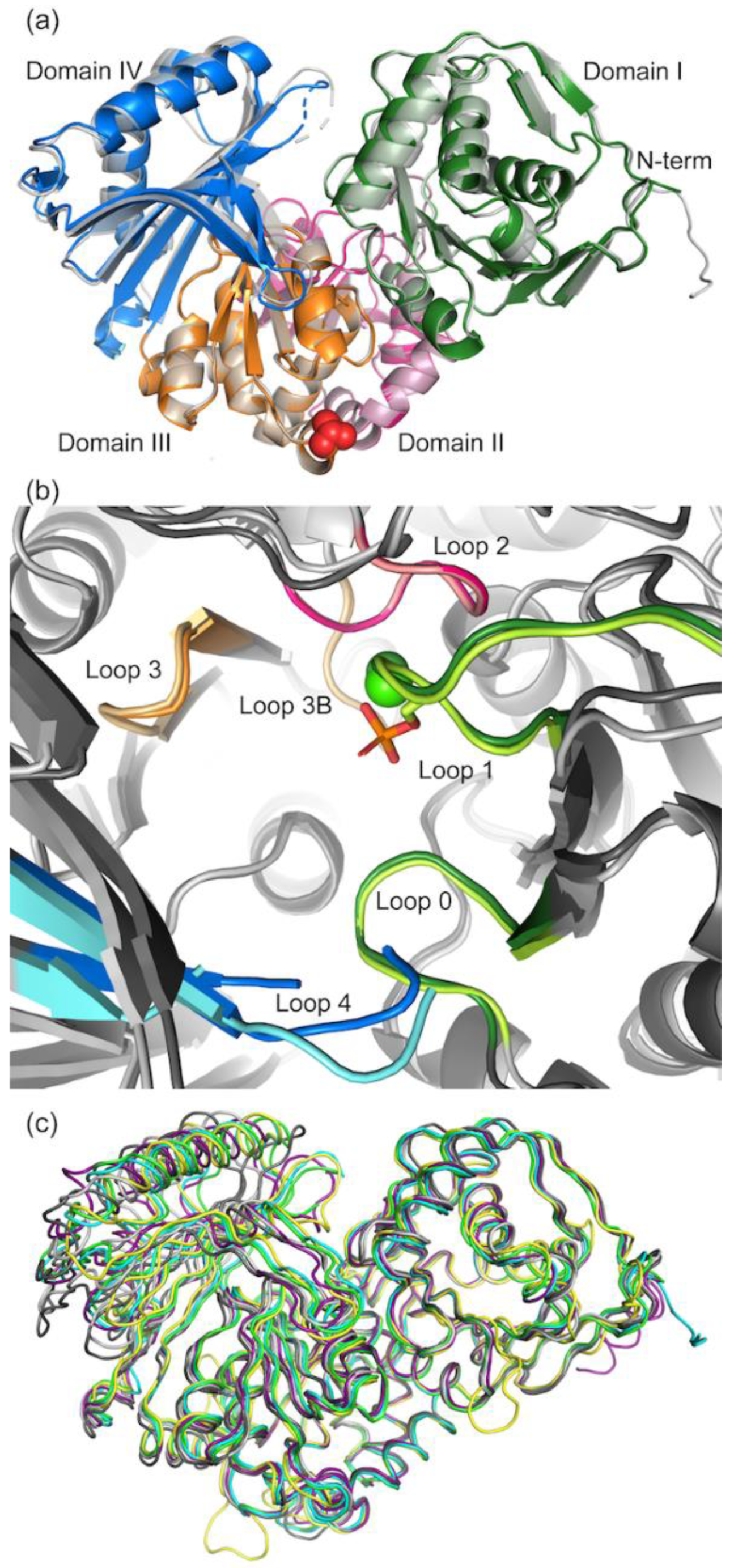
3.2.1. Overall Structure of PGM5
3.2.2. Comparison of Crystal Packing in aPGM5 and bPGM5
3.3. Structure of bPGM5 in Complex with Glucose-1-Phosphate
3.4. Glucose-1,6-Bisphosphate Stabilizes both aPGM5 and bPGM5
3.5. Enzymatic Activity of PGM5 Is Negligible
3.6. Sequence Analysis Shows the Main Difference between PGM5 and PGM1 Is in Loop 4
4. Discussion
4.1. Why Does PGM5 Lack PGM Activity?
4.2. Does PGM5 Have the Same Function in Fish as in Humans?
4.3. Why Is PGM5 Different in the Atlantic and Baltic Herring?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martinez Barrio, A.; Lamichhaney, S.; Fan, G.; Rafati, N.; Pettersson, M.; Zhang, H.; Dainat, J.; Ekman, D.; Höppner, M.; Jern, P.; et al. The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. Elife 2016, 5, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Linnaeus, C. Fauna Suecica; Salvius: Stockholm, Sweden, 1761. [Google Scholar]
- Ryman, N.; Lagercrantz, U.; Andersson, L.; Chakraborty, R.; Rosenberg, R. Lack of correspondence between genetic and morphologic variability patterns in Atlantic herring (Clupea harengus). Heredity (Edinb) 1984, 53, 687–704. [Google Scholar] [CrossRef]
- Pettersson, M.E.; Rochus, C.M.; Han, F.; Chen, J.; Hill, J.; Wallerman, O.; Fan, G.; Hong, X.; Xu, Q.; Zhang, H.; et al. A chromosome-level assembly of the Atlantic herring genome—detection of a supergene and other signals of selection. Genome Res. 2019, 29, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Jamsandekar, M.; Pettersson, M.E.; Su, L.; Fuentes-Pardo, A.; Davis, B.; Bekkevold, D.; Berg, F.; Cassini, M.; Dahle, G.; et al. The genetic architecture underlying ecological adaptation in Atlantic herring is not consistent with the infinitesimal model. bioRxiv 2020. [Google Scholar] [CrossRef]
- Andrén, T.; Björck, S.; Andrén, E.; Conley, D.; Zillén, L.; Anjar, J. The Development of the Baltic Sea Basin During the Last 130 ka. In The Baltic Sea Basin; Harff, J., Björck, S., Hoth, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 75–97. ISBN 978-3-642-17220-5. [Google Scholar]
- Hill, J.; Enbody, E.D.; Pettersson, M.E.; Sprehn, C.G.; Bekkevold, D.; Folkvord, A.; Laikre, L.; Kleinau, G.; Scheerer, P.; Andersson, L. Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc. Natl. Acad. Sci. USA 2019, 116, 18473–18478. [Google Scholar] [CrossRef]
- Muenks, A.G.; Stiers, K.M.; Beamer, L.J. Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1. PLoS ONE 2017, 12, e0183563. [Google Scholar] [CrossRef]
- Stiers, K.M.; Muenks, A.G.; Beamer, L.J. Biology, Mechanism, and Structure of Enzymes in the α- d -Phosphohexomutase Superfamily. In Advances in Protein Chemistry and Structural Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 109, pp. 265–304. [Google Scholar]
- Backe, P.H.; Laerdahl, J.K.; Kittelsen, L.S.; Dalhus, B.; Mørkrid, L.; Bjørås, M. Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the α-d-phosphohexomutase superfamily. Sci. Rep. 2020, 10, 5656. [Google Scholar] [CrossRef]
- Shackelford, G.S.; Regni, C.A.; Beamer, L.J. Evolutionary trace analysis of the α-D-phosphohexomutase superfamily. Protein Sci. 2004, 13, 2130–2138. [Google Scholar] [CrossRef]
- Stiers, K.M.; Kain, B.N.; Graham, A.C.; Beamer, L.J. Induced Structural Disorder as a Molecular Mechanism for Enzyme Dysfunction in Phosphoglucomutase 1 Deficiency. J. Mol. Biol. 2016, 428, 1493–1505. [Google Scholar] [CrossRef]
- Naught, L.E.; Tipton, P.A. Kinetic Mechanism and pH Dependence of the Kinetic Parameters of Pseudomonas aeruginosa Phosphomannomutase/Phosphoglucomutase. Arch. Biochem. Biophys. 2001, 396, 111–118. [Google Scholar] [CrossRef]
- Brautaset, T.; Petersen, S.B.; Valla, S. In Vitro Determined Kinetic Properties of Mutant Phosphoglucomutases and Their Effects on Sugar Catabolism in Escherichia coli. Metab. Eng. 2000, 2, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Mehra-Chaudhary, R.; Furdui, C.; Beamer, L.J. Identification of an essential active-site residue in the α-d-phosphohexomutase enzyme superfamily. FEBS J. 2013, 280, 2622–2632. [Google Scholar] [CrossRef] [PubMed]
- Regni, C.; Schramm, A.M.; Beamer, L.J. The Reaction of Phosphohexomutase from Pseudomonas aeruginosa. J. Biol. Chem. 2006, 281, 15564–15571. [Google Scholar] [CrossRef] [PubMed]
- Brás, N.F.; Fernandes, P.A.; Ramos, M.J.; Schwartz, S.D. Mechanistic Insights on Human Phosphoglucomutase Revealed by Transition Path Sampling and Molecular Dynamics Calculations. Chem. A Eur. J. 2018, 24, 1978–1987. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ray, W.J.; Baranidharan, S. Structure of Rabbit Muscle Phosphoglucomutase Refined at 2.4 Å Resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997, 53, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Stiers, K.M.; Beamer, L.J. A Hotspot for Disease-Associated Variants of Human PGM1 Is Associated with Impaired Ligand Binding and Loop Dynamics. Structure 2018, 26, 1337–1345.e3. [Google Scholar] [CrossRef]
- Luebbering, E.K.; Mick, J.; Singh, R.K.; Tanner, J.J.; Mehra-Chaudhary, R.; Beamer, L.J. Conservation of Functionally Important Global Motions in an Enzyme Superfamily across Varying Quaternary Structures. J. Mol. Biol. 2012, 423, 831–846. [Google Scholar] [CrossRef]
- Waugh, B.; Sen, U.; Banerjee, R. Crystal structure of phosphoglucomutase from Leishmania major at 3.5 Å resolution. Biochimie 2016, 121, 102–111. [Google Scholar] [CrossRef]
- Schramm, A.M.; Mehra-Chaudhary, R.; Furdui, C.M.; Beamer, L.J. Backbone Flexibility, Conformational Change, and Catalysis in a Phosphohexomutase from Pseudomonas aeruginosa. Biochemistry 2008, 47, 9154–9162. [Google Scholar] [CrossRef]
- Belkin, A.M.; Burridge, K. Expression and localization of the phosphoglucomutase-related cytoskeletal protein, aciculin, in skeletal muscle. J. Cell Sci. 1994, 107 Pt 7, 1993–2003. [Google Scholar]
- Belkin, A.M.; Klimanskaya, I.V.; Lukashev, M.E.; Lilley, K.; Critchley, D.R.; Koteliansky, V.E. A novel phosphoglucomutase-related protein is concentrated in adherens junctions of muscle and nonmuscle cells. J. Cell Sci. 1994, 107 Pt 1, 159–173. [Google Scholar]
- Moiseeva, E.P.; Belkin, A.M.; Spurr, N.K.; Koteliansky, V.E.; Critchley, D.R. A Novel Dystrophin/Utrophin-Associated Protein is an Enzymatically Inactive Member of the Phosphoglucomutase Superfamily. Eur. J. Biochem. 1996, 235, 103–113. [Google Scholar] [CrossRef]
- Belkin, A.M.; Burridge, K. Localization of Utrophin and Aciculin at Sites of Cell-Matrix and Cell-Cell Adhesion in Cultured Cells. Exp. Cell Res. 1995, 221, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Belkin, A.M.; Burridge, K. Association of aciculin with dystrophin and utrophin. J. Biol. Chem. 1995. [Google Scholar] [CrossRef] [PubMed]
- Molt, S.; Buhrdel, J.B.; Yakovlev, S.; Schein, P.; Orfanos, Z.; Kirfel, G.; Winter, L.; Wiche, G.; van der Ven, P.F.M.; Rottbauer, W.; et al. Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. J. Cell Sci. 2014, 127, 3578–3592. [Google Scholar] [CrossRef] [PubMed]
- Leber, Y.; Ruparelia, A.A.; Kirfel, G.; van der Ven, P.F.M.; Hoffmann, B.; Merkel, R.; Bryson-Richardson, R.J.; Fürst, D.O. Filamin C is a highly dynamic protein associated with fast repair of myofibrillar microdamage. Hum. Mol. Genet. 2016, 25, 2776–2788. [Google Scholar] [CrossRef]
- Zhang, Q.; Ding, Z.; Wan, L.; Tong, W.; Mao, J.; Li, L.; Hu, J.; Yang, M.; Liu, B.; Qian, X. Comprehensive analysis of the long noncoding RNA expression profile and construction of the lncRNA-mRNA co-expression network in colorectal cancer. Cancer Biol. Ther. 2020, 21, 157–169. [Google Scholar] [CrossRef]
- Sun, Y.; Long, H.; Sun, L.; Sun, X.; Pang, L.; Chen, J.; Yi, Q.; Liang, T.; Shen, Y. PGM5 is a promising biomarker and may predict the prognosis of colorectal cancer patients. Cancer Cell Int. 2019, 19, 253. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Y.; Jiang, P.; Han, W.; Liu, Y. PGM5: A novel diagnostic and prognostic biomarker for liver cancer. PeerJ 2019, 7, e7070. [Google Scholar] [CrossRef]
- Uzozie, A.C.; Selevsek, N.; Wahlander, A.; Nanni, P.; Grossmann, J.; Weber, A.; Buffoli, F.; Marra, G. Targeted Proteomics for Multiplexed Verification of Markers of Colorectal Tumorigenesis. Mol. Cell. Proteom. 2017, 16, 407–427. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Q.; Wu, R.; Li, B.; Chen, Q.; Zhang, X.; Shi, C. A Comprehensive Survey of Genomic Alterations in Gastric Cancer Reveals Recurrent Neoantigens as Potential Therapeutic Targets. Biomed Res. Int. 2019, 2019, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.M.; Zhang, B.; Hornbeck, P.V.; Gnad, F. Systematic analysis of the intersection of disease mutations with protein modifications. BMC Med. Genomics 2019, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, Z.; Yang, F.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; Lan, Q.; Wang, J.; et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J. Neurooncol. 2018, 136, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Yu, J.; Zhu, H.; Guo, Y.; Feng, S. Identification of key lncRNAs in colorectal cancer progression based on associated protein–protein interaction analysis. World J. Surg. Oncol. 2017, 15, 153. [Google Scholar] [CrossRef]
- Van Den Berg, S.; Löfdahl, P.Å.; Härd, T.; Berglund, H. Improved solubility of TEV protease by directed evolution. J. Biotechnol. 2006, 121, 291–298. [Google Scholar] [CrossRef]
- Stiers, K.M.; Xu, J.; Lee, Y.; Addison, Z.R.; Van Doren, S.R.; Beamer, L.J. Phosphorylation-Dependent Effects on the Structural Flexibility of Phosphoglucosamine Mutase from Bacillus anthracis. ACS Omega 2017, 2, 8445–8452. [Google Scholar] [CrossRef] [PubMed]
- Powell, H.R.; Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Leslie, A.G.W. Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm. Nat. Protoc. 2017. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Lebedev, A. MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, S19. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.L.; Van der Verren, S.E.; Kanchugal, S.P.; Näsvall, J.; Gutiérrez-de-Terán, H.; Selmer, M. Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase from Salmonella enterica. J. Biol. Chem. 2018, 293, 11481–11490. [Google Scholar] [CrossRef] [PubMed]
- Grøftehauge, M.K.; Hajizadeh, N.R.; Swann, M.J.; Pohl, E. Protein–ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI). Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 36–44. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Fersht, A. Structure and Mechanism in Protein Science; W.H. Freeman and Company: New York, NY, USA, 1999; ISBN 0716732688. [Google Scholar]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 2.2; Schrödinger LLC.: New York, NY, USA, 2020. [Google Scholar]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Namboori, S.C.; Graham, D.E. Acetamido Sugar Biosynthesis in the Euryarchaea. J. Bacteriol. 2008, 190, 2987–2996. [Google Scholar] [CrossRef]
- Akutsu, J.; Zhang, Z.; Tsujimura, M.; Sasaki, M.; Yohda, M.; Kawarabayasi, Y. Characterization of a Thermostable Enzyme with Phosphomannomutase/Phosphoglucomutase Activities from the Hyperthermophilic Archaeon Pyrococcus horikoshii OT3. J. Biochem. 2005, 138, 159–166. [Google Scholar] [CrossRef]
- Ye, R.W.; Zielinski, N.A.; Chakrabarty, A.M. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J. Bacteriol. 1994, 176, 4851–4857. [Google Scholar] [CrossRef]
- Ray, W.J. Role of bivalent cations in the phosphoglucomutase system. I. Characterization of enzyme-metal complexes. J. Biol. Chem. 1969, 244, 3740–3747. [Google Scholar]
- Ray, W.J.; Post, C.B.; Liu, Y.; Rhyu, G.I. Structural changes at the metal ion binding site during the phosphoglucomutase reaction. Biochemistry 1993, 32, 48–57. [Google Scholar] [CrossRef]
- Müller, S.; Diederichs, K.; Breed, J.; Kissmehl, R.; Hauser, K.; Plattner, H.; Welte, W. Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from paramecium reveals significant conformational variability. J. Mol. Biol. 2002, 315, 141–153. [Google Scholar] [CrossRef][Green Version]
- Sarma, A.V.S.; Anbanandam, A.; Kelm, A.; Mehra-Chaudhary, R.; Wei, Y.; Qin, P.; Lee, Y.; Berjanskii, M.V.; Mick, J.A.; Beamer, L.J.; et al. Solution NMR of a 463-Residue Phosphohexomutase: Domain 4 Mobility, Substates, and Phosphoryl Transfer Defect. Biochemistry 2012, 51, 807–819. [Google Scholar] [CrossRef]
- Xu, J.; Lee, Y.; Beamer, L.J.; Van Doren, S.R. Phosphorylation in the Catalytic Cleft Stabilizes and Attracts Domains of a Phosphohexomutase. Biophys. J. 2015, 108, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Ninfali, P.; Accorsi, A.; Palma, F.; Fazi, A.; Piatti, E.; Chiarantini, L.; Fornaini, G. Human erythrocyte phosphoglucomutase: Comparison of the kinetic properties of PGM1 and PGM2 isoenzymes. Biochimie 1984, 66, 617–623. [Google Scholar] [CrossRef]
- Lee, Y.; Stiers, K.M.; Kain, B.N.; Beamer, L.J. Compromised Catalysis and Potential Folding Defects in in Vitro Studies of Missense Mutants Associated with Hereditary Phosphoglucomutase 1 Deficiency. J. Biol. Chem. 2014, 289, 32010–32019. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Naught, L.E.; Regni, C.; Beamer, L.J.; Tipton, P.A. Roles of Active Site Residues in Pseudomonas aeruginosa Phosphomannomutase/Phosphoglucomutase. Biochemistry 2003, 42, 9946–9951. [Google Scholar] [CrossRef]
- Regni, C.; Naught, L.; Tipton, P.A.; Beamer, L.J. Structural Basis of Diverse Substrate Recognition by the Enzyme PMM/PGM from P. aeruginosa. Structure 2004, 12, 55–63. [Google Scholar] [CrossRef]
- Nishitani, Y.; Maruyama, D.; Nonaka, T.; Kita, A.; Fukami, T.A.; Mio, T.; Yamada-Okabe, H.; Yamada-Okabe, T.; Miki, K. Crystal Structures of N -Acetylglucosamine-phosphate Mutase, a Member of the α-d-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes. J. Biol. Chem. 2006, 281, 19740–19747. [Google Scholar] [CrossRef]
- Beamer, L.J. Mutations in hereditary phosphoglucomutase 1 deficiency map to key regions of enzyme structure and function. J. Inherit. Metab. Dis. 2015, 38, 243–256. [Google Scholar] [CrossRef]
- Ray, W.J.; Roscelli, G.A. A Kinetic Study of the Phosphoglucomutase Pathway. J. Biol. Chem. 1964, 239, 1228–1236. [Google Scholar]
- Naught, L.E.; Tipton, P.A. Formation and Reorientation of Glucose 1,6-Bisphosphate in the PMM/PGM Reaction: Transient-State Kinetic Studies. Biochemistry 2005, 44, 6831–6836. [Google Scholar] [CrossRef]
- Lee, Y.; Villar, M.T.; Artigues, A.; Beamer, L.J. Promotion of Enzyme Flexibility by Dephosphorylation and Coupling to the Catalytic Mechanism of a Phosphohexomutase. J. Biol. Chem. 2014, 289, 4674–4682. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, E.P.; Critchley, D.R. Characterisation of the Promoter which Regulates Expression of a Phosphoglucomutase-Related Protein, a Component of the Dystrophidutrophin Cytoskeleton Predominantly Expressed in Smooth Muscle. Eur. J. Biochem. 1997, 248, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Kumai, Y.; Bernier, N.J.; Perry, S.F. Angiotensin-II promotes Na+ uptake in larval zebrafish, Danio rerio, in acidic and ion-poor water. J. Endocrinol. 2014, 220, 195–205. [Google Scholar] [CrossRef]
- Takei, Y.; Hiroi, J.; Takahashi, H.; Sakamoto, T. Diverse mechanisms for body fluid regulation in teleost fishes. Am. J. Physiol. Integr. Comp. Physiol. 2014, 307, R778–R792. [Google Scholar] [CrossRef]
- Wakayama, Y.; Inoue, M.; Kojima, H.; Murahashi, M.; Shibuya, S.; Yamashita, S.; Oniki, H. Aciculin and its relation to dystrophin: Immunocytochemical studies in human normal and Duchenne dystrophy quadriceps muscles. Acta Neuropathol. 2000, 99, 654–662. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 34, 1260419. [Google Scholar] [CrossRef]
- Satir, B.H.; Wyroba, E.; Liu, L.; Lethan, M.; Satir, P.; Christensen, S.T. Evolutionary implications of localization of the signaling scaffold protein Parafusin to both cilia and the nucleus. Cell Biol. Int. 2015, 39, 136–145. [Google Scholar] [CrossRef]
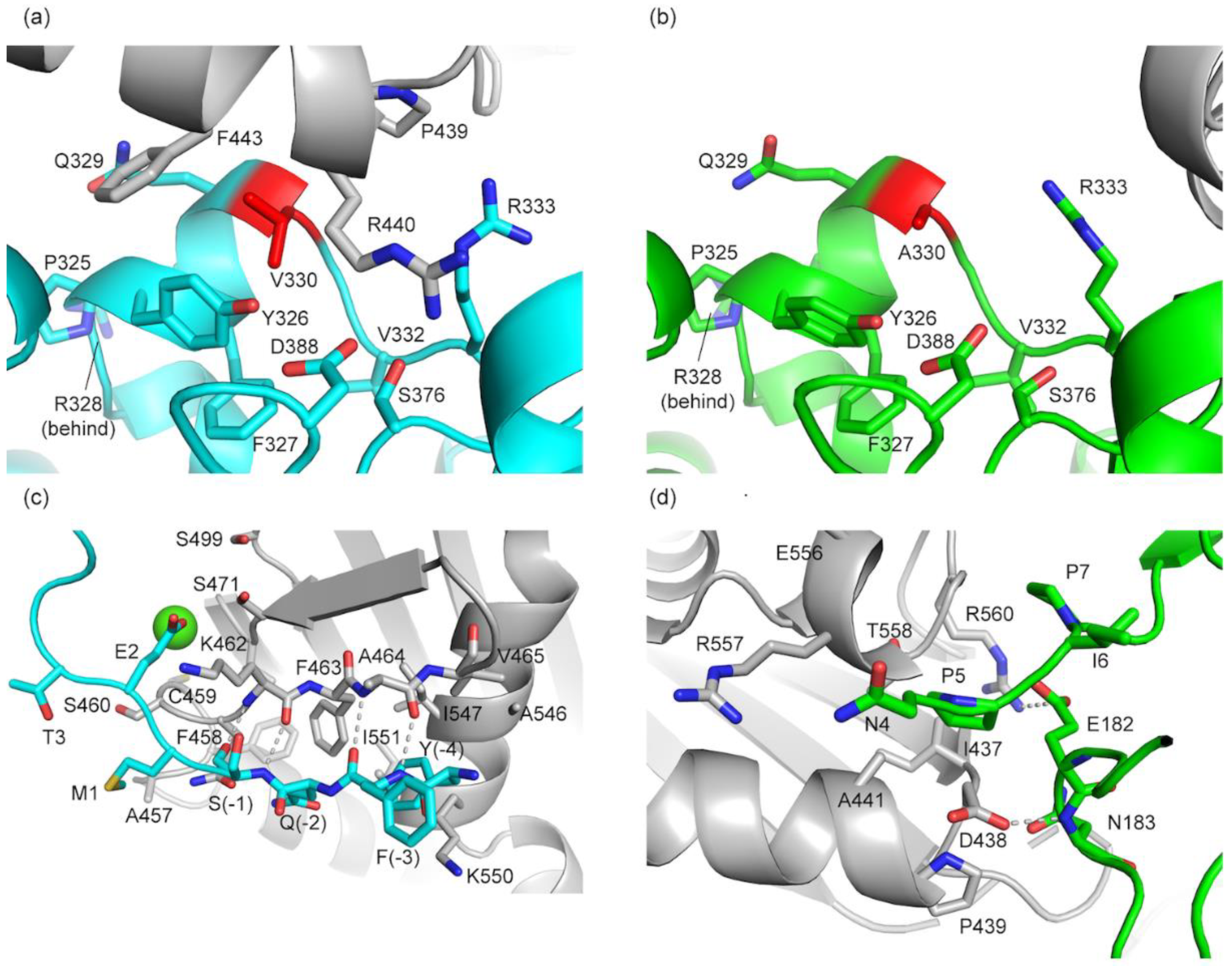
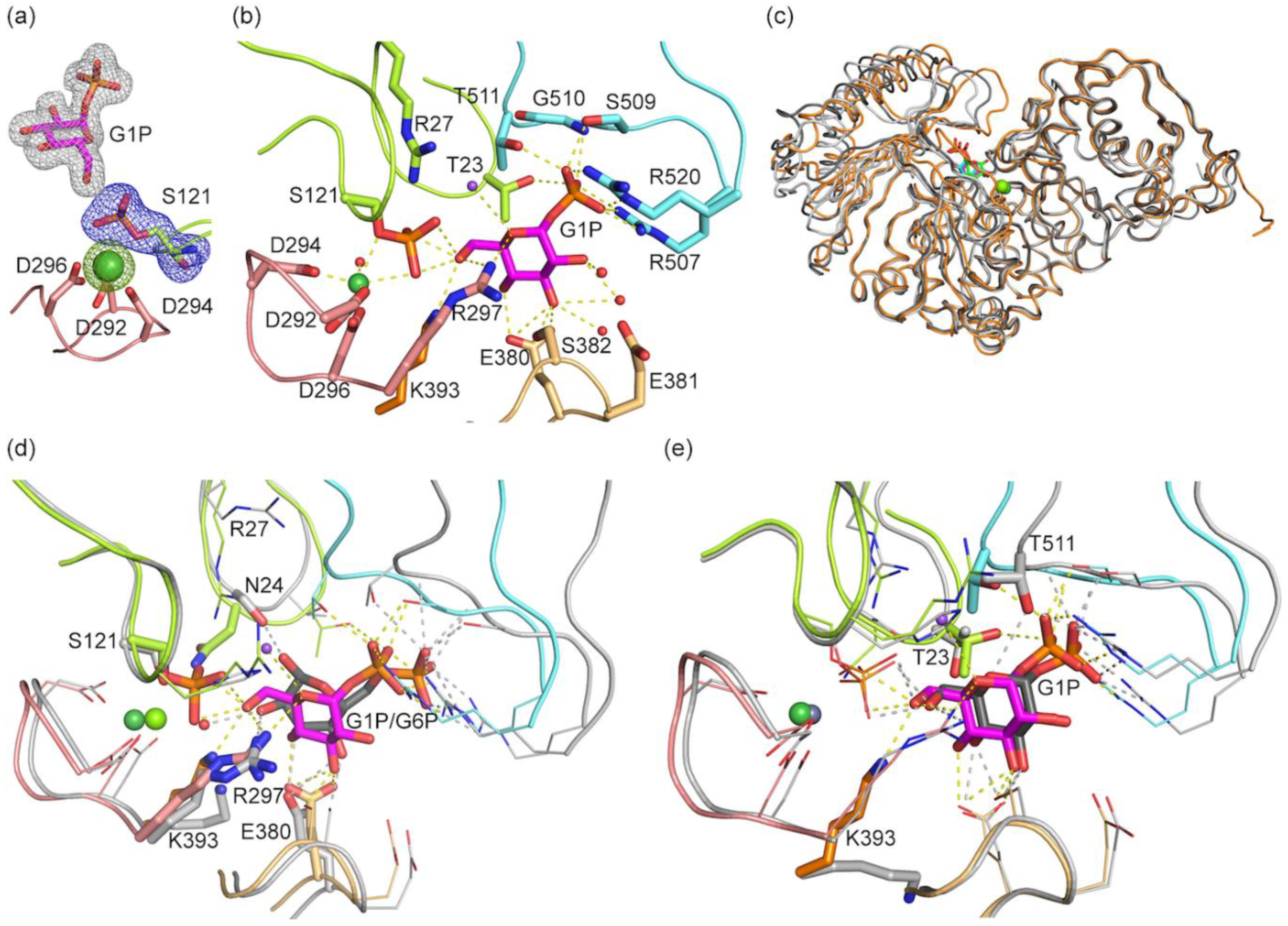
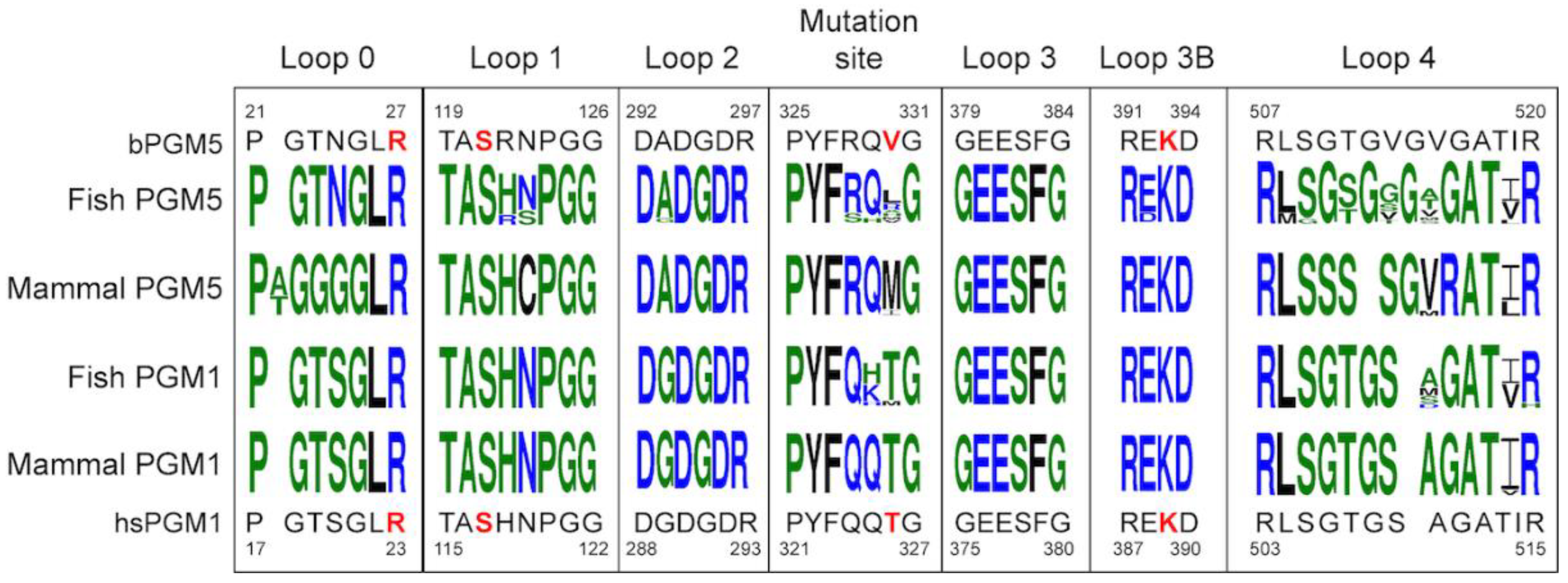
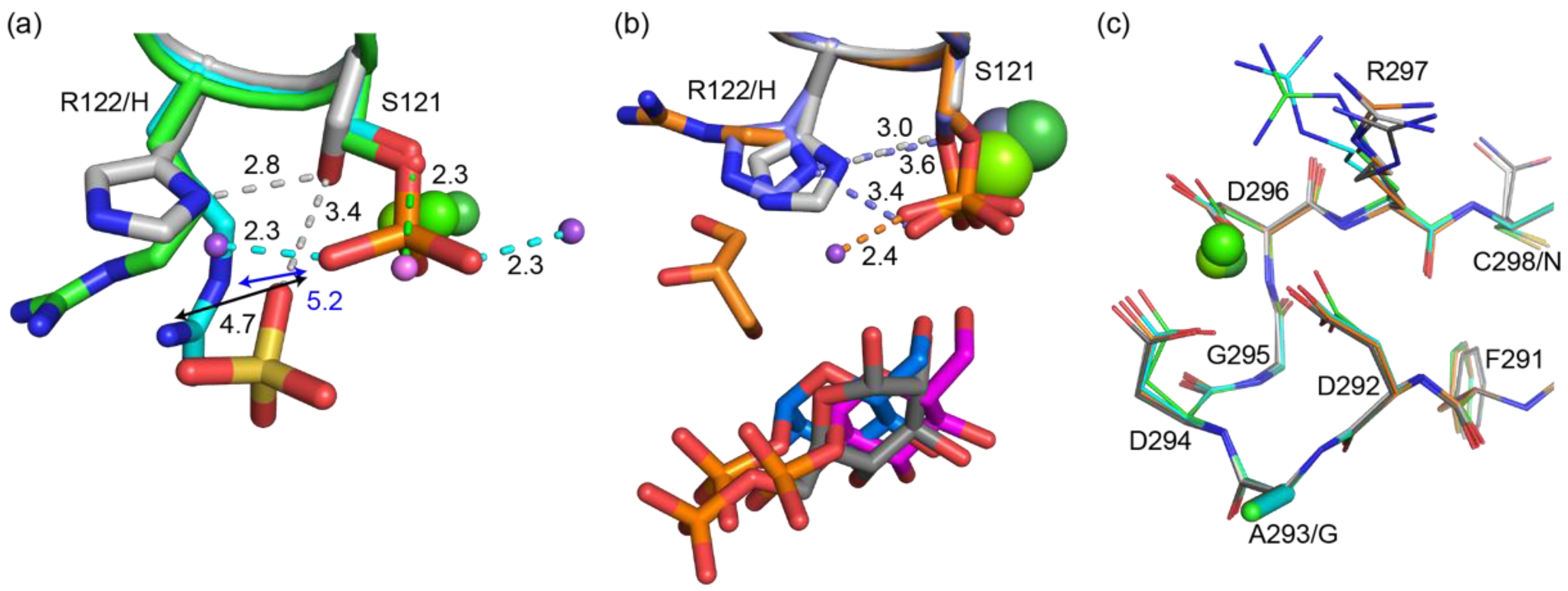
| Structure | aPGM5 apo | bPGM5 apo | bPGM5 + Ligand |
|---|---|---|---|
| PDB code | 6Y8X | 6Y8Z | 6Y8Y |
| Space Group | P21 | P 2 21 21 | P 2 21 21 |
| Unit cell parameters a, b, c (Å) | 59.21, 86.22, 63.52 | 47.20 94.21 146.69 | 47.23 94.68 146.97 |
| Unit cell angles α, β, γ (°) | 90, 109, 90 | 90 90 90 | 90 90 90 |
| Resolution (Å) | 55.98–2.25 (2.32–2.25) 2 | 48.90–2.05 (2.11–2.05) | 48.99–1.95 (2.00–1.95) |
| Observations 1 | 72197 (6868) | 183748 (9739) | 296805 (19464) |
| Unique reflections 1 | 28235 (2607) | 38168 (2184) | 48980 (3394) |
| Rmerge (I) 1 | 0.232 (0.885) | 0.124 (1.023) | 0.116 (0.816) |
| Rmeas (I) 1 | 0.289 (1.102) | 0.140 (1.222) | 0.127 (0.898) |
| Rpim (I) 1 | 0.171 (0.648) | 0.062 (0.636) | 0.051 (0.368) |
| Mean I/sigma(I) 1 | 3.3 (1.2) | 7.5 (2.0) | 9.2 (2.0) |
| CC1/2 1 | 0.966 (0.221) 2 | 0.948 (0.429) | 0.996 (0.454) |
| Completeness (%) 1 | 98.3(98.5) | 91.2 (69.0) | 99.9 (99.6) |
| Multiplicity 1 | 2.6 (2.6) | 4.8 (4.5) | 6.1 (5.7) |
| Resolution used in refinement (Å) | 55.98–2.25 (2.33–2.25) | 44.93–2.05 (2.10–2.05) | 47.34–1.95 (1.99–1.95) |
| No. atoms protein | 4337 | 4423 | 4482 |
| No. atoms ligands | 2 | 34 | 63 |
| No atoms waters | 216 | 379 | 447 |
| Rwork 1 | 0.2368 (0.3345) | 0.1871 (0.4744) | 0.1682 (0.2620) |
| Rfree 1 | 0.2884 (0.3504) | 0.2355 (0.4785) | 0.2097 (0.2790) |
| RMSD bond lengths (Å) | 0.002 | 0.002 | 0.004 |
| RMSD bond angles (°) | 0.473 | 0.496 | 0.622 |
| Ramachandran plot (%) | |||
| Favored | 95.33 | 97.50 | 97.71 |
| Additionally allowed | 4.67 | 2.50 | 2.29 |
| Wilson B factor (Å2) | 25.58 | 26.54 | 28.60 |
| Protein average B (Å2) | 35.85 | 35.31 | 32.48 |
| Ligands average B (Å2) | 39.99 | 49.97 | 43.01 |
| Water average B (Å2) | 30.29 | 39.68 | 39.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gustafsson, R.; Eckhard, U.; Ye, W.; Enbody, E.D.; Pettersson, M.; Jemth, P.; Andersson, L.; Selmer, M. Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring—An Inactive Enzyme with Intact Substrate Binding. Biomolecules 2020, 10, 1631. https://doi.org/10.3390/biom10121631
Gustafsson R, Eckhard U, Ye W, Enbody ED, Pettersson M, Jemth P, Andersson L, Selmer M. Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring—An Inactive Enzyme with Intact Substrate Binding. Biomolecules. 2020; 10(12):1631. https://doi.org/10.3390/biom10121631
Chicago/Turabian StyleGustafsson, Robert, Ulrich Eckhard, Weihua Ye, Erik D. Enbody, Mats Pettersson, Per Jemth, Leif Andersson, and Maria Selmer. 2020. "Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring—An Inactive Enzyme with Intact Substrate Binding" Biomolecules 10, no. 12: 1631. https://doi.org/10.3390/biom10121631
APA StyleGustafsson, R., Eckhard, U., Ye, W., Enbody, E. D., Pettersson, M., Jemth, P., Andersson, L., & Selmer, M. (2020). Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring—An Inactive Enzyme with Intact Substrate Binding. Biomolecules, 10(12), 1631. https://doi.org/10.3390/biom10121631






