Structural and Functional Analyses of Human ChaC2 in Glutathione Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Cloning and Protein Production
2.3. Mutagenesis
2.4. Crystallization
2.5. X-ray Diffraction Data Collection, Structure Determination, and Refinement
2.6. Cell Culture and Preparation of ChaC2-Overexpressing Cells
2.7. Quantification of GSH
2.8. Viability Assay (MTT Assay)
2.9. Colony-Forming Assay
2.10. Immunoblotting
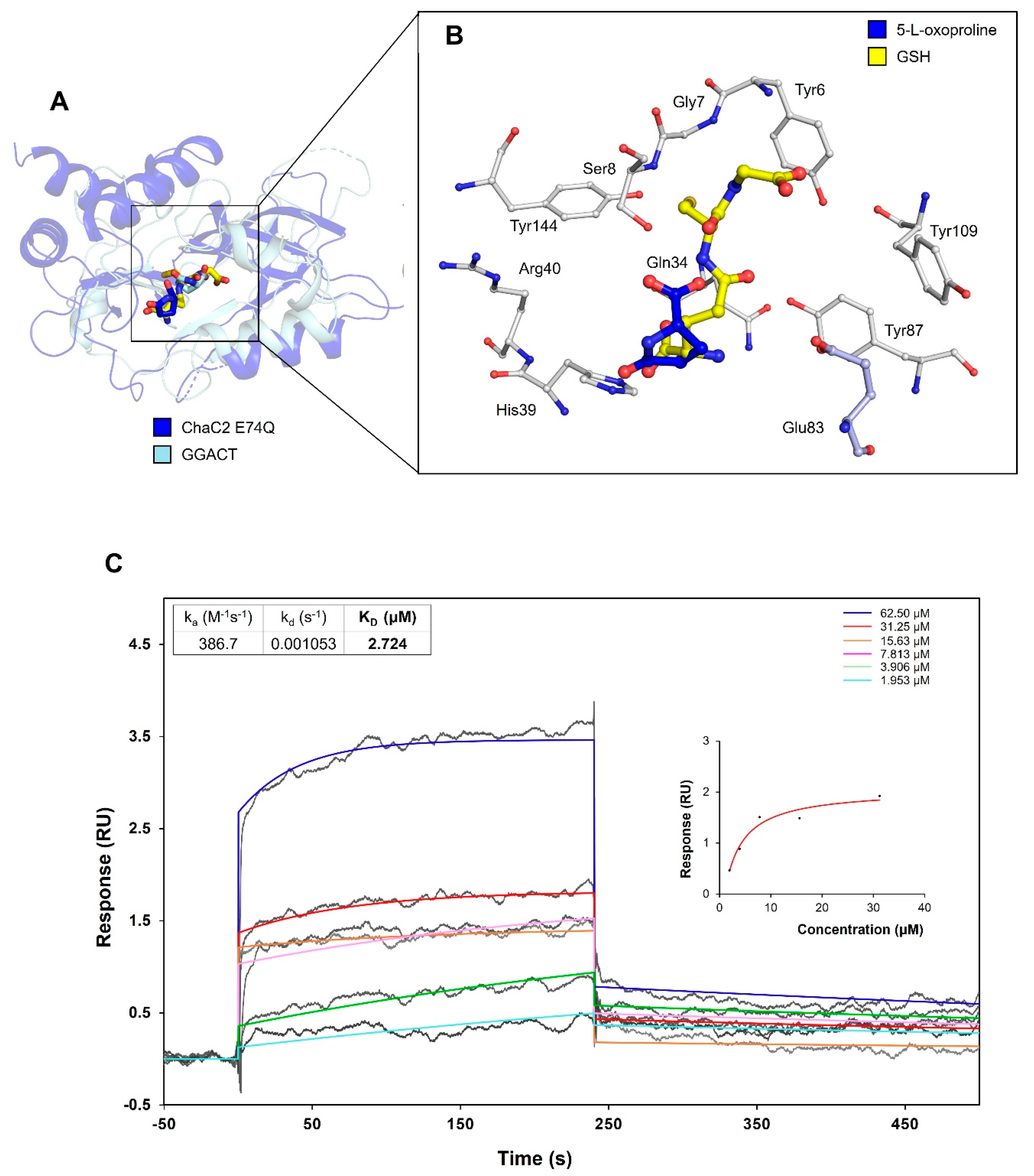
2.11. Docking Study
2.12. Surface Plasmon Resonance Experiment
2.13. Statistical Analysis
2.14. Accession Numbers
3. Results
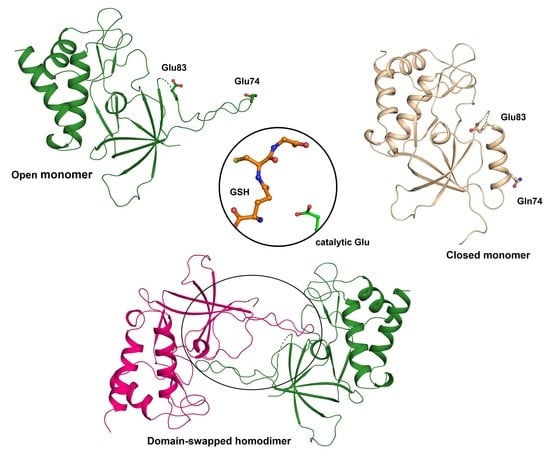
3.1. The ChaC2 Structure was Determined Through Rational Met-Substitution for MAD Phasing
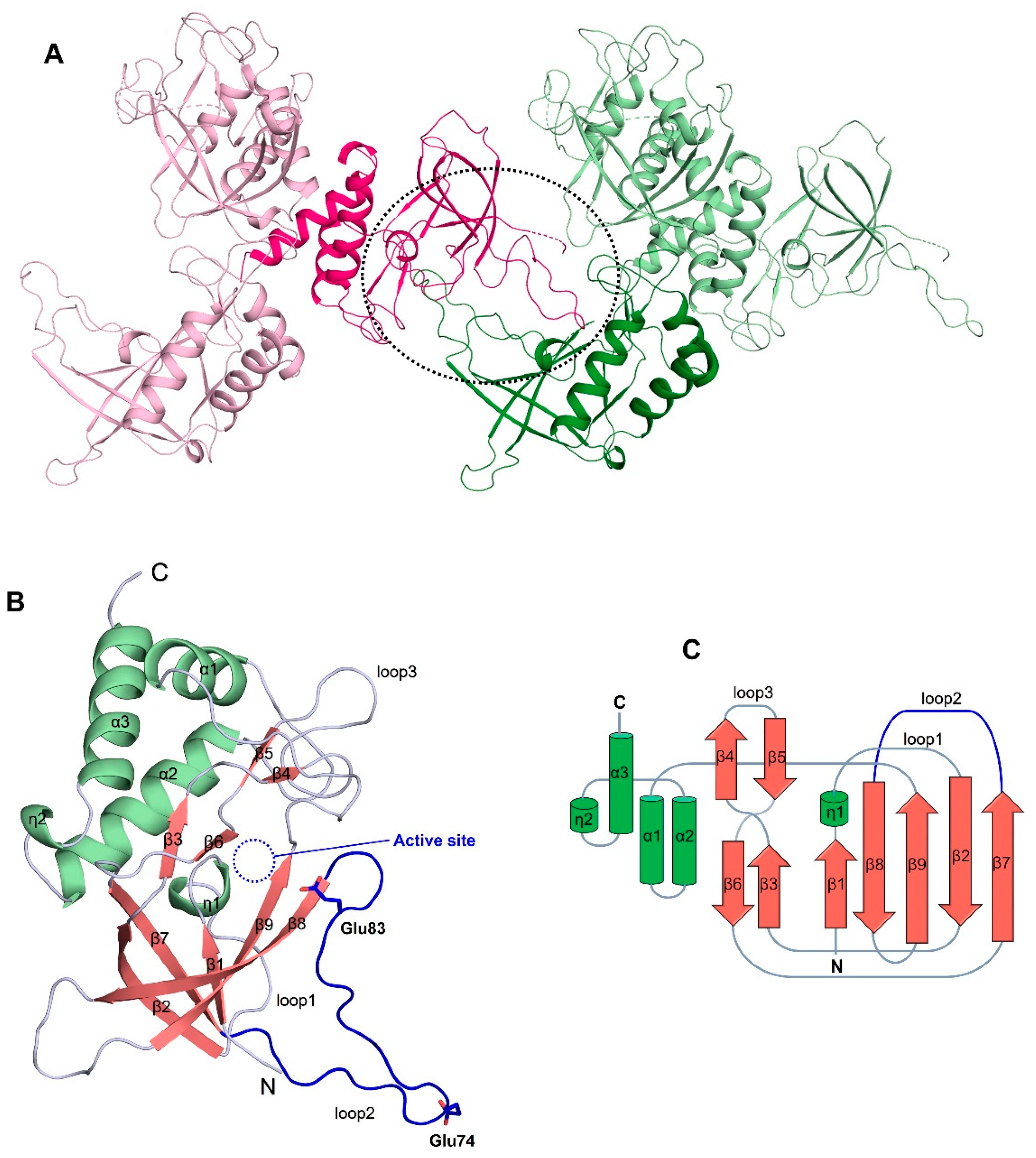
3.2. The Overall Structure of ChaC2 Adopts the GGCT Fold
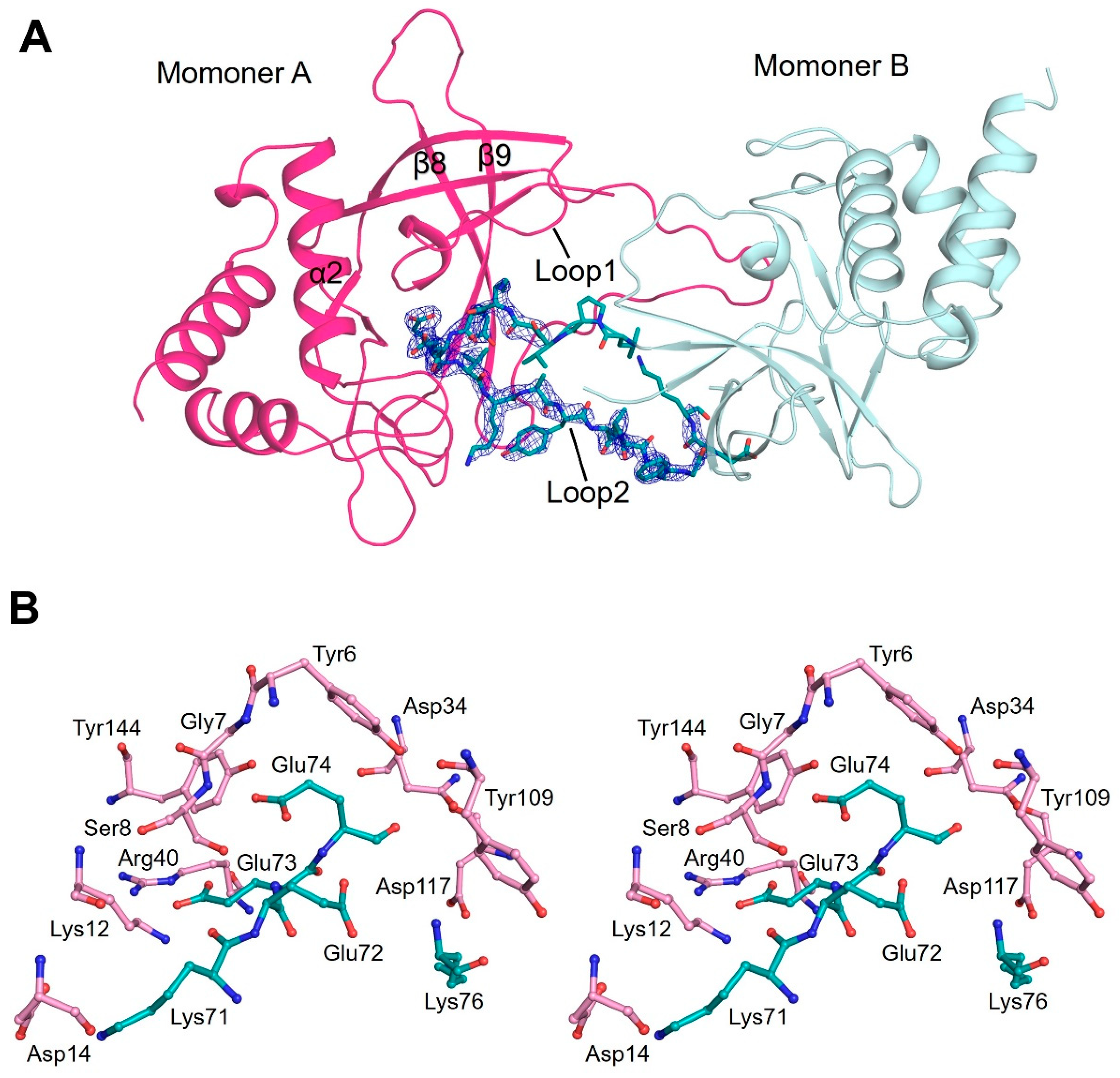
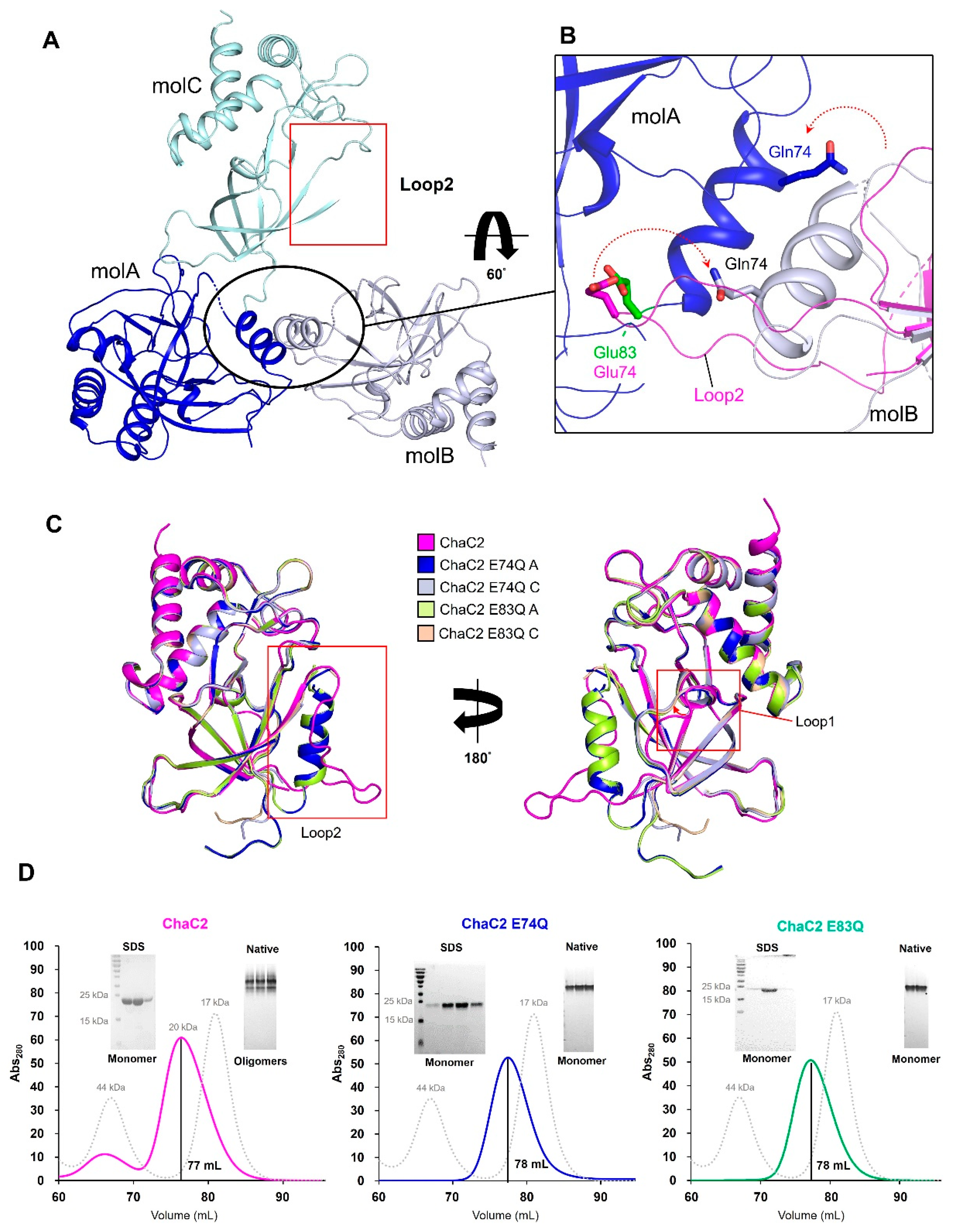
3.3. Flexible Loop2 in the Crystallographic ChaC2 Dimer Represents an Open Conformation
3.4. Single Mutations of E74Q and E83Q Induced Conformational Changes in the Flexible Loop2 Region of ChaC2 and Resulted in a Closed Conformation
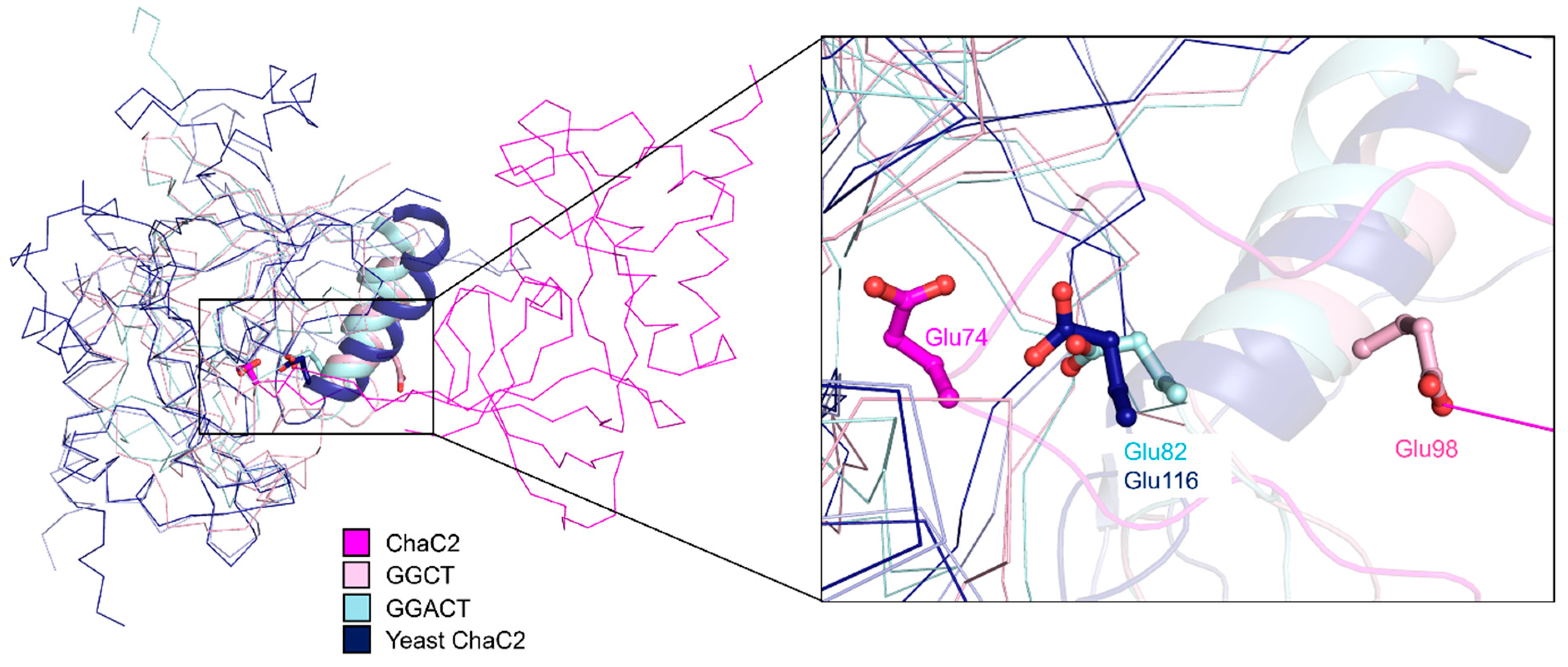
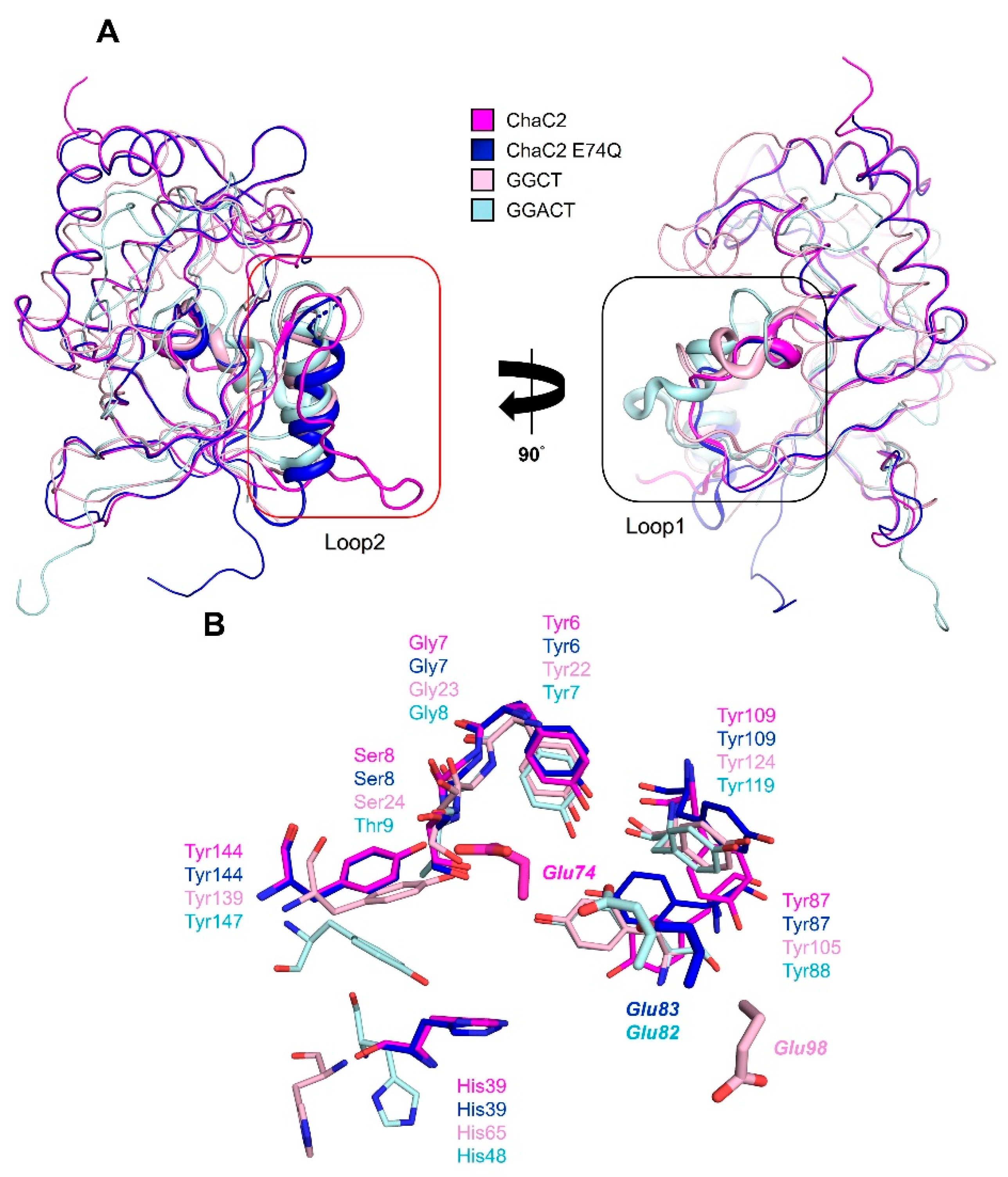
3.5. Structural Comparison of Human ChaC2 and other GGCT Enzymes Reveals Flexibility in the Active Site Region
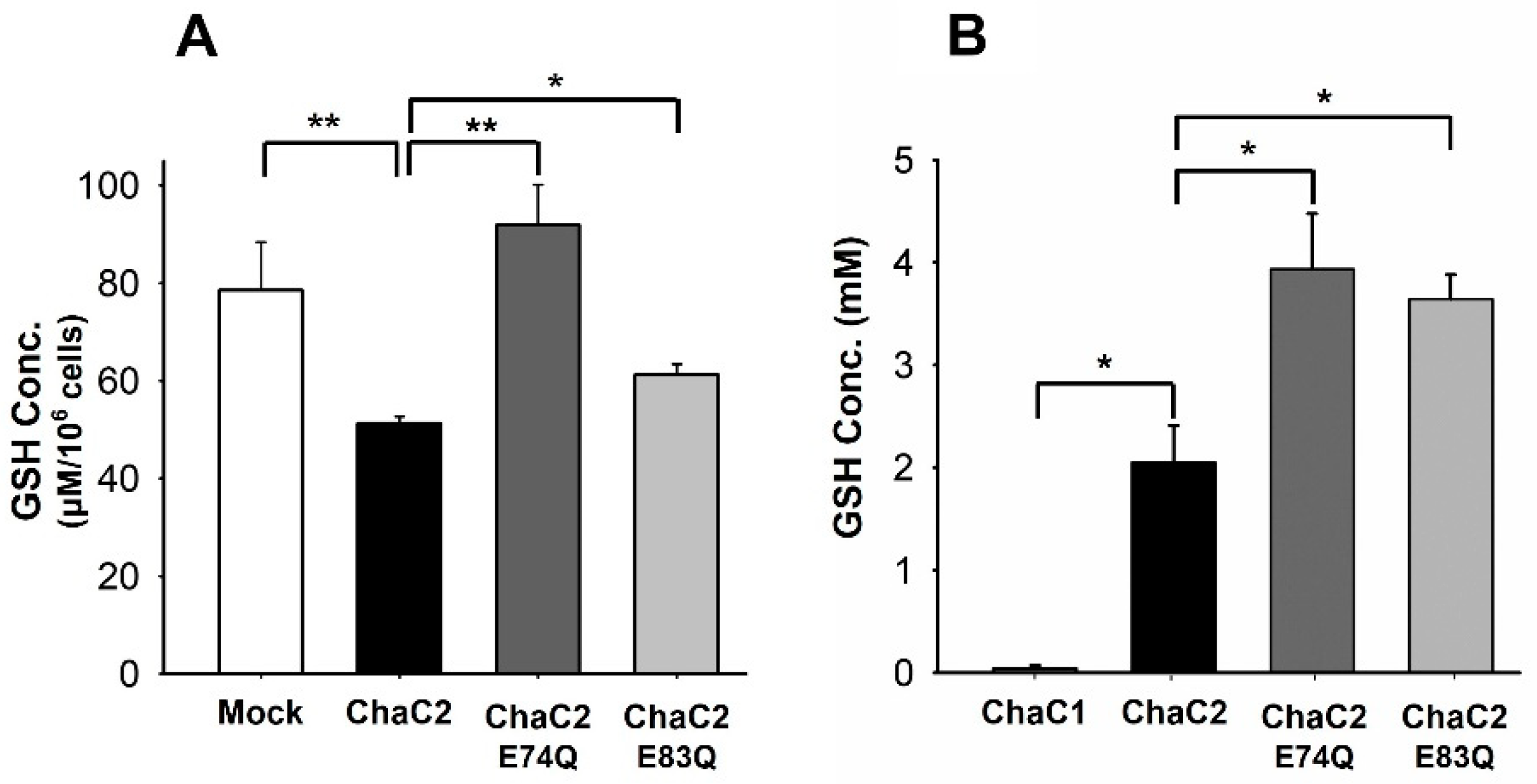
3.6. ChaC2 E74Q and ChaC2 E83Q Mutations Significantly Reduced GSH-Degradation Activities in Cell and In Vitro
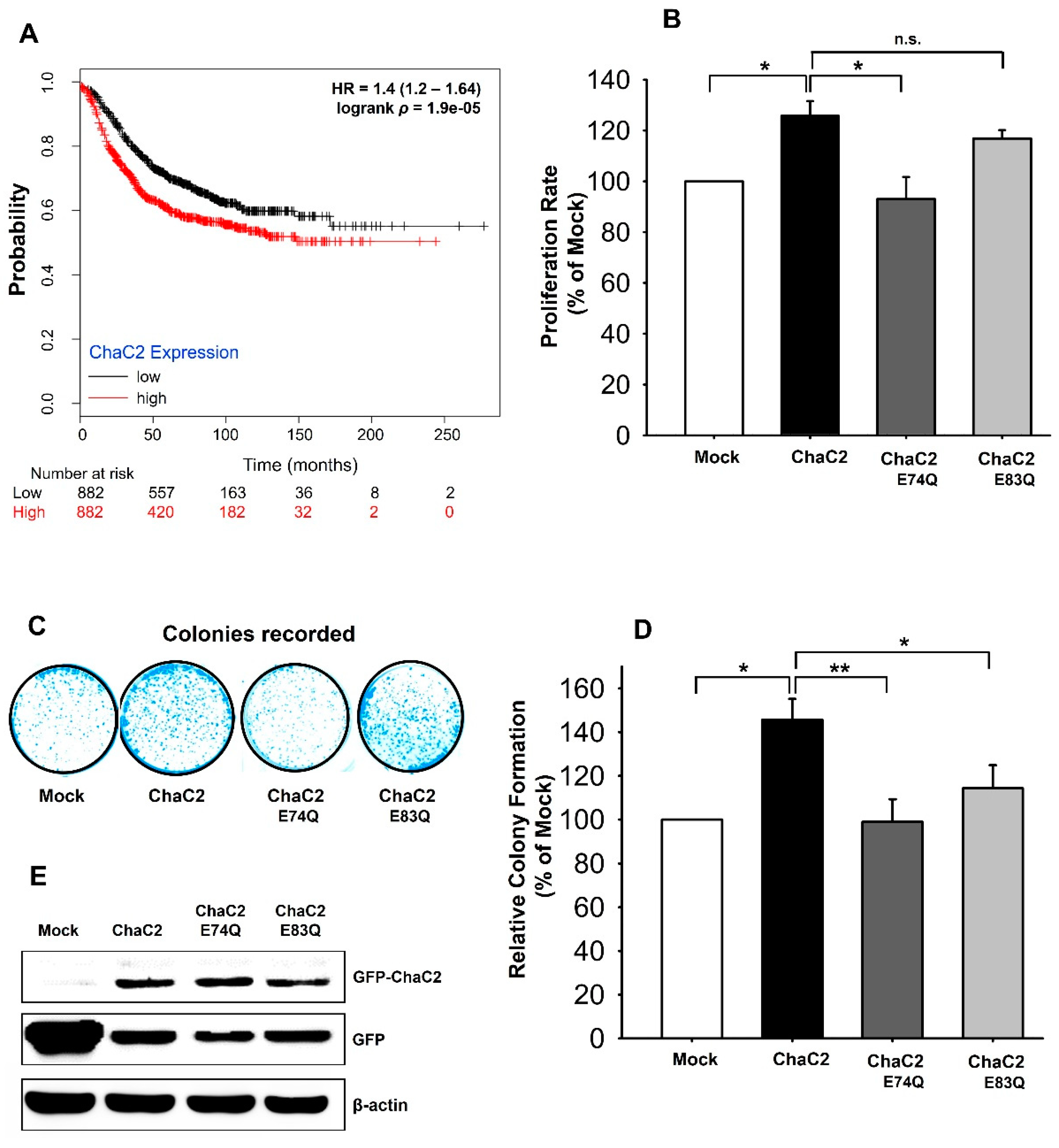
3.7. ChaC2 Overexpression Promotes Cell Proliferation in the MCF-7 Breast Cancer Cell Line
3.8. Proposed Mechanism of Substrate Recognition and GSH Degradation of ChaC2
4. Discussion
4.1. Role of the Flexible Loop2 Region of ChaC2 and Its Effects on Catalysis
4.2. Correlation of ChaC2, GSH Degradation, and Breast Cancer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Hyde, A.S.; Simpson, M.A.; Barycki, J.J. Chapter Two—Emerging Regulatory Paradigms in Glutathione Metabolism. In Advances in Cancer Research; Townsend, D.M., Tew, K.D., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 122, pp. 69–101. [Google Scholar]
- Messina, J.P.; Lawrence, D.A. Cell cycle progression of glutathione-depleted human peripheral blood mononuclear cells is inhibited at S phase. J. Immunol. 1989, 143, 1974–1981. [Google Scholar] [PubMed]
- Townsend, D.M.; Tew, K.D.; Tapiero, H. The importance of glutathione in human disease. Biomed. Pharmacother. 2003, 57, 145–155. [Google Scholar] [CrossRef]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of glutathione in cancer progression and chemoresistance. Oxidative Med. Cell. Longev. 2013, 2013, 972913. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.A.; Amandeep, K. Glutathione Degradation. Antioxid. Redox Signal. 2017, 27, 1200–1216. [Google Scholar] [CrossRef]
- Martin, M.N.; Saladores, P.H.; Lambert, E.; Hudson, A.O.; Leustek, T. Localization of Members of the γ-Glutamyl Transpeptidase Family Identifies Sites of Glutathione and Glutathione S-Conjugate Hydrolysis. Plant. Physiol. 2007, 144, 1715–1732. [Google Scholar] [CrossRef]
- Han, B.; Luo, G.; Zheng-Zheng, S.; Barrios, R. Gamma-glutamyl leukotrienase, a novel endothelial membrane protein, is specifically responsible for leukotriene D(4) formation in vivo. Am. J. Pathol. 2002, 161, 481. [Google Scholar] [CrossRef]
- Wickham, S.; West, M.B.; Cook, P.F.; Hanigan, M.H. Gamma-glutamyl compounds: Substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal. Biochem. 2011, 414, 208–214. [Google Scholar] [CrossRef]
- Wang, C.-K.; Yang, S.-C.; Hsu, S.-C.; Chang, F.-P.; Lin, Y.-T.; Chen, S.-F.; Cheng, C.-L.; Hsiao, M.; Lu, F.L.; Lu, J. CHAC2 is essential for self-renewal and glutathione maintenance in human embryonic stem cells. Free Radic. Biol. Med. 2017, 113, 439–451. [Google Scholar] [CrossRef]
- Oakley, A.J.; Yamada, T.; Liu, D.; Coggan, M.; Clark, A.G.; Board, P.G. The Identification and Structural Characterization of C7orf24 as γ-Glutamyl Cyclotransferase: An Essential Enzyme in the γ-Glutamyl Cycle. J. Biol. Chem. 2008, 283, 22031–22042. [Google Scholar] [CrossRef]
- Oakley, A.J.; Coggan, M.; Board, P.G. Identification and Characterization of γ-Glutamylamine Cyclotransferase, an Enzyme Responsible for γ-Glutamyl-ϵ-lysine Catabolism. J. Biol. Chem. 2010, 285, 9642–9648. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Gautam, R.; Srivastava, R.; Chandel, A.; Kumar, A.; Karthikeyan, S.; Bachhawat, A.K. ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione. J. Biol. Chem. 2017, 292, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Byrne, S.T.; Dolinko, A.; Harraz, M.M.; Kim, M.-S.; Umanah, G.; Zhong, J.; Chen, R.; Zhang, J.; Xu, J.; et al. Botch Is a γ-Glutamyl Cyclotransferase that Deglycinates and Antagonizes Notch. Cell Rep. 2014, 7, 681–688. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, A.; Tikoo, S.; Maity, S.; Sengupta, S.; Sengupta, S.; Kaur, A.; Kumar Bachhawat, A. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012, 13, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Joo, N.E.; Ritchie, K.; Kamarajan, P.; Miao, D.; Kapila, Y.L. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 2012, 1, 295–305. [Google Scholar] [CrossRef]
- Mungrue, I.N.; Pagnon, J.; Kohannim, O.; Gargalovic, P.S.; Lusis, A.J. CHAC1/MGC4504 Is a Novel Proapoptotic Component of the Unfolded Protein Response, Downstream of the ATF4-ATF3-CHOP Cascade. J. Immunol. 2009, 182, 466–476. [Google Scholar] [CrossRef]
- Goebel, G.; Berger, R.; Strasak, A.M.; Egle, D.; Müller-Holzner, E.; Schmidt, S.; Rainer, J.; Presul, E.; Parson, W.; Lang, S.; et al. Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br. J. Cancer 2012, 106, 189–198. [Google Scholar] [CrossRef]
- Crawford, R.R.; Prescott, E.T.; Sylvester, C.F.; Higdon, A.N.; Shan, J.; Kilberg, M.S.; Mungrue, I.N. Human CHAC1 Protein Degrades Glutathione, and mRNA Induction Is Regulated by the Transcription Factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element. J. Biol. Chem. 2015, 290, 15878–15891. [Google Scholar] [CrossRef]
- Nomura, Y.; Hirata, Y.; Kiuchi, K.; Oh-hashi, K. Translational and post-translational regulation of mouse cation transport regulator homolog 1. Sci. Rep. 2016, 6, 28016. [Google Scholar] [CrossRef]
- Liu, S.; Fei, W.; Shi, Q.; Li, Q.; Kuang, Y.; Wang, C.; He, C.; Hu, X. CHAC2, downregulated in gastric and colorectal cancers, acted as a tumor suppressor inducing apoptosis and autophagy through unfolded protein response. Cell Death Dis. 2017, 8, e3009. [Google Scholar] [CrossRef]
- L’Espérance, S.; Bachvarova, M.; Tetu, B.; Mes-Masson, A.-M.; Bachvarov, D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genom. 2008, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Bailey, H.H.; Ripple, G.; Tutsch, K.D.; Arzoomanian, R.Z.; Alberti, D.; Feierabend, C.; Mahvi, D.; Schink, J.; Pomplun, M.; Mulcahy, R.T.; et al. Phase I Study of Continuous-Infusion L-S,R-Buthionine Sulfoximine With Intravenous Melphalan. JNCI J. Natl. Cancer Inst. 1997, 89, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Courcelle, E.; Stuart, D.; Métoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef]
- Walden, H. Selenium incorporation using recombinant techniques. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 352–357. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Rafał, A.; Aleksey, P.; Jarosław, M. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins Struct. Funct. Bioinform. 2005, 59, 467–475. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. [20] Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1997; Volume 276, pp. 307–326. [Google Scholar]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Joosten, R.P.; Salzemann, J.; Bloch, V.; Stockinger, H.; Berglund, A.-C.; Blanchet, C.; Bongcam-Rudloff, E.; Combet, C.; Da Costa, A.L.; Deleage, G.; et al. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009, 42, 376–384. [Google Scholar] [CrossRef]
- Rahman, I.; Kode, A.; Biswas, S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2007, 1, 3159. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Wu, H.-Y.; Liu, M.-S.; Lin, T.-P.; Cheng, Y.-S. Structural and Functional Assays of AtTLP18.3 Identify Its Novel Acid Phosphatase Activity in Thylakoid Lumen. Plant Physiol. 2011, 157, 1015–1025. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Detection of Protein Assemblies in Crystals; Springer: Berlin/Heidelberg, Germany, 2005; pp. 163–174. [Google Scholar]
- Holm, L.; Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Ran, R.; Liu, Y.; Gao, H.; Kuang, Q.; Zhang, Q.; Tang, J.; Fu, H.; Zhang, Z.; He, Q. PEGylated Hyaluronic Acid-Modified Liposomal Delivery System with Anti-γ-Glutamylcyclotransferase siRNA for Drug-Resistant MCF-7 Breast Cancer Therapy. J. Pharm. Sci. 2015, 104, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, K.; Nakata, S.; Taniguchi, K.; Ii, H.; Ashihara, E.; Kageyama, S.; Kawauchi, A.; Yoshiki, T. Depletion of γ-glutamylcyclotransferase inhibits breast cancer cell growth via cellular senescence induction mediated by CDK inhibitor upregulation. BMC Cancer 2016, 16, 748. [Google Scholar] [CrossRef] [PubMed]
- Hiromi, I.; Taku, Y.; Susumu, N.; Keiko, T.; Koushi, H.; Shugo, T.; Masayoshi, M.; Yuji, N.; Yuko, T.; Kosei, I.; et al. A Novel Prodrug of a γ-Glutamylcyclotransferase Inhibitor Suppresses Cancer Cell Proliferation in vitro and Inhibits Tumor Growth in a Xenograft Mouse Model of Prostate Cancer. Chem. Med. Chem. 2018, 13, 155–163. [Google Scholar] [CrossRef]
- Jahn, B.; Arvandi, M.; Rochau, U.; Fiegl, H.; Goebel, G.; Marth, C.; Siebert, U. Development of a novel prognostic score for breast cancer patients using mRNA expression of CHAC1. J. Comp. Eff. Res. 2017, 6, 563–574. [Google Scholar] [CrossRef]
- Chen, M.-S.; Wang, S.-F.; Hsu, C.-Y.; Yin, P.-H.; Yeh, T.-S.; Lee, H.-C.; Tseng, L.-M. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2α-ATF4 pathway. Oncotarget 2017, 8, 114588–114602. [Google Scholar] [CrossRef]
- Taylor, C.A.t.; Juang, Y.-C.; Earnest, S.; Sengupta, S.; Goldsmith, E.J.; Cobb, M.H. Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK. Biochemistry 2015, 54, 5063–5071. [Google Scholar] [CrossRef]
- Golczak, M.; Sears, A.E.; Kiser, P.D.; Palczewski, K. LRAT-specific domain facilitates vitamin A metabolism by domain swapping in HRASLS3. Nat. Chem. Biol. 2015, 11, 26–32. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.-J.; Bao, Q.-C.; Wang, L.; Guo, T.-K.; Chen, W.-L.; Xu, L.-L.; Zhou, H.-S.; Bian, J.-L.; Yang, Y.-R.; et al. NRF2 promotes breast cancer cell proliferation and metastasis by increasing RhoA/ROCK pathway signal transduction. Oncotarget 2016, 7, 73593–73606. [Google Scholar] [CrossRef]
- Zhang, H.-S.; Du, G.-Y.; Zhang, Z.-G.; Zhou, Z.; Sun, H.-L.; Yu, X.-Y.; Shi, Y.-T.; Xiong, D.-N.; Li, H.; Huang, Y.-H. NRF2 facilitates breast cancer cell growth via HIF1ɑ-mediated metabolic reprogramming. Int. J. Biochem. Cell Biol. 2018, 95, 85–92. [Google Scholar] [CrossRef]
| SeMet-λ1 (Peak) | SeMet-λ2 (Edge) | SeMet-λ3 (Remote) | |
|---|---|---|---|
| Data Collection | |||
| Wavelength (Å) | 0.9793 | 0.9795 | 0.9717 |
| Space Group | P3121 | ||
| Cell dimensions | |||
| a, b, c (Å) | 64.12, 64.12, 103.78 90.00, 90.00, 120.00 | ||
| α, ß, γ (°) | |||
| Resolution (Å) * | 50.00–2.80 (2.85–2.80) | 50.00–2.95 (3.00–2.95) | 50.00–3.00 (3.05–3.00) |
| Rsym * | 0.130 (0.948) | 0.130 (0.854) | 0.148 (0.145) |
| Total no. observations | 136,727 | 117,519 | 111,444 |
| Total no. unique reflections | 11,761 | 10,110 | 9597 |
| I/σ(I) * | 14.7 (3.1) | 14.7 (3.1) | 47.7 (3.5) |
| Completeness (%) * | 99.9 (100.0) | 99.9 (100.0) | 99.9 (100.0) |
| Multiplicity * | 21.2 (22.3) | 21.1 (22.0) | 21.0 (21.9) |
| CC1/2 | 0.997 (0.990) | 0.996 (0.985) | 0.996 (0.980) |
| Structures | ChaC2 (PDB ID: 6K95) | ChaC2 E74Q (PDB ID: 6KYO) | ChaC2 E83Q (PDB ID: 6KY1) |
|---|---|---|---|
| Data Collection | |||
| Beam line | Spring–8 | PAL–7A | PAL–5C |
| Wavelength (Å) | 0.9000 | 0.9793 | 0.9795 |
| Space Group | C2 | P31 | P31 |
| Cell dimensions | |||
| a, b, c (Å) | 108.84, 62.16, 103.62 | 72.92, 72.92,104.25 | 72.64, 72.64, 104.13 |
| α, ß, γ (°) | 90.00, 90.01, 90.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Resolution (Å) * | 50.00–2.30 (2.34–2.30) | 50.00–2.06 (2.10–2.06) | 50.00–2.04 (2.08–2.04) |
| Rsym * | 0.056 (0.403) | 0.079 (0.826) | 0.070 (0.563) |
| Total no. reflections | 164,479 | 363,215 | 241,362 |
| Total no. unique reflections | 31,202 | 38,639 | 38,900 |
| I/σ(I) * | 23.0 (5.2) | 22.7 (8.1) | 42.5 (4.2) |
| Completeness (%) * | 99.5 (99.9) | 99.7 (99.8) | 99.9 (100.0) |
| Multiplicity * | 5.3 (5.2) | 9.4 (9.4) | 6.2 (6.0) |
| CC1/2 | 0.995 (0.993) | 0.996 (0.95) | 0.99 (0.96) |
| Refinement | |||
| Resolution range (Å) | 37.41–2.30 | 31.59–2.06 | 31.48–2.04 |
| Rwork/Rfree † | 0.239/0.268 | 0.224/0.258 | 0.225/0.255 |
| No. atoms | |||
| Protein | 4177 | 4144 | 4188 |
| Water | 16 | 77 | 85 |
| Average B factors (Å2) | 47.36 | 36.84 | 35.56 |
| Protein | 47.30 | 36.6 | 35.07 |
| Water | 69.26 | 51.00 | 59.08 |
| R.m.s. deviations | |||
| Bond length (Å) | 0.011 | 0.008 | 0.009 |
| Bond angles (°) | 1.440 | 1.184 | 1.300 |
| Ramachandran Favored/outlier | 97.45/0.0 | 98.22/0.0 | 96.8/0.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, Y.T.K.; Park, J.S.; Jang, J.Y.; Kim, K.R.; Vo, T.T.L.; Kim, K.-W.; Han, B.W. Structural and Functional Analyses of Human ChaC2 in Glutathione Metabolism. Biomolecules 2020, 10, 31. https://doi.org/10.3390/biom10010031
Nguyen YTK, Park JS, Jang JY, Kim KR, Vo TTL, Kim K-W, Han BW. Structural and Functional Analyses of Human ChaC2 in Glutathione Metabolism. Biomolecules. 2020; 10(1):31. https://doi.org/10.3390/biom10010031
Chicago/Turabian StyleNguyen, Yen T. K., Joon Sung Park, Jun Young Jang, Kyung Rok Kim, Tam T. L. Vo, Kyu-Won Kim, and Byung Woo Han. 2020. "Structural and Functional Analyses of Human ChaC2 in Glutathione Metabolism" Biomolecules 10, no. 1: 31. https://doi.org/10.3390/biom10010031
APA StyleNguyen, Y. T. K., Park, J. S., Jang, J. Y., Kim, K. R., Vo, T. T. L., Kim, K.-W., & Han, B. W. (2020). Structural and Functional Analyses of Human ChaC2 in Glutathione Metabolism. Biomolecules, 10(1), 31. https://doi.org/10.3390/biom10010031