Molecular Insight into Stereoselective ADME Characteristics of C20-24 Epimeric Epoxides of Protopanaxadiol by Docking Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of ADME-Related Protein Targets
2.2. Homology Modeling and Model Validation of UGT1A8
2.3. Protein Preparation and Optimization
2.4. Ligands Preparation and Optimization
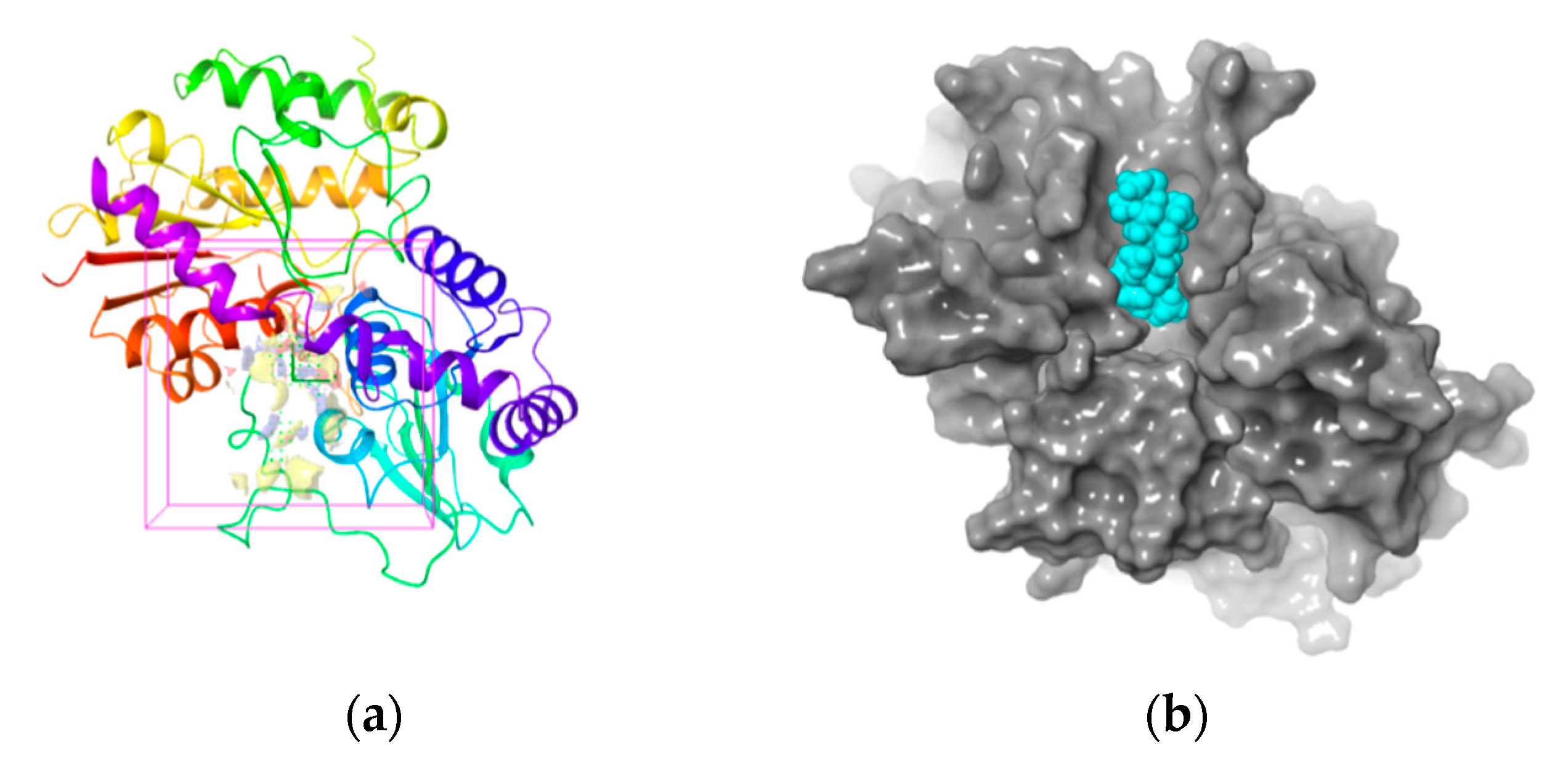
2.5. Binding Site Selection
2.6. Molecular Docking
3. Results
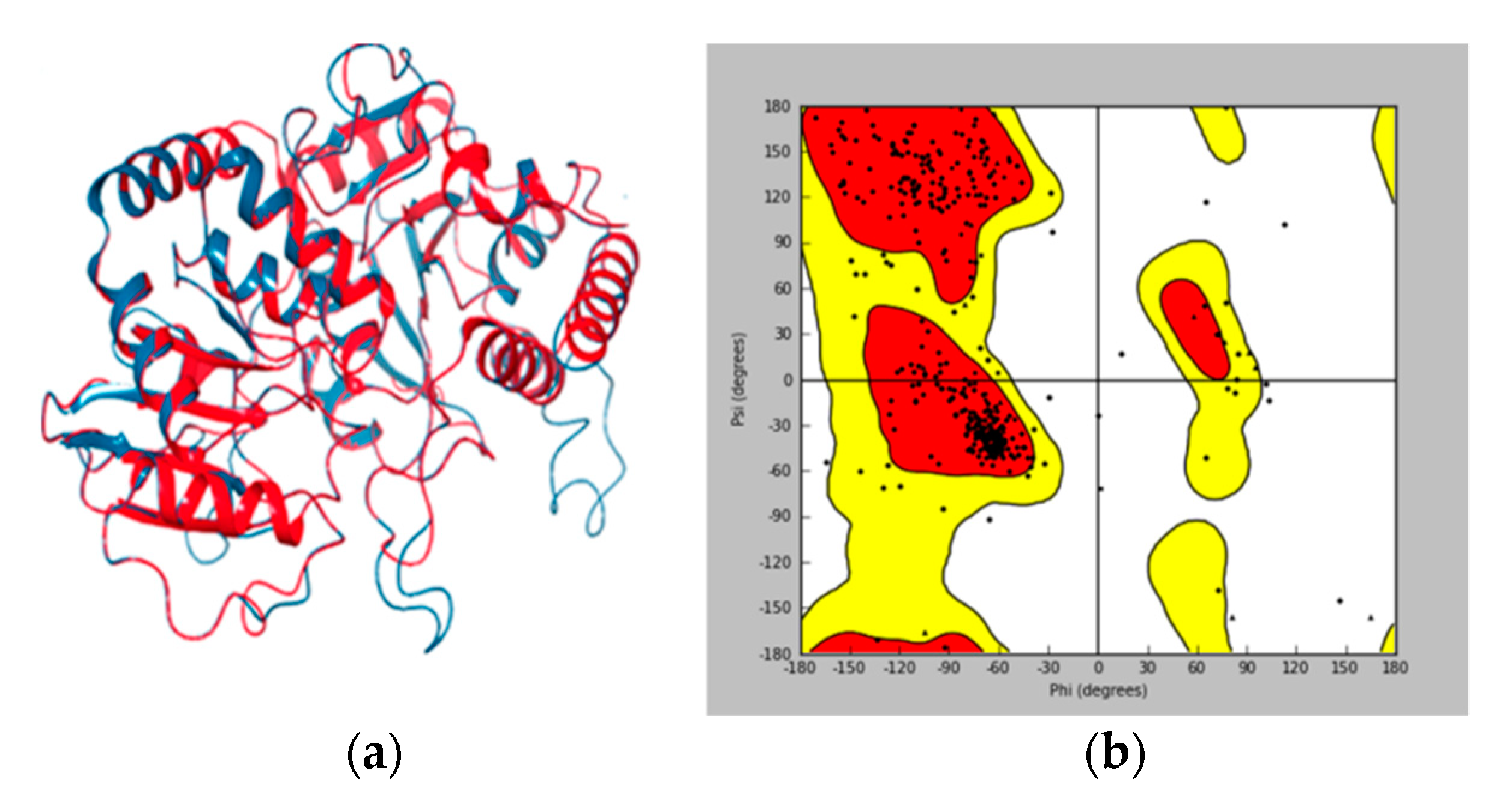
3.1. Homology Modeling and Validation of UGT1A8
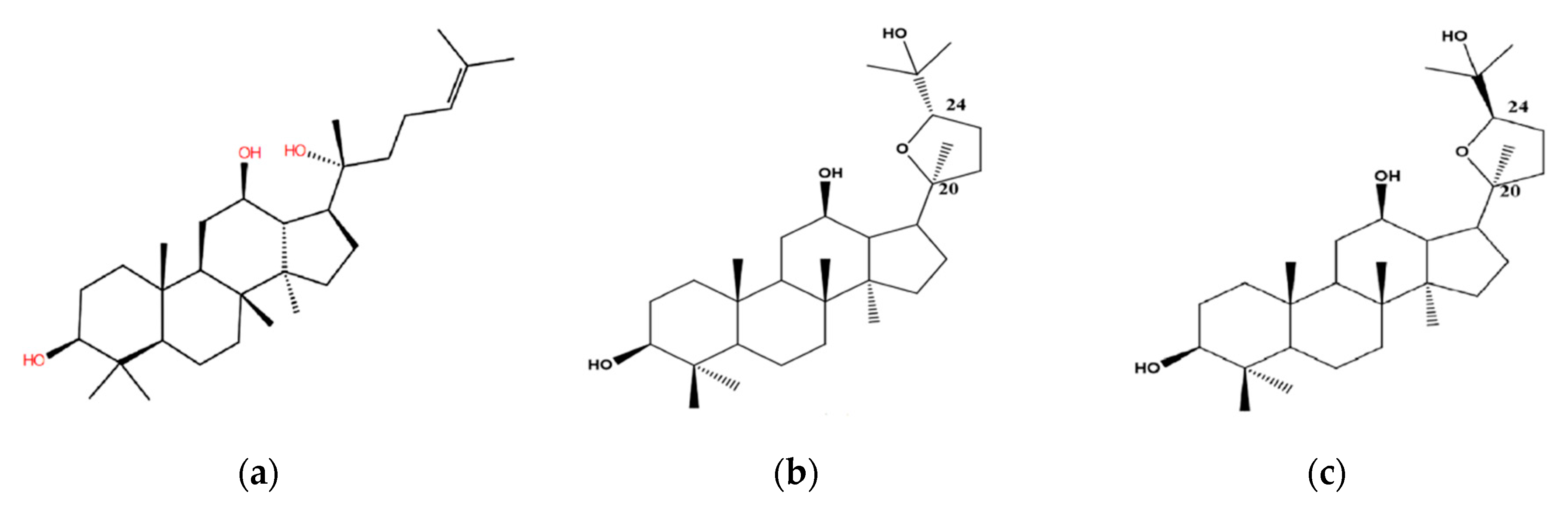
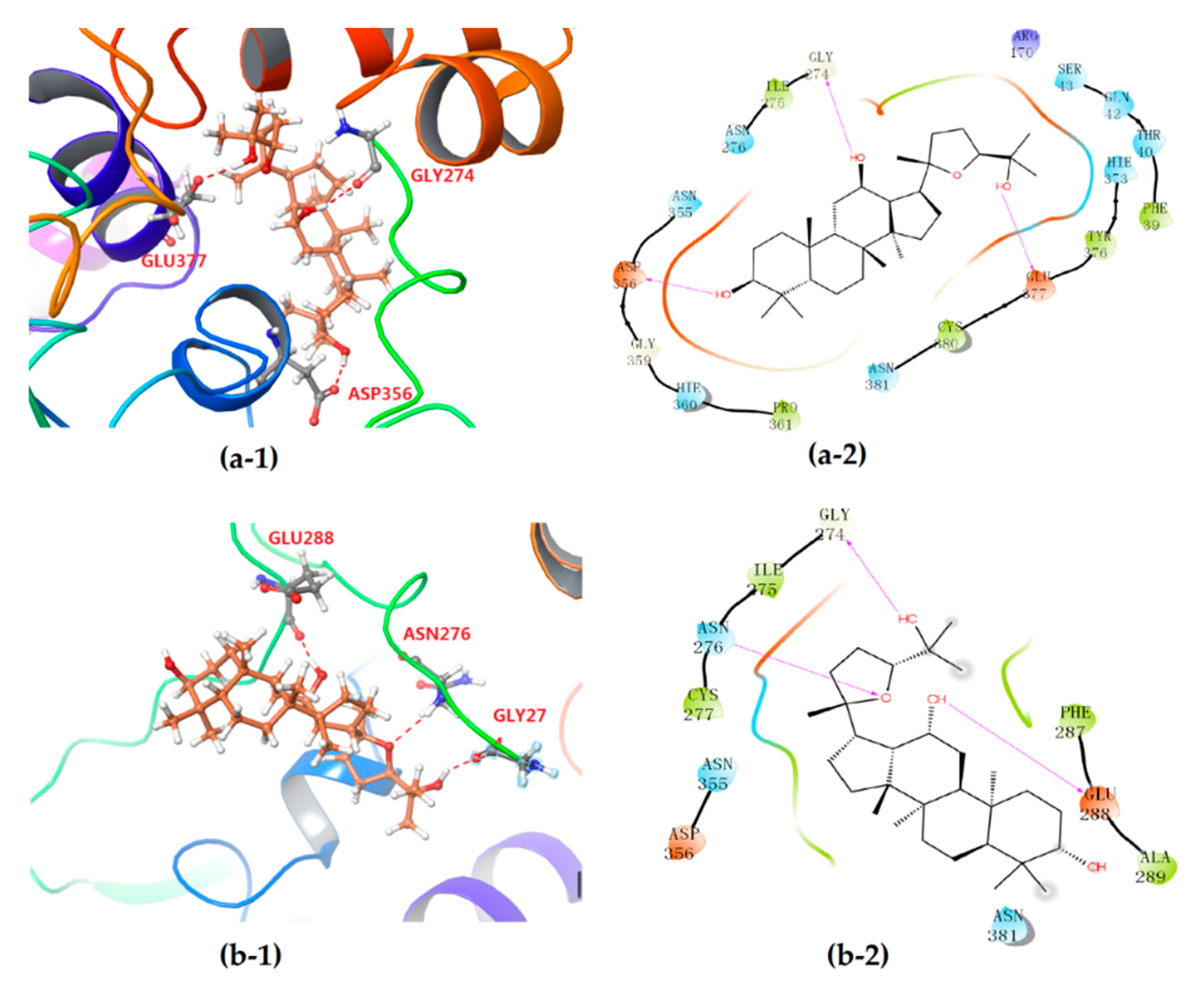
3.2. Stereoselective Interaction Between PDQ Epimers and UGT1A8
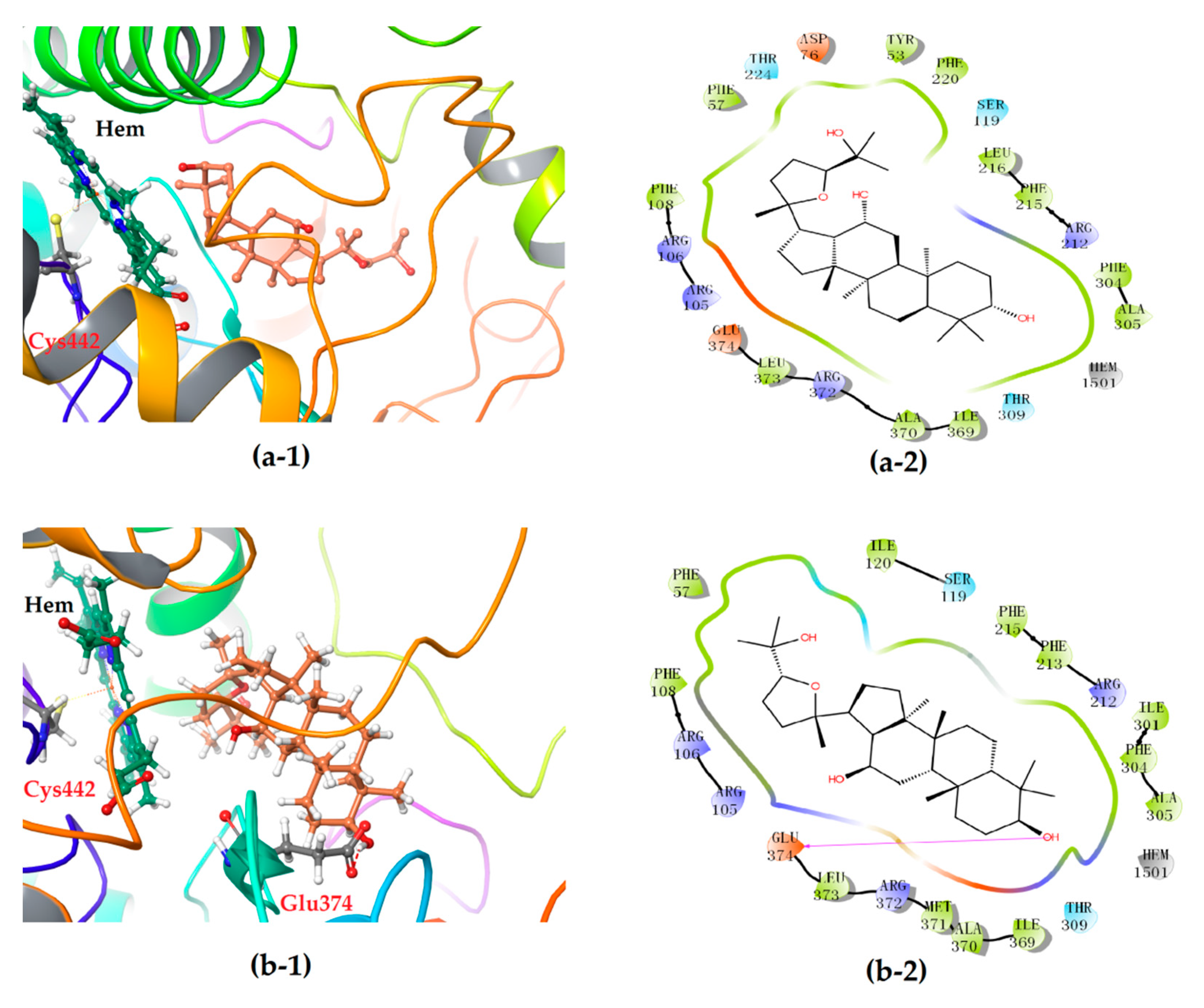
3.3. Stereoselective Interaction between PDQ Epimers and CYP3A4
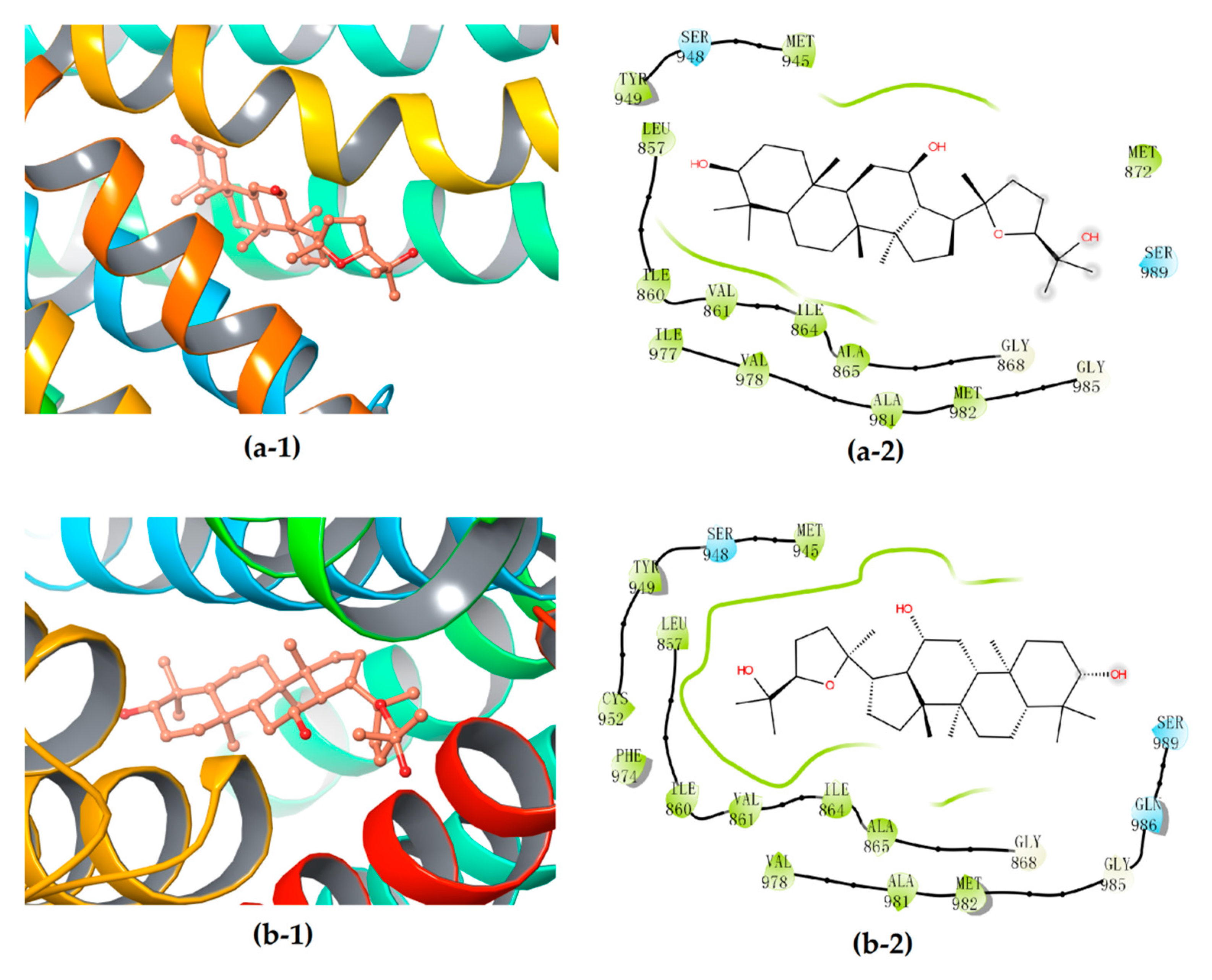
3.4. Stereoselective Interaction between PDQ Epimers and P-gp
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, Y.; De, B.K.C.; Kirchmair, J. Data resources for the computer-guided discovery of bioactive natural products. J. Chem. Inf. Model. 2017, 57, 9. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, J.H. A review on the medicinal potentials of ginseng and ginsenosides on cardiovascular diseases. J. Ginseng Res. 2014, 38, 161–166. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D.; Yang, D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 2015, 33, 717–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.R.; Yang, J.J.; Wang, W.Z.; Zhang, J.Q.; Zhang, R.M.; Meng, Q.G. Discovery, semisynthesis, biological activities, and metabolism of ocotillol-type saponins. J. Ginseng Res. 2017, 41, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.W.; Wang, N.; Zhang, J.; Chen, G.; Xu, H.; Meng, Q.G.; Du, Y.; Yang, X.; Fan, H.Y. In vitro and in silico evaluation of stereoselective effect of ginsenoside isomers on platelet p2y12 receptor. Phytomedicine 2019, 64, 152899. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.K.; Kwon, S.W.; Park, J.H. Chemical diversity of ginseng saponins from Panax ginseng. J. Ginseng Res. 2015, 39, 287–298. [Google Scholar] [CrossRef]
- Wu, W.; Sun, L.; Zhang, Z.; Guo, Y.Y.; Liu, S.Y. Profiling and multivariate statistical analysis of Panax ginseng based on ultra-high-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2015, 107, 141–150. [Google Scholar] [CrossRef]
- Wang, W.Y.; Ni, Y.Y.; Wang, L.; Che, X.; Liu, W.H.; Meng, Q.G. Stereoselective oxidation metabolism of 20(S)–protopanaxatriol in human liver microsomes and in rats. Xenobiotica 2015, 45, 385–395. [Google Scholar] [CrossRef]
- Yang, Z.; Gao, S.; Wang, J.R.; Yin, T.J.; Teng, Y.; Wu, B.J.; You, M.; Jiang, Z.H.; Hu, M. Enhancement of oral bioavailability of 20(S)-ginsenoside Rh2 through improved understanding of its absorption and efflux mechanisms. Drug Metab. Dispos. 2011, 39, 1866–1872. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.H.; Lee, P.Y.; Bae, K.H.; Cho, S.; Park, B.C.; Shin, H.; Park, S.G. Ginsenoside Rb1 is transformed into Rd and Rh2 by microbacterium trichothecenolyticum. J. Microbiol. Biotechnol. 2013, 23, 1802–1805. [Google Scholar] [CrossRef]
- Meng, Q.G.; Tan, W.J.; Hou, G.G.; Zhang, X.Y.; Hu, X.Y.; Yang, F.; Bai, G.J.; Zhu, W.W.; Cai, Y.; Bi, Y. Synthesis and structural characterization of two epimers driven from 20(S)–protopanaxadiol. J. Mol. Struct. 2013, 51, 1054–1055. [Google Scholar] [CrossRef]
- Chiu, N.T.C.; Guns, E.S.T.; Hans, A.; William, J.; Subrata, D. Identification of human cytochrome P450 enzymes involved in the hepatic and intestinal biotransformation of 20(S)–protopanaxadiol. Biopharm. Drug Dispos. 2014, 35, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Meng, Q.G.; Zhang, J.F.; Bi, Y.; Jiang, N.C. Study on the structure-function relationship of 20(S)-panaxadiol and its epimeric derivatives in myocardial injury induced by isoproterenol. Fitoterapia 2010, 81, 783–787. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Wu, X.; Xu, L.; Meng, Q.; Liu, W. Stereoselective formation and metabolism of 20(S)–protopanaxadiol ocotillol type epimers in vivo and in vitro. Chirality 2015, 27, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Shao, Y.; Ma, S.; Wu, X.; Meng, Q.G. Determination of 20(S)–protopanaxadiol ocotillol type epimers in rat plasma by liquid chromatography tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2012, 3, 887–888. [Google Scholar] [CrossRef]
- Morris, G.M.; Limwilby, M. Molecular docking. Methods Mol. Biol. 2008, 443, 365–382. [Google Scholar]
- Zhang, J.; Zhou, F.; Niu, F.; Lu, M.; Wu, X.; Sun, J.; Wang, G. Stereoselective regulations of P-glycoprotein by ginsenoside Rh2 epimers and the potential mechanisms from the view of pharmacokinetics. PLoS ONE 2012, 7, e35768. [Google Scholar] [CrossRef]
- Kim, D.; Yu, F.Z.; Min, J.S.; Park, J.B.; Bae, S.H.; Yoon, K.D.; Chin, Y.W.; Oh, E.; Bae, S.K. In vitro stereoselective inhibition of ginsenosides toward UDP-glucuronosyltransferase (UGT) isoforms. Toxicol. Lett. 2016, 259, 1–10. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef]
- Halgren, T.A. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Ohkura, K.; Kawaguchi, Y.; Watanabe, Y.; Masubuchi, Y.; Hori, H. Flexible structure of cytochrome p450: Promiscuity of ligand binding in the cyp3a4 heme pocket. Anticancer Res. 2009, 29, 935–942. [Google Scholar] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Mee, S.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Cho, A.E. Using reverse docking to identify potential targets for ginsenosides. J. Ginseng Res. 2017, 41, 534–539. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Kumar, D.H.; Barch, H.P.; Buolamwini, J.K. Homology modeling of human concentrative nucleoside transporters (hCNTs) and validation by virtual screening and experimental testing to identify novel hCNT1 inhibitors. Drug Des. 2017, 146, 6. [Google Scholar] [CrossRef]
- Kiang, T.K.; Ensom, M.H.; Chang, T.K. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol. Ther. 2005, 106, 97–132. [Google Scholar] [CrossRef]
- Oda, S.; Fukami, T.; Yokoi, T.; Nakajima, M. A comprehensive review of UDP-glucuronosyltransferase and esterases for drug development. Drug Metab. Pharmacokinet. 2015, 30, 30–51. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Nakajin, S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab. Dispos. 2009, 37, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Strassburg, C.P.; Manns, M.P.; Tukey, R.H. Expression of the UDP-glucuronosyltransferase 1A locus in human colon. Identification and characterization of the novel extrahepatic UGT1A8. J. Biol. Chem. 1998, 273, 8719–8726. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.Z.; Cao, Y.F.; Hu, C.M.; Hong, M.; Sun, X.Y.; Ge, G.B.; Liu, Y.; Zhang, Y.Y.; Yang, L.; Sun, H.Z. Structure-inhibition relationship of ginsenosides towards UDP-glucuronosyltransferases (UGTs). Toxicol. Appl. Pharmacol. 2013, 267, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Bohnuud, T.; Luo, L.; Wodak, S.J.; Bonvin, A.M.; Weng, Z.; Vajda, S.; Schueler-Furman, O.; Kozakov, D. A benchmark testing ground for integrating homology modeling and protein docking. Proteins 2016, 85, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.C.D.; Siqueira, A.S.; Lima, A.R.J.; Lima, A.D.M.; Santos, A.S.; Aguiar, D.C.F.; Gonçalves, E.C. In silico characterization of a cyanobacterial plant-type isoaspartyl aminopeptidase/asparaginase. J. Mol. Model. 2018, 24, 108. [Google Scholar] [CrossRef]
- Geoffrey, M.; Matthew, B.; Cadd, V.A.; Chaddock, J.A.; Acharya, K.R. Structure and activity of a functional derivative of Clostridium botulinum neurotoxin B. J. Struct. Biol. 2011, 174, 52–57. [Google Scholar]
- Williams, P.A.; Cosme, J.; Vinkovic, D.M.; Ward, A.; Jhoti, H. Crystal structures of human cytochrome p450 3a4 bound to metyrapone and progesterone. Science 2004, 305, 683–686. [Google Scholar] [CrossRef]
- Engidawork, E.; Roberts, J.C.; Hardmeier, R.; Scheper, R.J.; Lubec, G. Expression of the multidrug resistance P glycoprotein (Pgp) and multidrug resistance associated protein (MRP1) in down syndrome brains. J. Neural. Transm. Suppl. 2001, 61, 35–45. [Google Scholar]
- Wang, W.Y.; Wu, X.M.; Wang, L.; Meng, Q.G.; Liu, W.H. Stereoselective property of 20(S)–protopanaxadiol ocotillol type epimers affects its absorption and also the inhibition of P-glycoprotein. PLoS ONE 2014, 9, e98887. [Google Scholar] [CrossRef]

| Protein-Ligand Interaction | Docking Score | ΔGbind 2 | Residues for Hydrogen Bonding | |
|---|---|---|---|---|
| UGT1A8 | 24S-PDQ | −(4.268 ± 0.174) | −(49.89 ± 2.06) | Gly274, Asp356, Glu377 |
| 24R-epimer | −(3.794 ± 0.208) | −(45.11 ± 1.58) | Gly274, Asn276, Glu288 | |
| CYP3A4 | 24S-PDQ | −(5.737 ± 1.350) | −(54.95 ± 5.27) | / |
| 24R-epimer | −(7.162 ± 0.0395) | −(61.65 ± 0.572) | Glu374 | |
| P-gp | 24S-PDQ | −(5.636 ± 0.0326) | −(42.73 ± 0.577) | / |
| 24R-epimer | −(5.879 ± 0.380) | −(40.73 ± 0.774) | / | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Li, Z.; Yuan, M.; Chen, G.; Li, Q.; Xu, H.; Yang, X. Molecular Insight into Stereoselective ADME Characteristics of C20-24 Epimeric Epoxides of Protopanaxadiol by Docking Analysis. Biomolecules 2020, 10, 112. https://doi.org/10.3390/biom10010112
Guo W, Li Z, Yuan M, Chen G, Li Q, Xu H, Yang X. Molecular Insight into Stereoselective ADME Characteristics of C20-24 Epimeric Epoxides of Protopanaxadiol by Docking Analysis. Biomolecules. 2020; 10(1):112. https://doi.org/10.3390/biom10010112
Chicago/Turabian StyleGuo, Wenna, Zhiyong Li, Meng Yuan, Geng Chen, Qiao Li, Hui Xu, and Xin Yang. 2020. "Molecular Insight into Stereoselective ADME Characteristics of C20-24 Epimeric Epoxides of Protopanaxadiol by Docking Analysis" Biomolecules 10, no. 1: 112. https://doi.org/10.3390/biom10010112
APA StyleGuo, W., Li, Z., Yuan, M., Chen, G., Li, Q., Xu, H., & Yang, X. (2020). Molecular Insight into Stereoselective ADME Characteristics of C20-24 Epimeric Epoxides of Protopanaxadiol by Docking Analysis. Biomolecules, 10(1), 112. https://doi.org/10.3390/biom10010112





