Autophagy Stimulus-Dependent Role of the Small GTPase Ras2 in Peroxisome Degradation
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Plasmids
2.3. Cell Culture Conditions
2.4. Fluorescence Microscopy
2.5. Immunodetection
2.6. Pexophagy Assay
2.7. Peroxisome Degradation in Constitutive Presence of Glucose Induced by Rapamycin
2.8. Statistical Analysis
3. Results and Discussion
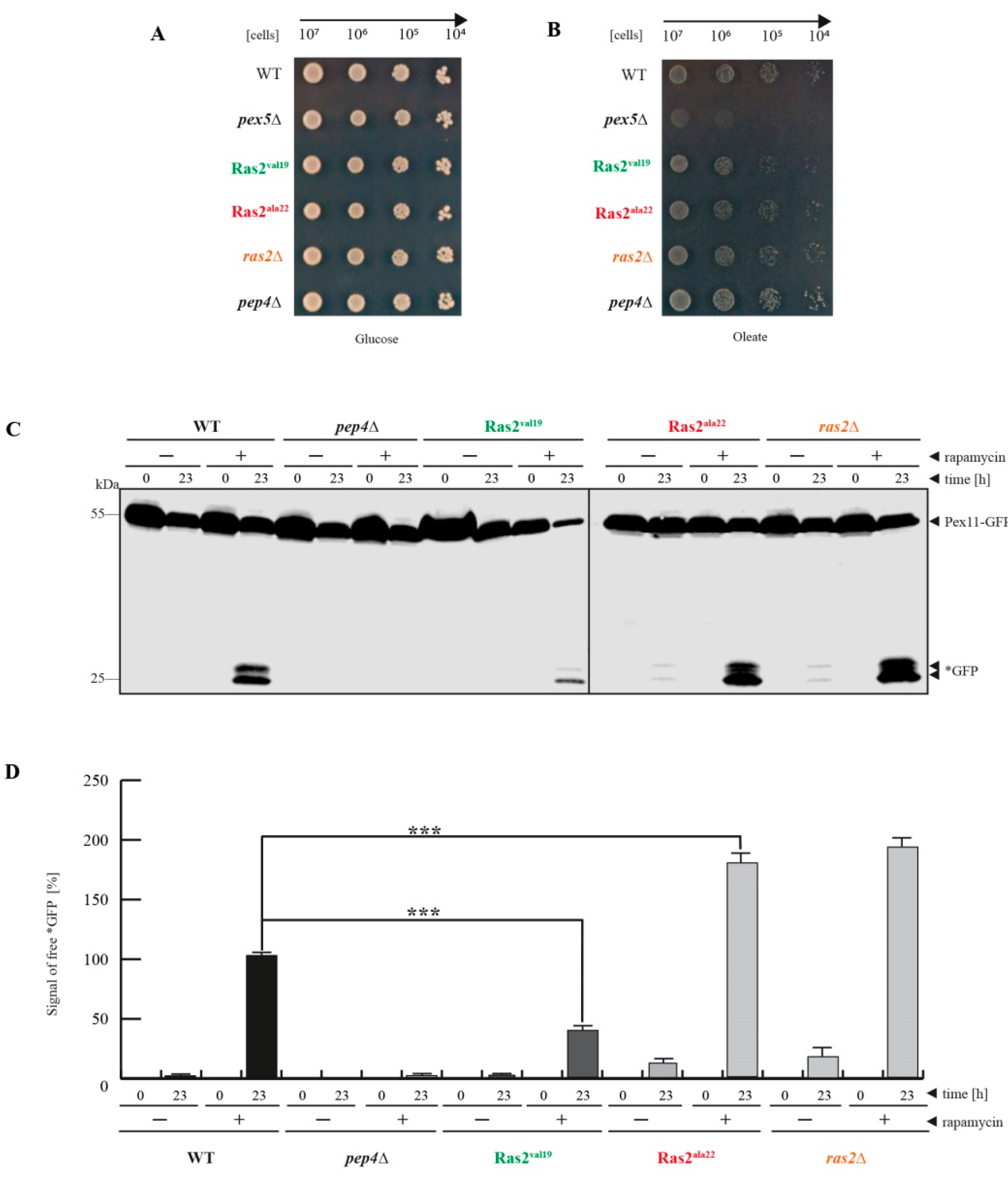
3.1. Ras2ON Suppresses and Ras2OFF Supports Peroxisome Degradation after Rapamycin-Treatment of Glucose-Grown Cells
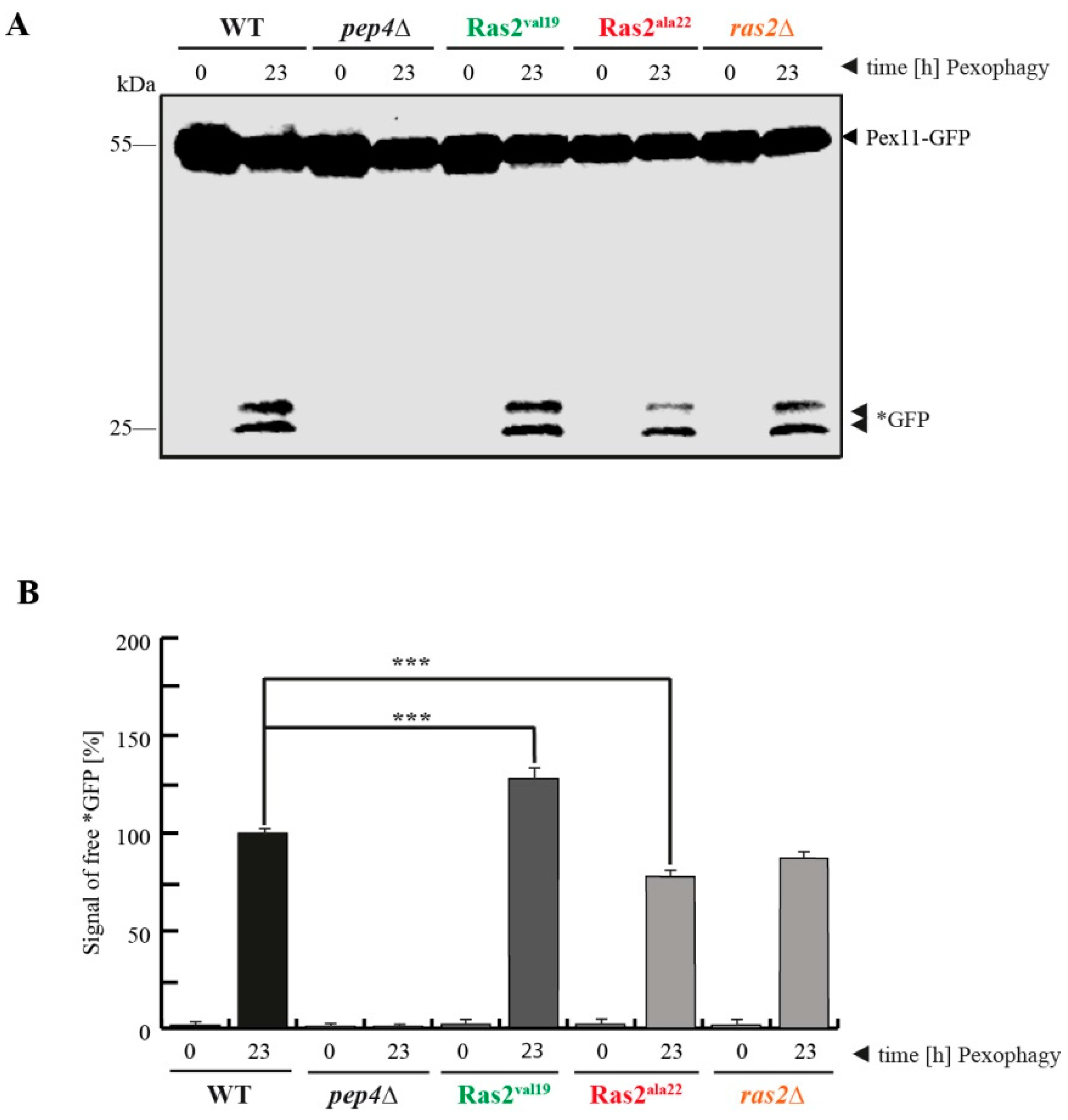
3.2. Ras2ON Supports and Ras2OFF Suppresses Pexophagy after the Shift of Glucose-Lacking to Glucose-Containing Medium
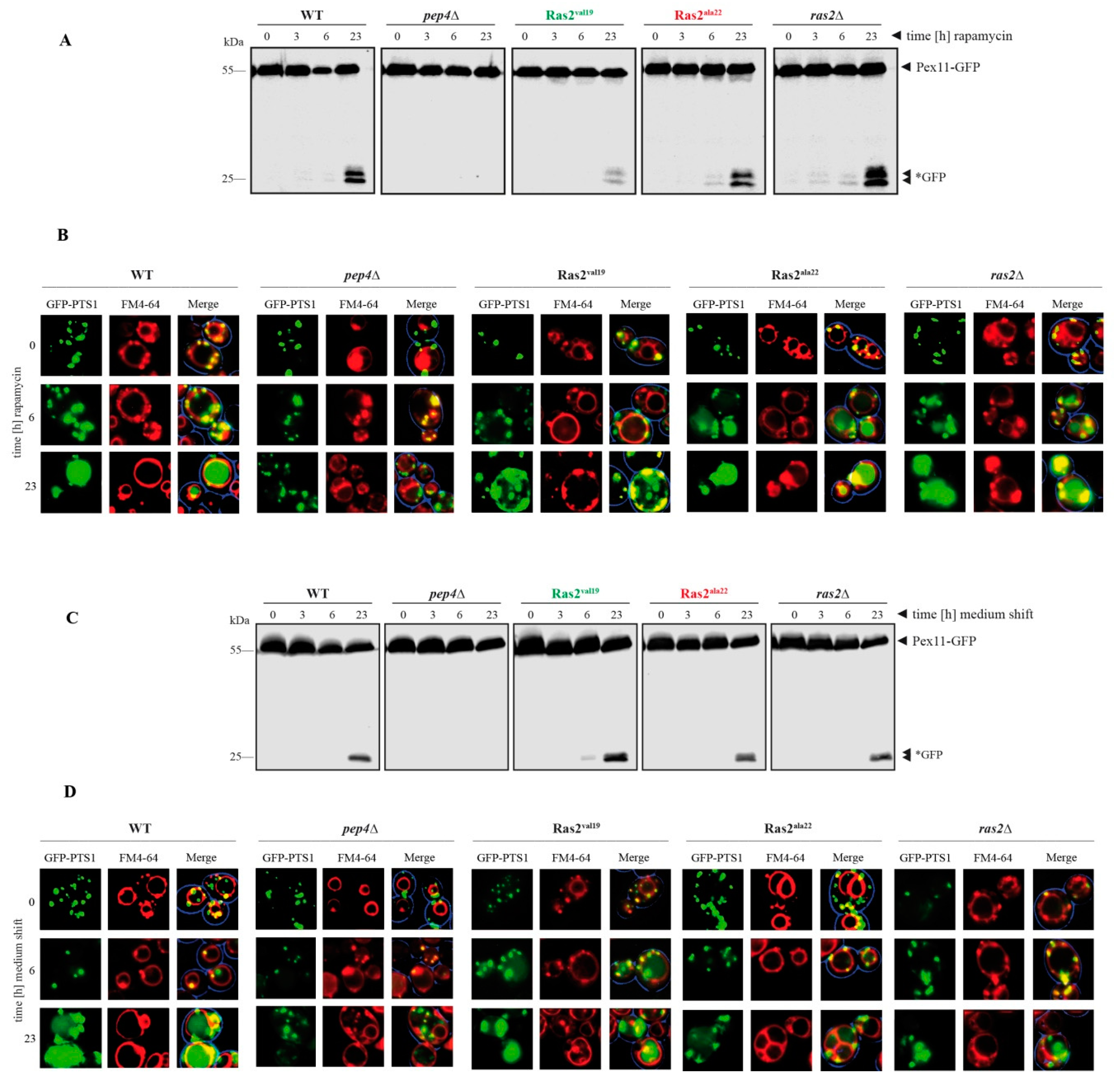
3.3. Time-Dependent Analysis of the Ras2 Activity Effects on Peroxisome Breakdown
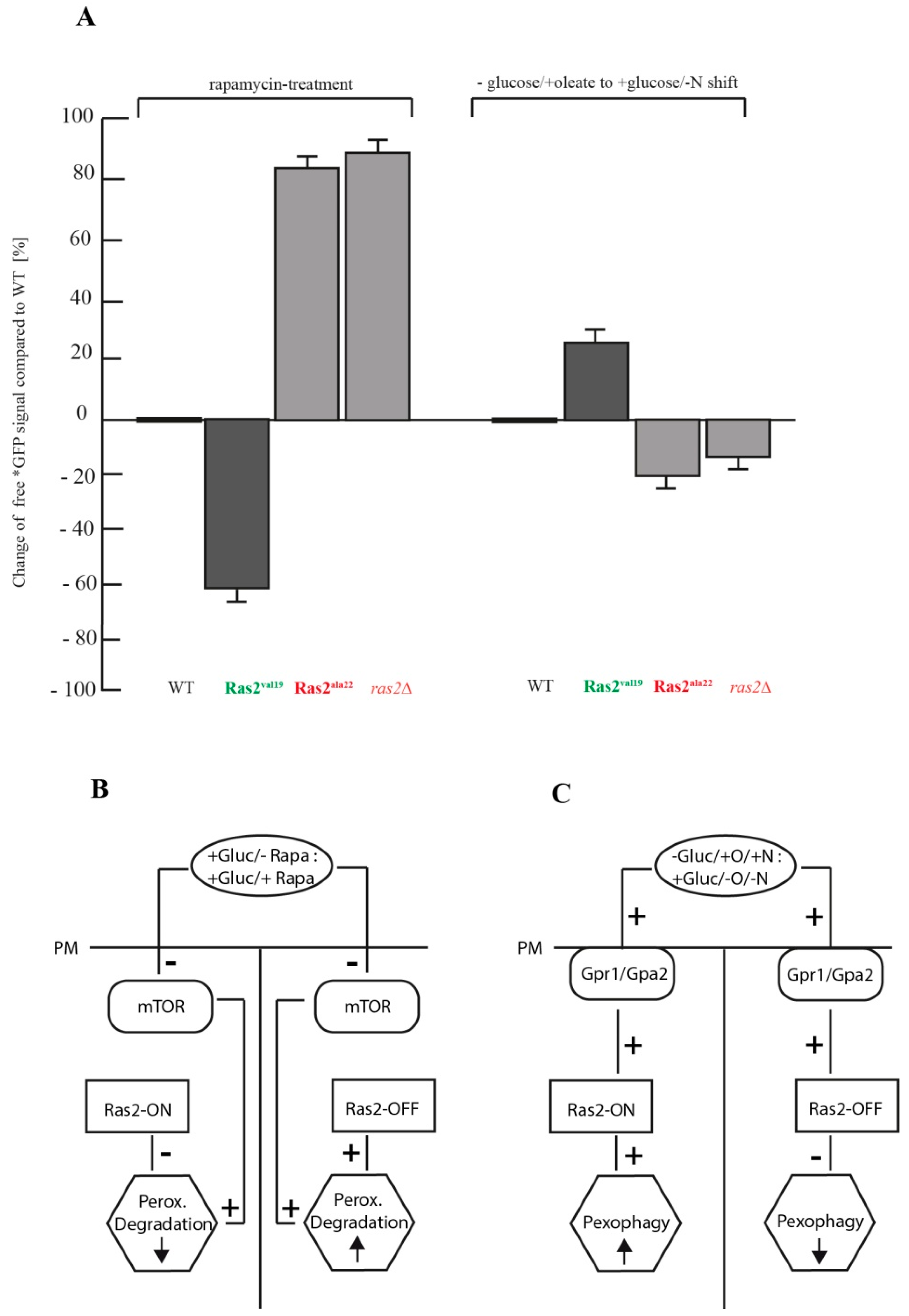
3.4. Comparison of the Autophagy Stimulus-Dependent Role of Ras2 Activity Mutants on Different Modes of Peroxisome Breakdown
4. Conclusions
- (1)
- Ras2 supports mTOR in suppressing peroxisome degradation via bulk autophagy under standard glucose-conditions. After inhibition of mTOR with rapamycin, the Ras2ON version allows only a small rate of peroxisome degradation. The additional inactivation Ras2OFF results in a strongly enhanced peroxisome degradation under glucose conditions.
- (2)
- Ras2 supports glucose-sensing to enhance selective pexophagy. In cells shifted from an oleate-containing/glucose-deficient medium (oleate medium) to a glucose-containing/nitrogen- reduced medium (pexophagy medium), Ras2ON enhances pexophagy, most likely via its central role in glucose-sensing signaling. In contrast, the Ras2OFF displays a reduced rate of pexophagy, most likely because the mutant is unable to fully sense the presence of glucose in the medium and therefore cannot support the breakdown of peroxisomes in presence of glucose.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dechant, R.; Peter, M. Nutrient signals driving cell growth. Curr. Opin. Cell Biol. 2008, 20, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed]
- Boutouja, F.; Stiehm, C.M.; Platta, H.W. mTOR: A Cellular Regulator Interface in Health and Disease. Cells 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef]
- Boutouja, F.; Stiehm, C.M.; Mastalski, T.; Brinkmeier, R.; Reidick, C.; El Magraoui, F.; Platta, H.W. Vps10-mediated targeting of Pep4 determines the activity of the vacuole in a substrate-dependent manner. Sci. Rep. 2019, 9, 10557. [Google Scholar] [CrossRef]
- Kataoka, T.; Powers, S.; McGill, C.; Fasano, O.; Strathern, J.; Broach, J.; Wigler, M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 1984, 37, 437–445. [Google Scholar] [CrossRef]
- Broek, D.; Samiy, N.; Fasano, O.; Fujiyama, A.; Tamanoi, F.; Northup, J.; Wigler, M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell 1985, 41, 763–769. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Gasper, R.; Wittinghofer, F. The Ras switch in structural and historical perspective. Biol. Chem. 2019, 401, 143–163. [Google Scholar] [CrossRef]
- Brunsveld, L.; Kuhlmann, J.; Alexandrov, K.; Wittinghofer, A.; Goody, R.S.; Waldmann, H. Lipidated ras and rab peptides and proteins--synthesis, structure, and function. Angew. Chem. Int. Ed. Engl. 2006, 45, 6622–6646. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Weinberg, R.A. Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 2008, 9, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.A.; Mattos, C. The K-Ras, N-Ras, and H-Ras Isoforms: Unique Conformational Preferences and Implications for Targeting Oncogenic Mutants. Cold Spring Harb. Perspect. Med. 2018, 8, a031427. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.M.; Choi, D.K.; Jung, K.; Bae, J.; Kim, J.S.; Park, S.W.; Song, K.H.; Kim, Y.S. Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration. Nat. Commun. 2017, 8, 15090. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.; Powers, S.; Cameron, S.; Fasano, O.; Goldfarb, M.; Broach, J.; Wigler, M. Functional homology of mammalian and yeast RAS genes. Cell 1985, 40, 19–26. [Google Scholar] [CrossRef]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Boutouja, F.; Stiehm, C.M.; Reidick, C.; Mastalski, T.; Brinkmeier, R.; El Magraoui, F.; Platta, H.W. Vac8 Controls Vacuolar Membrane Dynamics during Different Autophagy Pathways in Saccharomyces cerevisiae. Cells 2019, 8, 661. [Google Scholar] [CrossRef]
- Erdmann, R.; Veenhuis, M.; Mertens, D.; Kunau, W.-H. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1989, 86, 5419–5423. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adel, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Motley, A.M.; Nuttall, J.M.; Hettema, E.H. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012, 31, 2852–2868. [Google Scholar] [CrossRef]
- Platta, H.W.; Girzalsky, W.; Erdmann, R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 2004, 384, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Brinkmeier, R.; Reidick, C.; Galiani, S.; Clausen, M.P.; Eggeling, C. Regulation of peroxisomal matrix protein import by ubiquitination. Biochim. Biophys. Acta 2016, 1863, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Reidick, C.; Boutouja, F.; Platta, H.W. The class III phosphatidylinositol 3-kinase Vps34 in Saccharomyces cerevisiae. Biol. Chem. 2017, 398, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Mastalski, T.; Brinkmeier, R.; Platta, H.W. The peroxisomal PTS1-import defect of PEX1-deficient cells is independent of pexophagy in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2020, 21, 867. [Google Scholar] [CrossRef]
- Stephan, J.S.; Yeh, Y.Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Kraft, C. Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 2020, 65, 50–57. [Google Scholar] [CrossRef]
- Nazarko, V.Y.; Futej, K.O.; Thevelein, J.M.; Sibirny, A.A. Differences in glucose sensing and signaling for pexophagy between the baker’s yeast Saccharomyces cerevisiae and the methylotrophic yeast Pichia pastoris. Autophagy 2008, 4, 381–384. [Google Scholar] [CrossRef][Green Version]
- Nazarko, V.Y.; Thevelein, J.M.; Sibirny, A.A. G-protein-coupled receptor Gpr1 and G-protein Gpa2 of cAMP-dependent signaling pathway are involved in glucose-induced pexophagy in the yeast Saccharomyces cerevisiae. Cell Biol. Int. 2008, 32, 502–504. [Google Scholar] [CrossRef]
- Colombo, S.; Ronchetti, D.; Thevelein, J.M.; Winderickx, J.; Martegani, E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 46715–46722. [Google Scholar] [CrossRef]
- Peeters, K.; Van Leemputte, F.; Fischer, B.; Bonini, B.M.; Quezada, H.; Tsytlonok, M.; Haesen, D.; Vanthienen, W.; Bernardes, N.; Gonzalez-Blas, C.B.; et al. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 2017, 8, 922. [Google Scholar] [CrossRef]
- Milanesi, R.; Coccetti, P.; Tripodi, F. The Regulatory Role of Key Metabolites in the Control of Cell Signaling. Biomolecules 2020, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Hung, G.C.; Brown, C.R.; Wolfe, A.B.; Liu, J.; Chiang, H.L. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J. Biol. Chem. 2004, 279, 49138–49150. [Google Scholar] [CrossRef] [PubMed]
- Hlavatá, L.; Aguilaniu, H.; Pichová, A.; Nyström, T. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J. 2003, 22, 3337–3345. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, C.; Tan, L.J.; Mochida, K.; Kirisako, H.; Koizumi, M.; Asai, E.; Sakoh-Nakatogawa, M.; Ohsumi, Y.; Nakatogawa, H. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J. Cell Biol. 2014, 207, 91–105. [Google Scholar] [CrossRef]
- Wang, L.; Shang, Z.; Zhou, Y.; Hu, X.; Chen, Y.; Fan, Y.; Wei, X.; Wu, L.; Liang, Q.; Zhang, J.; et al. Autophagy mediates glucose starvation-induced glioblastoma cell quiescence and chemoresistance through coordinating cell metabolism, cell cycle, and survival. Cell Death Dis. 2018, 12, 213. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boutouja, F.; Platta, H.W. Autophagy Stimulus-Dependent Role of the Small GTPase Ras2 in Peroxisome Degradation. Biomolecules 2020, 10, 1553. https://doi.org/10.3390/biom10111553
Boutouja F, Platta HW. Autophagy Stimulus-Dependent Role of the Small GTPase Ras2 in Peroxisome Degradation. Biomolecules. 2020; 10(11):1553. https://doi.org/10.3390/biom10111553
Chicago/Turabian StyleBoutouja, Fahd, and Harald W. Platta. 2020. "Autophagy Stimulus-Dependent Role of the Small GTPase Ras2 in Peroxisome Degradation" Biomolecules 10, no. 11: 1553. https://doi.org/10.3390/biom10111553
APA StyleBoutouja, F., & Platta, H. W. (2020). Autophagy Stimulus-Dependent Role of the Small GTPase Ras2 in Peroxisome Degradation. Biomolecules, 10(11), 1553. https://doi.org/10.3390/biom10111553




