Genetic Factors Associated with Increased Host Defense Antimicrobial Peptide Resistance in Sequence Type 5 Healthcare-Associated MRSA Clinical Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. S. aureus Strains and Culture
2.2. Sequencing and Cloning of mprF and graRS
2.3. Quantification of Ttranscriptional Expression by RT-qPCR
2.4. Net Cell Surface Charge in MRSA Strains
2.5. Statistical Analyses
3. Results
3.1. SNPs within mprF and graRS ORFs
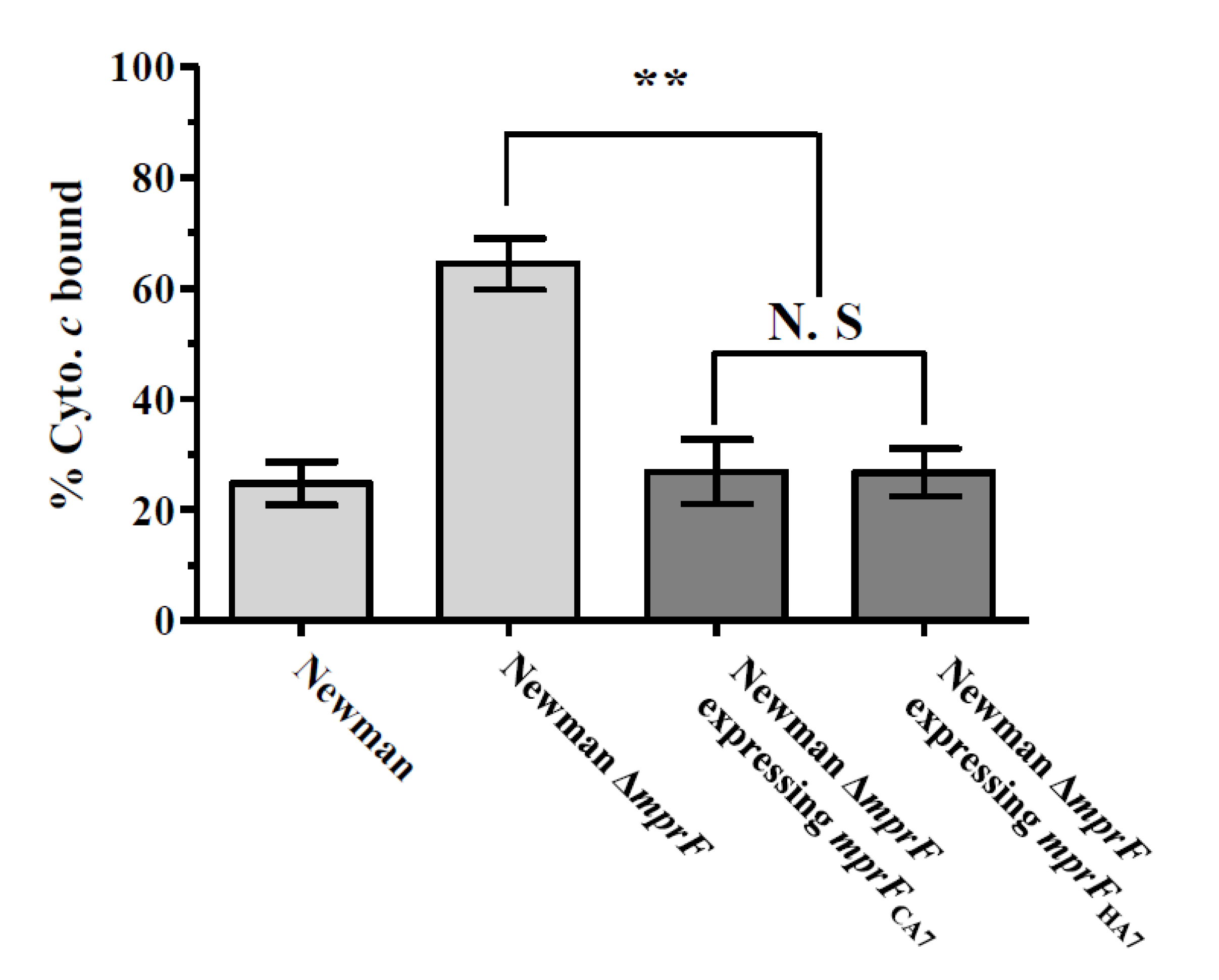
3.2. Effect of mprF SNPs on The Net Cell Surface Positive Charge
3.3. Expression of mprF, dltA, and graS in MRSA Strains
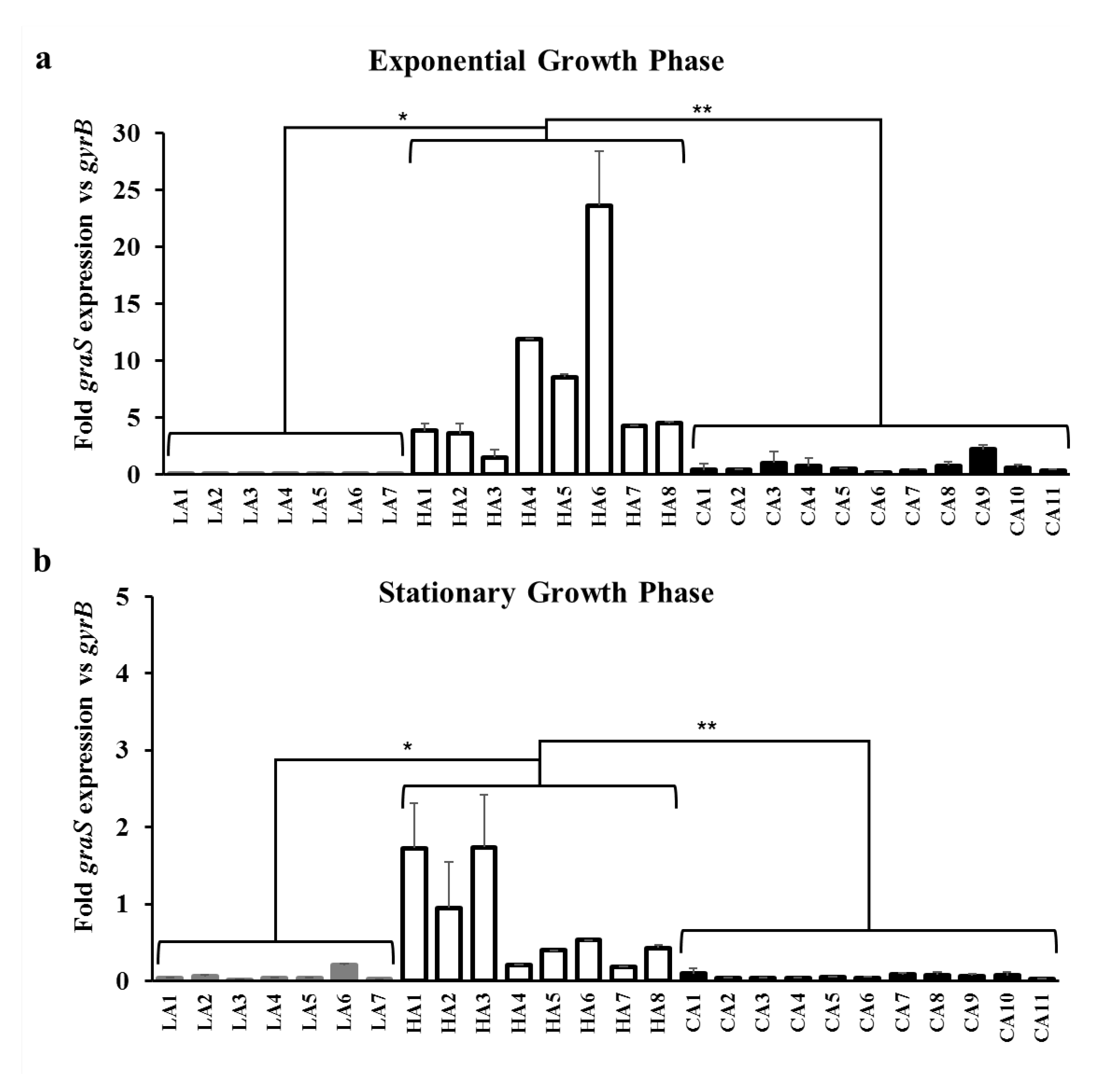
3.4. Effect of GraS SNPs on the Regulation of mprF and dltABCD Transcription
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Graffunder, E.M.; Venezia, R.A. Risk factors associated with nosocomial methicillin-resistant Staphylococcus aureus (MRSA) infection including previous use of antimicrobials. J. Antimicrob. Chemother. 2002, 49, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Soo Ko, K.; Kim, Y.S.; Song, J.H.; Yeom, J.S.; Lee, H.; Jung, S.I.; Jeong, D.R.; Kim, S.W.; Chang, H.H.; Ki, H.K.; et al. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob. Agents Chemother. 2005, 49, 3583–3585. [Google Scholar] [CrossRef] [PubMed]
- Okuma, K.; Iwakawa, K.; Turnidge, J.D.; Grubb, W.B.; Bell, J.M.; O’Brien, F.G.; Coombs, G.W.; Pearman, J.W.; Tenover, F.C.; Kapi, M.; et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 2002, 40, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio 2012, 3. [Google Scholar] [CrossRef]
- Song, J.-H.; Hsueh, P.-R.; Chung, D.R.; Ko, K.S.; Kang, C.-I.; Peck, K.R.; Yeom, J.-S.; Kim, S.-W.; Chang, H.-H.; Kim, Y.-S. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: An ANSORP study. J. Antimicrob. Chemother. 2011, 66, 1061–1069. [Google Scholar] [CrossRef]
- Kim, E.S.; Song, J.S.; Lee, H.J.; Choe, P.G.; Park, K.H.; Cho, J.H.; Park, W.B.; Kim, S.-H.; Bang, J.-H.; Kim, D.-M. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J. Antimicrob. Chemother. 2007, 60, 1108–1114. [Google Scholar] [CrossRef]
- Joo, E.-J.; Chung, D.; Ha, Y.; Park, S.; Kang, S.-J.; Kim, S.; Kang, C.-I.; Peck, K.; Lee, N.; Ko, K. Community-associated Panton–Valentine leukocidin-negative meticillin-resistant Staphylococcus aureus clone (ST72-MRSA-IV) causing healthcare-associated pneumonia and surgical site infection in Korea. J. Hosp. Infect. 2012, 81, 149–155. [Google Scholar] [CrossRef]
- Mishra, N.N.; Bayer, A.S.; Moise, P.A.; Yeaman, M.R.; Sakoulas, G. Reduced susceptibility to host-defense cationic peptides and daptomycin coemerge in methicillin-resistant Staphylococcus aureus from daptomycin-naive bacteremic patients. J. Infect. Dis. 2012, 206, 1160–1167. [Google Scholar] [CrossRef]
- Kang, K.M.; Park, J.H.; Kim, S.H.; Yang, S.J. Potential role of host defense antimicrobial peptide resistance in increased virulence of health care-associated MRSA strains of sequence type (ST) 5 versus livestock-associated and community-associated MRSA strains of ST72. Comp. Immunol. Microbiol. Infect. Dis. 2019, 62, 13–18. [Google Scholar] [CrossRef]
- Mishra, N.N.; Yang, S.-J.; Sawa, A.; Rubio, A.; Nast, C.C.; Yeaman, M.R.; Bayer, A.S. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 2312–2318. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Kreiswirth, B.N.; Sakoulas, G.; Yeaman, M.R.; Xiong, Y.Q.; Sawa, A.; Bayer, A.S. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 2009, 200, 1916–1920. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Xiong, Y.Q.; Dunman, P.M.; Schrenzel, J.; Francois, P.; Peschel, A.; Bayer, A.S. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2009, 53, 2636–2637. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Yang, S.J.; Chen, L.; Muller, C.; Saleh-Mghir, A.; Kuhn, S.; Peschel, A.; Yeaman, M.R.; Nast, C.C.; Kreiswirth, B.N.; et al. Emergence of daptomycin resistance in daptomycin-naive rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS ONE 2013, 8, e71151. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.J.; Mishra, N.N.; Kang, K.M.; Lee, G.Y.; Park, J.H.; Bayer, A.S. Impact of multiple single-nucleotide polymorphisms within mprF on daptomycin resistance in Staphylococcus aureus. Microb. Drug Resist. 2018, 24, 1075–1081. [Google Scholar] [CrossRef]
- Yang, S.-J.; Mishra, N.N.; Rubio, A.; Bayer, A.S. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob. Agents Chemother. 2013, 57, 5658–5664. [Google Scholar] [CrossRef]
- Bayer, A.S.; Mishra, N.N.; Chen, L.; Kreiswirth, B.N.; Rubio, A.; Yang, S.J. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-resistant Staphylococcus aureus clinical isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2015, 59, 4930–4937. [Google Scholar] [CrossRef]
- Yang, S.J.; Bayer, A.S.; Mishra, N.N.; Meehl, M.; Ledala, N.; Yeaman, M.R.; Xiong, Y.Q.; Cheung, A.L. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun. 2012, 80, 74–81. [Google Scholar] [CrossRef]
- Chaili, S.; Cheung, A.L.; Bayer, A.S.; Xiong, Y.Q.; Waring, A.J.; Memmi, G.; Donegan, N.; Yang, S.J.; Yeaman, M.R. The GraS sensor in Staphylococcus aureus mediates resistance to host defense peptides differing in mechanisms of action. Infect Immun. 2016, 84, 459–466. [Google Scholar] [CrossRef]
- Cheung, A.L.; Bayer, A.S.; Yeaman, M.R.; Xiong, Y.Q.; Waring, A.J.; Memmi, G.; Donegan, N.; Chaili, S.; Yang, S.J. Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect Immun. 2014, 82, 5336–5345. [Google Scholar] [CrossRef]
- Kim, E.S.; Bae, I.-G.; Cho, J.E.; Choi, Y.J.; Kim, I.-H.; Kang, G.-S.; Sin, H.-y.; Song, K.-H.; Park, C.; Lee, D.-G. Clinical and molecular characterization of invasive heteroresistant vancomycin-intermediate Staphylococcus aureus infections in Korean hospitals. J. Clin. Microbiol. 2016, 54, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Song, J.W.; Yang, S.J.; Shin, S.; Seo, K.S.; Park, Y.H.; Park, K.T. Genotypic and phenotypic characterization of methicillin-resistant Staphylococcus aureus isolated from bovine mastitic milk in Korea. J. Food Prot. 2016, 79, 1725–1732. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.W.; Iandolo, J.J. Rapid isolation of DNA from Staphylococcus aureus. Appl. Environ. Microbiol. 1983, 46, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Jack, R.W.; Otto, M.; Collins, L.V.; Staubitz, P.; Nicholson, G.; Kalbacher, H.; Nieuwenhuizen, W.F.; Jung, G.; Tarkowski, A.; et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 2001, 193, 1067–1076. [Google Scholar] [CrossRef]
- Bruckner, R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 1992, 122, 187–192. [Google Scholar] [CrossRef]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Schenk, S.; Laddaga, R.A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 1992, 73, 133–138. [Google Scholar] [CrossRef]
- Lan, L.; Cheng, A.; Dunman, P.M.; Missiakas, D.; He, C. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J. Bacteriol. 2010, 192, 3068–3077. [Google Scholar] [CrossRef]
- Bertsche, U.; Weidenmaier, C.; Kuehner, D.; Yang, S.J.; Baur, S.; Wanner, S.; Francois, P.; Schrenzel, J.; Yeaman, M.R.; Bayer, A.S. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation. Antimicrob. Agents Chemother. 2011, 55, 3922–3928. [Google Scholar] [CrossRef]
- Bayer, A.S.; Mishra, N.N.; Cheung, A.L.; Rubio, A.; Yang, S.J. Dysregulation of mprF and dltABCD expression among daptomycin-non-susceptible MRSA clinical isolates. J. Antimicrob. Chemother. 2016, 71, 2100–2104. [Google Scholar] [CrossRef]
- Yang, S.J.; Nast, C.C.; Mishra, N.N.; Yeaman, M.R.; Fey, P.D.; Bayer, A.S. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: Evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 2010, 54, 3079–3085. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.J.; Proctor, R.A.; Sahl, H.G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Murthy, M.H.; Olson, M.E.; Wickert, R.W.; Fey, P.D.; Jalali, Z. Daptomycin non-susceptible meticillin-resistant Staphylococcus aureus USA 300 isolate. J. Med. Microbiol. 2008, 57, 1036–1038. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009, 5, e1000660. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.M.; Kuhn, S.; Slavetinsky, C.J.; Krismer, B.; Heilbronner, S.; Gekeler, C.; Kraus, D.; Wagner, S.; Peschel, A. The lipid-modifying multiple peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Götz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [PubMed]
- Meehl, M.; Herbert, S.; Gotz, F.; Cheung, A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 2679–2689. [Google Scholar] [CrossRef]
- Mishra, N.N.; Bayer, A.S.; Tran, T.T.; Shamoo, Y.; Mileykovskaya, E.; Dowhan, W.; Guan, Z.; Arias, C.A. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS ONE 2012, 7, e43958. [Google Scholar] [CrossRef]
- Nam, E.Y.; Yang, S.J.; Kim, E.S.; Cho, J.E.; Park, K.H.; Jung, S.I.; Yoon, N.; Kim, D.M.; Lee, C.S.; Jang, H.C.; et al. Emergence of daptomycin-nonsusceptible methicillin-resistant Staphylococcus aureus clinical isolates among daptomycin-naive patients in Korea. Microb. Drug Resist. 2018, 24, 534–541. [Google Scholar] [CrossRef]
- Bayer, A.S.; Mishra, N.N.; Sakoulas, G.; Nonejuie, P.; Nast, C.C.; Pogliano, J.; Chen, K.T.; Ellison, S.N.; Yeaman, M.R.; Yang, S.J. Heterogeneity of mprF sequences in methicillin-resistant Staphylococcus aureus clinical isolates: Role in cross-resistance between daptomycin and host defense antimicrobial peptides. Antimicrob. Agents Chemother. 2014, 58, 7462–7467. [Google Scholar] [CrossRef]
- Mishra, N.N.; Bayer, A.S.; Weidenmaier, C.; Grau, T.; Wanner, S.; Stefani, S.; Cafiso, V.; Bertuccio, T.; Yeaman, M.R.; Nast, C.C.; et al. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: Relative roles of mprF and dlt operons. PLoS ONE 2014, 9, e107426. [Google Scholar] [CrossRef] [PubMed]



| mprF SNPs a | graS SNPs a | graR SNPs a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | MLST | SCC mec | Codon Change | b Nucleotide Position | Amino Acid Change | Codon Change | b Nucleotide Position | Amino Acid Change | Codon Change | b Nucleotide Position | Amino Acid Change |
| LA1 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | c NA | NA | No change | NA | NA |
| LA2 | ST72 | IV | ATT→ATG ATT→ACT TTC→TGC | 1125 1382 1649 | I375M I461T F550C | No change | NA | NA | No change | NA | NA |
| LA3 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| LA4 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| LA5 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| LA6 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| LA7 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| HA1 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | AAA→AAT | 300 | K100N |
| HA2 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA3 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA4 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA5 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA6 | ST5 | II | CAT→CCT GAG→AAG | 362 802 | H121P E268K | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA7 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| HA8 | ST5 | II | No change | NA | NA | ATA→ACA | 671 | I224T | No change | NA | NA |
| CA1 | ST72 | IV | ATT→ATG ATT→ACT ACT→ATT | 1125 1382 1997 | I375M I461T T666I | No change | NA | NA | No change | NA | NA |
| CA2 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA3 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA4 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | GAT→AAT | 631 | D211N | No change | NA | NA |
| CA5 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA6 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA7 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA8 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA9 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| CA10 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | GTG→GGG | 260 | V87G |
| CA11 | ST72 | IV | ATT→ATG ATT→ACT | 1125 1382 | I375M I461T | No change | NA | NA | No change | NA | NA |
| Parameter | Groups | p Value for | |||
|---|---|---|---|---|---|
| ST72 LA-MRSA | ST5 HA-MRSA | ST72 CA-MRSA | ST72 LA-MRSA Vs. ST5 HA-MRSA | ST72 CA-MRSA Vs. ST5 HA-MRSA | |
| Fold mprF expression vs. gyrB in: | |||||
| Exponential growth phase | 2.12 ± 1.07 | 4.68 ± 1.92 | 4.21 ± 1.40 | <0.05 | a NS |
| Stationary growth phase | 3.54 ± 3.23 | 5.38 ± 4.64 | 1.63 ± 0.60 | a NS | <0.05 |
| Fold dltA expression vs. gyrB in: | |||||
| Exponential growth phase | 1.24 ± 1.65 | 9.80 ± 7.02 | 2.88 ± 0.81 | <0.01 | <0.01 |
| Stationary growth phase | 1.60 ± 1.63 | 9.65 ± 9.86 | 0.39 ± 0.16 | NS | <0.01 |
| Fold graS expression vs. gyrB in: | |||||
| Exponential growth phase | 0.03 ± 0.04 | 7.71 ± 7.22 | 0.67 ± 0.55 | <0.05 | <0.01 |
| Stationary growth phase | 0.06 ± 0.07 | 0.77 ± 0.64 | 0.05 ± 0.02 | <0.05 | <0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.-M.; Lee, G.Y.; Yang, S.-J. Genetic Factors Associated with Increased Host Defense Antimicrobial Peptide Resistance in Sequence Type 5 Healthcare-Associated MRSA Clinical Isolates. Biomolecules 2020, 10, 1415. https://doi.org/10.3390/biom10101415
Kang K-M, Lee GY, Yang S-J. Genetic Factors Associated with Increased Host Defense Antimicrobial Peptide Resistance in Sequence Type 5 Healthcare-Associated MRSA Clinical Isolates. Biomolecules. 2020; 10(10):1415. https://doi.org/10.3390/biom10101415
Chicago/Turabian StyleKang, Kyoung-Mi, Gi Yong Lee, and Soo-Jin Yang. 2020. "Genetic Factors Associated with Increased Host Defense Antimicrobial Peptide Resistance in Sequence Type 5 Healthcare-Associated MRSA Clinical Isolates" Biomolecules 10, no. 10: 1415. https://doi.org/10.3390/biom10101415
APA StyleKang, K.-M., Lee, G. Y., & Yang, S.-J. (2020). Genetic Factors Associated with Increased Host Defense Antimicrobial Peptide Resistance in Sequence Type 5 Healthcare-Associated MRSA Clinical Isolates. Biomolecules, 10(10), 1415. https://doi.org/10.3390/biom10101415





