Dissecting the Interaction of FGF8 with Receptor FGFRL1
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Constructs
2.2. Expression of Recombinant Proteins
2.3. ELISA
2.4. Surface Plasmon Resonance Analysis
3. Results
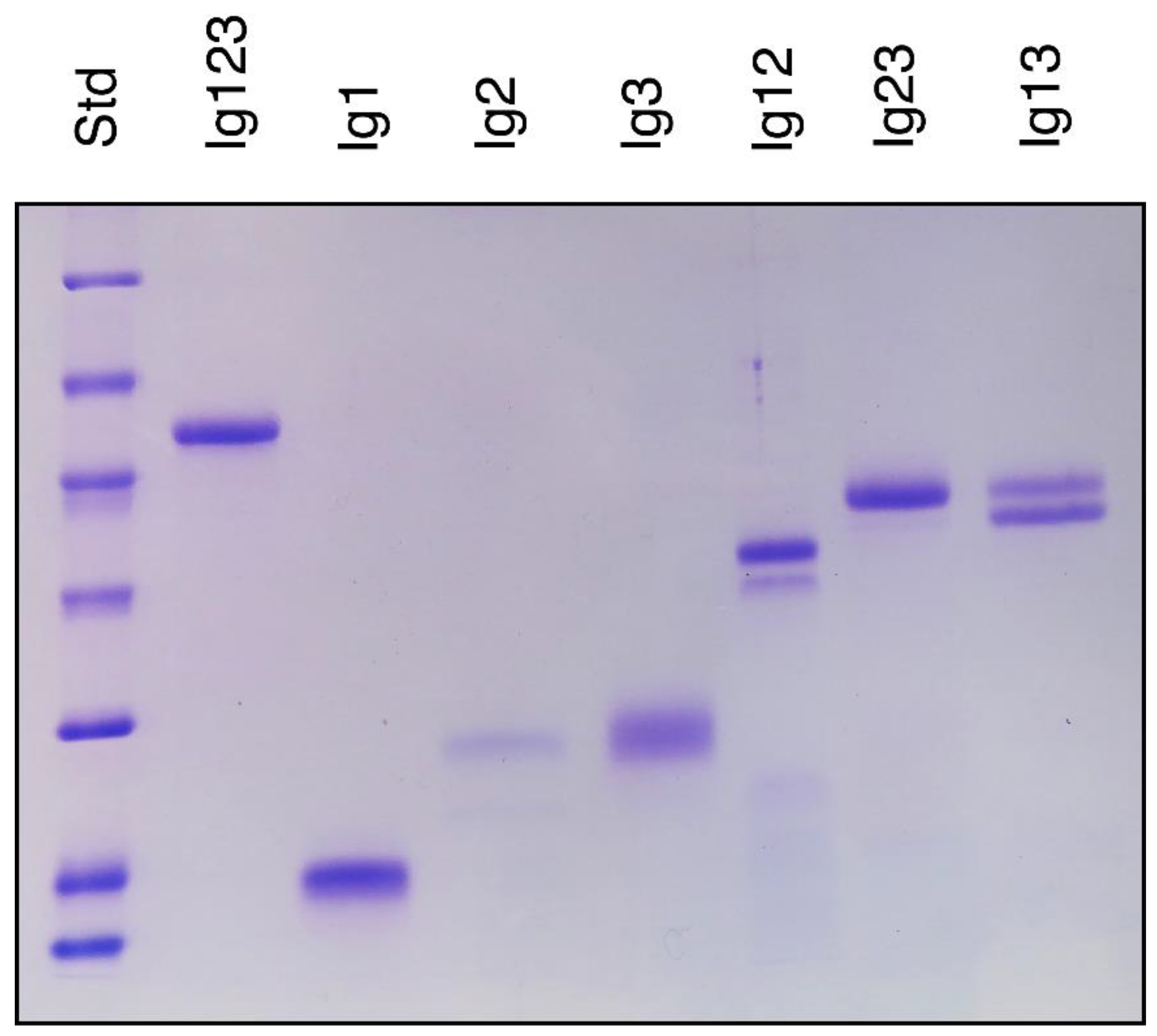
3.1. Protein Expression
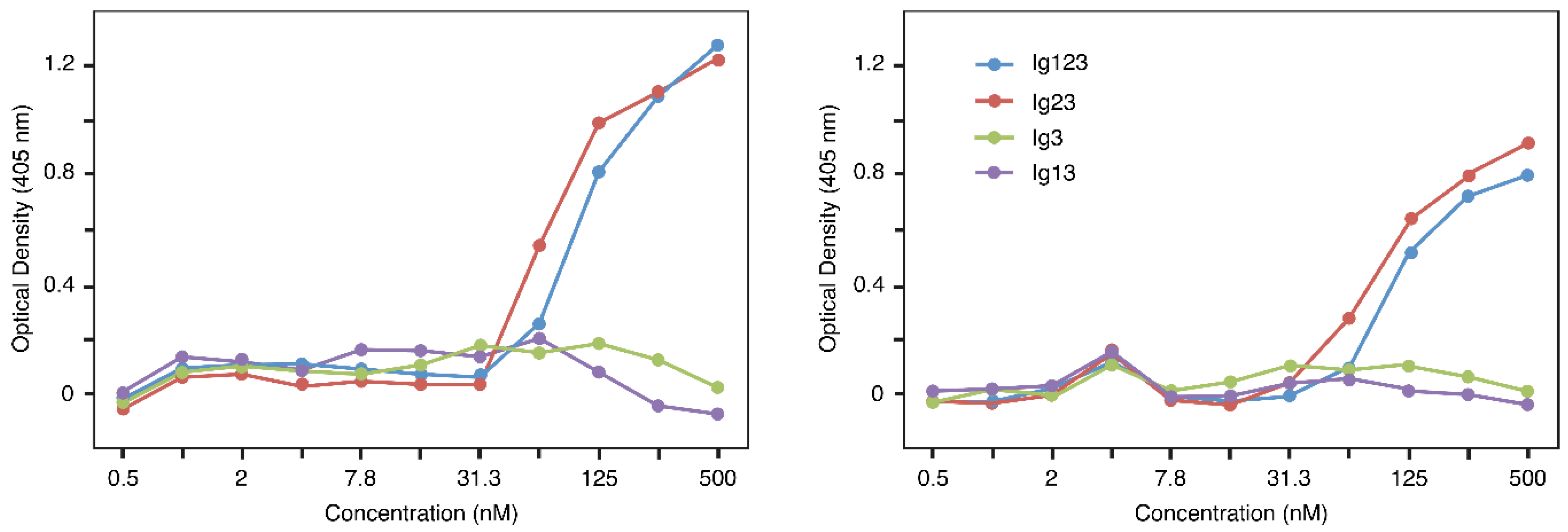
3.2. Interaction by ELISA
3.3. Biacore Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ornitz, D.M.; Marie, P.J. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015, 29, 1463–1486. [Google Scholar] [CrossRef]
- Beenken, A.; Mohammadi, M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 2009, 8, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Ornitz, D.M. Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J. Biochem. 2010, 149, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, M.; Trueb, B. Characterization of a Novel Protein (FGFRL1) from Human Cartilage Related to FGF Receptors. Genomics 2000, 69, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Trueb, B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell. Mol. Life Sci. 2010, 68, 951–964. [Google Scholar] [CrossRef]
- Sleeman, M.; Fraser, J.; McDonald, M.; Yuan, S.; White, D.; Grandison, P.; Kumble, K.; Watson, J.D.; Murison, J. Identification of a new fibroblast growth factor receptor, FGFR5. Gene 2001, 271, 171–182. [Google Scholar] [CrossRef]
- Bluteau, G.; Zhuang, L.; Amann, R.; Trueb, B. Targeted Disruption of the Intracellular Domain of Receptor FgfrL1 in Mice. PLoS ONE 2014, 9, e105210. [Google Scholar] [CrossRef]
- Trueb, B.; Zhuang, L.; Taeschler, S.; Wiedemann, M. Characterization of FGFRL1, a Novel Fibroblast Growth Factor (FGF) Receptor Preferentially Expressed in Skeletal Tissues. J. Biol. Chem. 2003, 278, 33857–33865. [Google Scholar] [CrossRef]
- Baertschi, S.; Zhuang, L.; Trueb, B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007, 274, 6241–6253. [Google Scholar] [CrossRef]
- Gerber, S.D.; Steinberg, F.; Beyeler, M.; Villiger, P.M.; Trueb, B. The murine Fgfrl1 receptor is essential for the development of the metanephric kidney. Dev. Biol. 2009, 335, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, I.; Vainio, S. Kidney Development: An Overview. Nephron Exp. Nephrol. 2014, 126, 40–44. [Google Scholar] [CrossRef]
- Combes, A.; Davies, J.A.; Little, M.H. Cell–Cell Interactions Driving Kidney Morphogenesis. Mech. Regen. 2015, 112, 467–508. [Google Scholar] [CrossRef]
- Tanigawa, S.; Perantoni, A.O. Modeling renal progenitors-defining the niche. Differentiation 2016, 91, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Walker, K.A.; Sims-Lucas, S.; Bates, C.M. Fibroblast growth factor receptor signaling in kidney and lower urinary tract development. Pediatr. Nephrol. 2015, 31, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.; Vainio, S.; Vassileva, G.; McMahon, A.P. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 1994, 372, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.D.; Beauchamp, P.; Zhuang, L.; Villiger, P.M.; Trueb, B. Functional domains of the FgfrL1 receptor. Dev. Biol. 2020, 461, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Dubrulle, J.; Pourquie, O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 2004, 427, 419–422. [Google Scholar] [CrossRef]
- Martinez, S.; Crossley, P.H.; Cobos, I.; Rubenstein, J.L.; Martin, G.R. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development 1999, 126, 1189–1200. [Google Scholar]
- Xu, J.; Liu, Z.; Ornitz, D.M. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development 2000, 127, 1833–1843. [Google Scholar]
- Grieshammer, U.; Cebrian, C.; Ilagan, R.; Meyers, E.; Herzlinger, D.; Martin, G.R. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development 2005, 132, 3847–3857. [Google Scholar] [CrossRef] [PubMed]
- Perantoni, A.O.; Timofeeva, O.; Naillat, F.; Richman, C.; Pajni-Underwood, S.; Wilson, C.; Vainio, S.; Dove, L.F.; Lewandoski, M. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 2005, 132, 3859–3871. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.D.; Amann, R.; Wyder, S.; Trueb, B. Comparison of the Gene Expression Profiles from Normal and Fgfrl1 Deficient Mouse Kidneys Reveals Downstream Targets of Fgfrl1 Signaling. PLoS ONE 2012, 7, e33457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trueb, B.; Amann, R.; Gerber, S.D. Role of FGFRL1 and other FGF signaling proteins in early kidney development. Cell. Mol. Life Sci. 2012, 70, 2505–2518. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, F.; Zhuang, L.; Beyeler, M.; Kälin, R.E.; Mullis, P.E.; Brandli, A.; Trueb, B. The FGFRL1 Receptor Is Shed from Cell Membranes, Binds Fibroblast Growth Factors (FGFs), and Antagonizes FGF Signaling inXenopusEmbryos. J. Biol. Chem. 2009, 285, 2193–2202. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Krummel, B.; Saiki, R. A general method ofin vitropreparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988, 16, 7351–7367. [Google Scholar] [CrossRef]
- Rieckmann, T.; Kotevic, I.; Trueb, B. The cell surface receptor FGFRL1 forms constitutive dimers that promote cell adhesion. Exp. Cell Res. 2008, 314, 1071–1081. [Google Scholar] [CrossRef]
- Rieckmann, T.; Zhuang, L.; Flück, C.E.; Trueb, B. Characterization of the first FGFRL1 mutation identified in a craniosynostosis patient. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2009, 1792, 112–121. [Google Scholar] [CrossRef][Green Version]
- Zhuang, L.; Pandey, A.; Villiger, P.M.; Trueb, B. Cell–cell fusion induced by the Ig3 domain of receptor FGFRL1 in CHO cells. Biochim. Biophys. Acta (BBA)-Bioenerg. 2015, 1853, 2273–2285. [Google Scholar] [CrossRef]
- Ibrahimi, O.A.; Zhang, F.; Eliseenkova, A.V.; Linhardt, R.J.; Mohammadi, M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum. Mol. Genet. 2003, 13, 69–78. [Google Scholar] [CrossRef]
- Olsen, S.K.; Li, J.Y.; Bromleigh, C.; Eliseenkova, A.V.; Ibrahimi, O.A.; Lao, Z.; Zhang, F.; Linhardt, R.J.; Joyner, A.L.; Mohammadi, M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006, 20, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.N.; Hubbard, S.R.; Schlessinger, J.; Mohammadi, M. Crystal Structures of Two FGF-FGFR Complexes Reveal the Determinants of Ligand-Receptor Specificity. Cell 2000, 101, 413–424. [Google Scholar] [CrossRef]
- Amann, R.; Trueb, B. Evidence that the novel receptor FGFRL1 signals indirectly via FGFR1. Int. J. Mol. Med. 2013, 32, 983–988. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Steinberg, F.; Gerber, S.D.; Rieckmann, T.; Trueb, B. Rapid Fusion and Syncytium Formation of Heterologous Cells upon Expression of the FGFRL1 Receptor. J. Biol. Chem. 2010, 285, 37704–37715. [Google Scholar] [CrossRef]


| Construct | ka (1/Ms) | kd (1/s) | KD (M) | Rmax (RU) | tc | Chi2 (RU2) | U−Value |
|---|---|---|---|---|---|---|---|
| Ig123 | 3.73 × 105 | 1.26 × 10−3 | 3.37 × 10−9 | 224.20 (0.33) | 6.257 × 1017 | 3.00 | 1 |
| (9.0 × 102) * | (6.8 × 10−6) | ||||||
| Ig12 | 5.49 × 106 | 1.07 × 10−2 | 1.94 × 10−9 | 192.10 (0.84) | 1.107 × 107 | 4.02 | 12 |
| (6.1 × 105) | (1.0 × 10−3) | ||||||
| Ig23 | 3.90 × 105 | 8.06 × 10−4 | 2.07 × 10−9 | 264.00 (0.60) | 7.533 × 107 | 1.43 | 1 |
| (4.5 × 103) | (5.7 × 10−6) | ||||||
| Ig13 | 1.59 × 106 | 2.04 × 10−3 | n.a. ** | 37.47 (0.31) | 8.027 × 1017 | 10.10 | 5 |
| (2.2 × 104) | (5.9 × 10−5) | ||||||
| Ig1 | 2.25 × 106 | 2.98 × 10−2 | n.a. | 17.02 (0.41) | 1.863 × 1014 | 1.18 | 7 |
| (1.1 × 105) | (7.2 × 10−4) | ||||||
| Ig2 | 5.32 × 106 | 9.20 × 10−3 | 1.73 × 10−9 | 143.50 (0.18) | 1.421 × 107 | 1.13 | 3 |
| (1.5 × 105) | (2.5 × 10−4) | ||||||
| Ig3 | 3.09 × 105 | 5.27 × 10−3 | n.a. | 14.75 (0.49) | 4.606 × 107 | 1.24 | 7 |
| (4.5 × 104) | (6.2 × 10−4) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, L.; Vogel, M.; Villiger, P.M.; Trueb, B. Dissecting the Interaction of FGF8 with Receptor FGFRL1. Biomolecules 2020, 10, 1399. https://doi.org/10.3390/biom10101399
Zhuang L, Vogel M, Villiger PM, Trueb B. Dissecting the Interaction of FGF8 with Receptor FGFRL1. Biomolecules. 2020; 10(10):1399. https://doi.org/10.3390/biom10101399
Chicago/Turabian StyleZhuang, Lei, Monique Vogel, Peter M. Villiger, and Beat Trueb. 2020. "Dissecting the Interaction of FGF8 with Receptor FGFRL1" Biomolecules 10, no. 10: 1399. https://doi.org/10.3390/biom10101399
APA StyleZhuang, L., Vogel, M., Villiger, P. M., & Trueb, B. (2020). Dissecting the Interaction of FGF8 with Receptor FGFRL1. Biomolecules, 10(10), 1399. https://doi.org/10.3390/biom10101399






