Abstract
The Atomic and Molecular Data Unit of the International Atomic Energy Agency has developed a new database, CollisionDB, to provide an open, free, robust and long-term repository of data on plasma collisional processes. The database contains data on cross sections and rate coefficients for collisions of electrons, photons and heavy particles with atomic and molecular species. A fundamental requirement for this database is the implementation of standardized metadata, which provide an unambiguous description of the collisional data available in peer-reviewed sources. CollisionDB offers both a browser-based search interface and an application programming interface (API) that allows users to filter, process and compare collisional datasets. For this purpose, a Python package PyCollisionDB has been developed to access the CollisionDB API. Here, we present an overview of the technical developments, including data schemas, standards and user interface underlying the CollisionDB application, with particular emphasis on the API developed to support the integration of data into modeling and other codes.
1. Introduction
Plasma processes are central to many areas of scientific and technical research and development. Modeling such processes requires accurate cross section and/or rate coefficient datasets for a wide variety of collisional processes. To this end, many experimental and theoretical studies of such processes are described in the peer-reviewed scientific literature each year. Although individual journals may make these data available in some form, the datasets are usually more conveniently obtained through one of several actively developed databases, most of which focus on a particular application, including LXCat (cold plasma) [1], the database of the Data Center for Plasma Properties at the Korea Institute of Fusion Energy (fusion energy research) [2], Phys4Entry (to model re-entry plasma in planetary atmospheres) [3], QuantemolDB (focused on plasma chemistry for technological applications) [4] and ADAS (rate coefficients and codes for astrophysics and fusion energy applications) [5]. These databases provide different online interfaces for searching and retrieving data and, generally, adopt different conventions for describing the species, states and processes involved.
The Atomic and Molecular Data (AMD) Unit of the International Atomic Energy Agency (IAEA) has, since 1988, maintained the ALADDIN database of evaluated data for fusion energy research [6]. It too has an online search interface (https://www-amdis.iaea.org/ALADDIN/, accessed on 23 March 2024) and provides the coefficients and details of functional fits to key processes. However, its datasets are not all described by unambiguous metadata, and it does not expose an Application Programming Interface (API) for the automated searching and retrieval of data.
CollisionDB is an open-source repository for cross section and rate coefficient data of plasma collisional processes, developed to support fusion research with an emphasis on the FAIR (Findable, Accessible, Interoperable and Reusable) principles [7] of database management. All data hosted in CollisionDB are associated with rich metadata needed to describe collisional datasets and are assigned a persistent, globally unique identifier (F). Data can be retrieved by searching both from the user interface and through the API (A). In order to facilitate the interpretation and exchange of datasets (I), data are provided in a formal, accessible and standardized format. Furthermore, including provenance information, such as a DOI, enhances the comprehensibility and traceability of the data. Data are made available under a clear license with relevant attributes and unambiguous metadata to ensure reusability (R).
While the data that CollisionDB contains are of most relevance to fusion energy research, the standards, formats and software stack it is built around, which are described in this article, should prove of use for similar databases of collisional and chemical reaction processes for other applications. In particular, the data model and API described below allow the automated querying and retrieval of data and metadata directly from code in a way that facilitates visualization, data exploration and machine learning.
2. Results
2.1. Search Interface
CollisionDB offers a user-friendly web interface to interact with the database, implemented with various features such as filtering, retrieval and easy access to datasets. Figure 1 shows the search form for querying the database based on a set of relevant metadata attributes describing the collisional datasets. Users can search for reactants or products as species (with or without states), and Table 1 provides examples of species and state notation used in CollisionDB. Other search options include method, data type and process types, which can be selected from the appropriate drop-down menu. The search can be further refined by filtering for author and publication DOI. Additionally, the users can retrieve the evaluated/recommended data from the search form. Since the datasets are timestamped, users can also retrieve deprecated datasets using the valid_on date field from the search form. In this way, a query on a known date can always be reproduced exactly, with the same datasets returned as were originally downloaded.
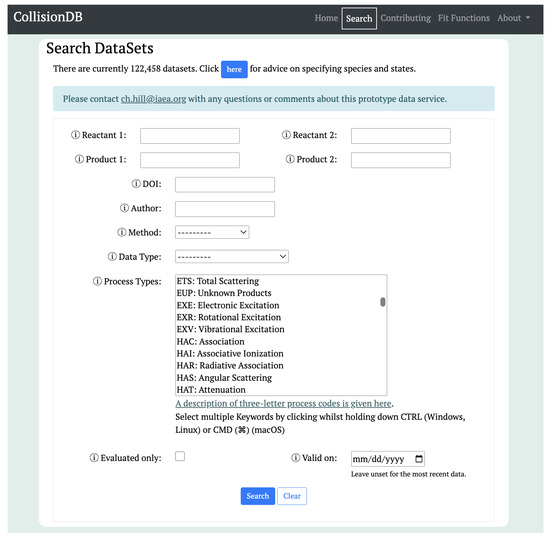
Figure 1.
The browser-based search form for querying data in CollisionDB, available online at https://amdis.iaea.org/db/collisiondb/search/ (accessed on 23 March 2024).

Table 1.
Examples of species and states identification using PyValem notation in CollisionDB.
The datasets that match the search query will be displayed in a paginated list on the search results page. Users can download individual datasets as plain-text files (.txt) (including the JSON metadata header) as shown in Listing 1. Alternatively, users can download all datasets as an archive, which includes individual dataset files, a manifest file and a bibliography file: see the archive structure in Figure 2. The manifest file provides the list of dataset files within the archive, identified by their “qualified ID” (qid; see Table 2). It also includes additional information about the download timestamp, a universally unique identifier for the archive name (uuid), the query string (GET_string) and number of datasets. A sample manifest file is shown in Listing 2, demonstrating a search for reactant1 (Be+4) as extracted from GET_string, along with information about the retrieved archive.

Figure 2.
Archive structure for downloaded data.

Table 2.
A description of the JSON metadata header used in each dataset file.
| Listing 1. An example dataset file for download. |
 |
| Listing 2. A sample manifest JSON file describing the dataset files in the archive. |
 |
The web interface also allows users to access detailed information about individual collisional datasets, along with an interactive graphical display of the data, as demonstrated in Figure 3 for a dataset with the primary key ID 15115. This collisional dataset contains the fit coefficients along with cross sections, which can also be viewed through the interface (see Figure 4). As can be seen from this figure, the rendered LaTeX representation of the corresponding fit function is shown along with its Python implementation. More information about these fitting functions is available online at https://amdis.iaea.org/db/collisiondb/fit-functions/ (accessed on 23 March 2024).
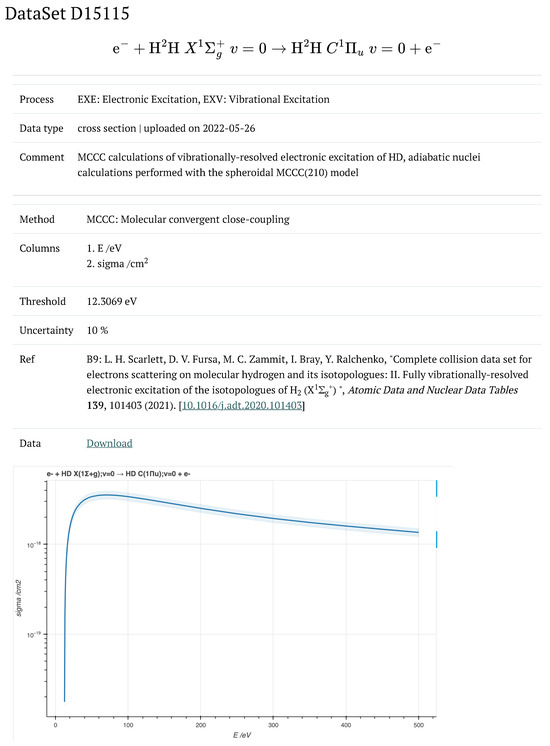
Figure 3.
An example showing the detailed information of a collisional dataset, identified by its primary key ID 15115, along with an interactive graphical representation of the collisional data. Available online at https://amdis.iaea.org/db/collisiondb/datasets/15115 (accessed on 23 March 2024).

Figure 4.
Fit coefficients for collisional dataset (ID=15115) along with the rendered LaTeX representation of the corresponding fit function and its Python implementation. Details of the fit function can be found online at https://amdis.iaea.org/db/collisiondb/fit-functions/singlet_singlet_H2 (accessed on 23 March 2024), identified by the fit function name.
2.2. PyCollisionDB Package for API
PyCollisionDB [11] is a Python package for interacting with the CollisionDB API to obtain collisional datasets from the database. Datasets can be retrieved in a standardized way, output in different formats, compared and manipulated using a number of pre-defined Python methods.
To use PyCollisionDB, users need to install and import the PyCollision module to access the available functions and attributes, as shown in Listing 3.
| Listing 3. Initialize the main instance of PyCollisionDB package to interact with the database in Python Shell. |
| >>> # import PyCollision module to access associated functions and attributes. >>> from pycollisiondb import PyCollision |
The main methods of the PyCollisionDB package, including query structures/schema and usage examples to efficiently explore interaction and data exchange, are described below:
- PyCollision.get_datasets(): This is the main class method for querying and retrieving the datasets in a standardized format from the server for a given query, which should be passed as a Python dictionary (dict object). Only valid metadata keys, such as ids (a list of dataset IDs) or reactants, are accepted in each query. The values are specified as either a string or a list of comma-separated strings depending upon the query key. A list of valid keys and examples are given in Table 3.
 Table 3. Examples of valid query keys for interacting with CollisionDB database over API.
Table 3. Examples of valid query keys for interacting with CollisionDB database over API.
An example of querying the database for the proton-impact ionization cross sections of hydrogen in its ground state using the PyCollisionDB package in a Python shell is given in Listing 4. In this example, the database is queried in two ways: (a) by passing a list of reactants ['H+', 'H 1s'] combined with process types ['HIN'], which refers to heavy-particle impact ionization, and (b) using the query key reaction_texts. Both queries returned the same number of datasets and a zip-compressed archive comprising individual dataset files, a manifest and bibliography file, as explained in the “Search interface” section above.
| Listing 4. Search and fetch datasets from the server over the API. |
 |
- PyCollision.summarize_datasets(): This method provides a summary of the retrieved datasets and groups them into different blocks based on the reaction text. The output includes pertinent information such as qualified ID, process types, data type and references for each collisional dataset, as shown in Listing 5.
| Listing 5. Summary of datasets in blocks for each distinct reaction text. |
 |
- PyCollision.resolve_refs(): Call this method to resolve the references for all retrieved datasets into a proper, citeable format. The method returns references (refs) as a Python dictionary, with bibliographic data identified by reference ID; see Listing 6.
| Listing 6. Resolving the references for all the datasets into a proper, citeable format. |
 |
- PyCollision.datasets: To access the details of a collisional dataset, users can use the dataset ID as a key for the datasets dict attribute. An example of retrieving the metadata and numerical data of a specific dataset 102737 is given in Listing 7.
| Listing 7. Retrieve details of an individual dataset. |
 |
- PyCollision.convert_units(): Use this method to change the units for any or all datasets. This function accesses the PyQn library [10] to perform unit conversions. Listing 8 returns all datasets with energy and cross sections in units of and Mb, respectively.
| Listing 8. Unit conversions of all datasets. |
 |
- PyCollision.plot_all_datasets(): The PyCollision module also provides visualization function for the retrieved datasets, which assists in data evaluation and quality assessment. The plot_all_datasets method can be used to create plots using the pyplot submodule of the Matplotlib library. An example representation of the retrieved datasets for the proton-impact ionization of H 1s is shown in Figure 5. One can see that the peak cross sections reported in the references identified within CollisionDB as B33 [12] and B34 [13] are about 20–30% lower than those in other works [14,15]. As is evident in Listing 9, data visualization allows users to intuitively identify inconsistencies in the data. This serves as a first step in evaluating the data quality and requires further in-depth analysis of the data [16].
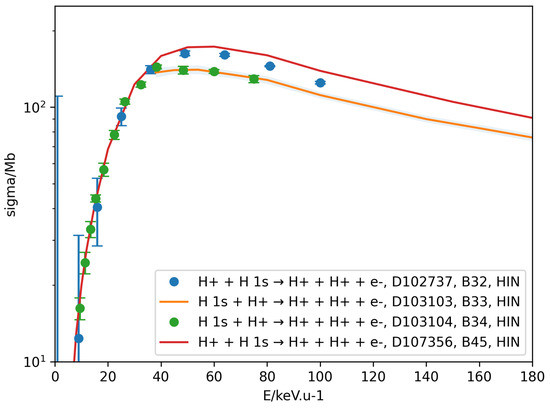 Figure 5. Comparison of the different datasets for proton-impact ionization cross sections of H 1s. Datasets were retrieved from the CollisionDB server using the API, and the legend includes reaction text, dataset qualified ID, reference ID and process code.
Figure 5. Comparison of the different datasets for proton-impact ionization cross sections of H 1s. Datasets were retrieved from the CollisionDB server using the API, and the legend includes reaction text, dataset qualified ID, reference ID and process code.
| Listing 9. An example of data visualization. |
 |
In the context of querying the database through the API, filtering is based on the search for the text representation of the objects, such as reactants and products, where applicable. However, filtering the datasets by reaction text looks for the ordered_text attribute stored in the rxn_reaction table, returning all reactions regardless of the order of reactants and products. This is implemented to ensure that there are no missing datasets in the query, as the cross sections typically do not depend on the order of the reactants if the relative velocities are the same. As can be seen in the proton-impact ionization example, the equivalent representations of the reactants "H+ + H 1s" or "H 1s + H+" represent the same collision process. This is ensured by the fact that energies for heavy-particle collisions are standardized in , giving the same relative velocity between the (non-)identical reactants. However, there may be cases where the order of reactants/products can affect the cross sections, in particular, for identical species but with different atomic states. For example, "H+ + H -> H+ + H 2p" and "H+ + H -> H 2p + H+" represent two different collisional processes, the former being the proton-impact excitation of hydrogen into the excited state and the latter representing the electron capture by protons into the 2p state. These processes can be distinguished by looking at the associated process types with the values "HEX" and "HCX", respectively. However, the reactions like "He 1s.2s 3S + He 1s2 1S -> He 1s.4s 3S + He" and "He 1s.2s 3S + He 1s2 1S -> He + He 1s4s 3S" belong to the same process type "HEX", but they represent two different collisional processes. The first reaction represents the excitation of the projectile He from 3 to 3 with a threshold energy of about 4 eV, while the latter is the target excitation from ground state 1 to 3 with a threshold energy of about 24 eV. We have preserved the canonical form of the reaction as well as the ordered text to search for all possible equivalent reactions. Additional metadata, such as process types, threshold and comments, can guide the users when affected by reactant/product order.
2.3. Current Status of CollisionDB
As of July 2023 there are 122,352 datasets in CollisionDB. Figure 6 summarizes the distribution of datasets for individual atoms and ions; as can be seen from this figure, there are some data for almost all charge states of atoms lighter than iron, but for heavier species there are data only for either neutral or low-charge states or for nearly fully stripped ions. There is better coverage for xenon and tungsten because of their importance for magnetic confinement fusion experiments.
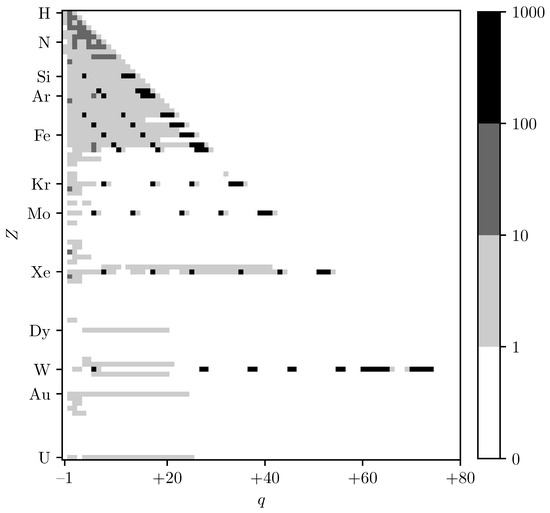
Figure 6.
A summary of the number of collisional datasets available in CollisionDB as of July 2023 for atomic species: Z is the nuclear charge and q the ion charge.
A breakdown of the classification of datasets in CollisionDB by process type and reactant type (total vs. molecular species) is given in Table 4. The database is dominated by the large number of electron-impact vibronic excitation cross sections and rate coefficients for molecular hydrogen and its isotopologues. At the time of writing, there are 115,773 cross sections and 6579 rate coefficient datasets in CollisionDB. Nearly 60% of the datasets have been fitted to functions described within CollisionDB, each of them with a Python implementation and some also in Fortran. These fit functions can be used on provided energy or temperature intervals to generate and return custom datasets.

Table 4.
Reactions categorized by collisional process type and by reactant as molecule. Abbreviations starting with the letter E denote “electron-impact” processes; those starting with the letter H are “heavy-particle” collisions.
A release of all datasets as a downloadable archive will be made on a periodic basis; the first such release, 2023.1, was issued on 31 August 2023 and is available from the CollisionDB website (see Data availability, below).
All data from CollisionDB are released under the Creative Commons Attribution 4.0 International (CC BY 4.0) license: data may be copied, shared, reused and adapted without restriction beyond the requirement to maintain appropriate attribution to the data providers.
3. Materials and Methods
3.1. Data Model
The CollisionDB database has been created to store and manage large amounts of atomic and molecular collisional data. It is built on a relational database management system backend: the main tables and relationships are described in the extended Entity Relationship Diagram (EER) shown in Figure 7. Here, we provide an overview of the main tables and their relationships, along with a description of the basic metadata stored in each table.
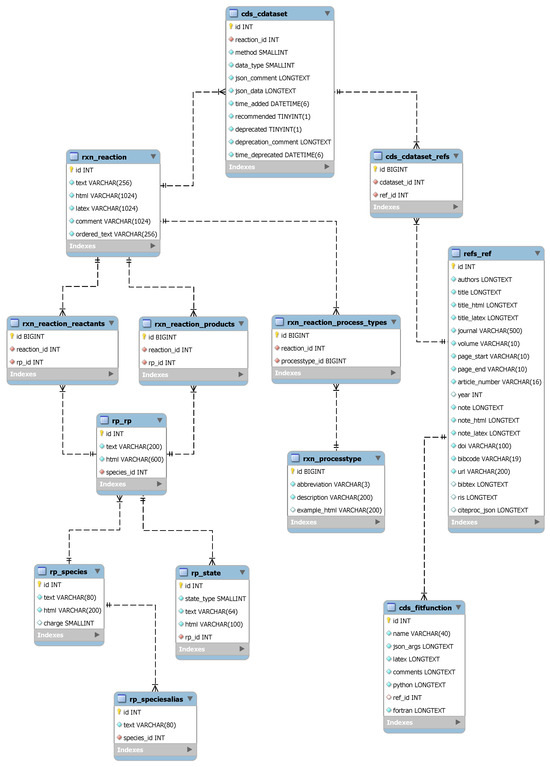
Figure 7.
EER diagram of the CollisionDB relational database schema.
Each collisional dataset in the database is described by the primary table cds_cdataset, which includes the following metadata attributes:
- id: a unique integer primary key identifying the dataset within the CollisionDB ecosystem.
- reaction_id: a foreign key identifying the single collisional process with which a dataset is associated (represented in the rxn_reaction table seen below).
- method: the method used to obtain the numerical data: one of "experiment", "theory", "semi-empirical" or "estimate". More precise details about the computational method used to calculate data can be specified using pre-defined abbreviations in the json_data attribute (see below).
- data_type: the type of collisional data, which can be one of the following: "cross section", "differential cross section" or "rate coefficient".
- comment: a free-text comment field providing additional information concerning the dataset.
- json_data: all other metadata such as threshold, uncertainty and units, stored as key–value pairs in a JSON (JavaScript Object Notation) object (see Section 3.2).
- recommended: A Boolean flag, indicating whether the dataset is evaluated and recommended (details may be provided in the comment field).
- deprecated: A Boolean flag, indicating whether the dataset is out of date due to identified errors or quality issues. Details can be provided in the deprecation_comment field. A new dataset may be available in the database.
- time_added, time_deprecated: timestamps indicating when a dataset was added or deprecated in the CollisionDB database.
- One or more bibliographic references for the data, identified through a many-to-many relationship with the refs_ref table.
This schema ensures that each dataset has appropriate and unambiguous provenance and contextual metadata to ensure its accurate use. It will often be the case that a single collisional process is associated with multiple datasets. For instance, a given reaction can be described by different methods, such as an experiment, theory or a different computational approach, or have a different energy range or data type, such as a cross section or rate coefficient.
The rxn_reaction table describes each collisional process (“reaction”) through the structure outlined below; each cds_cdataset dataset entry is associated with exactly one rxn_reaction entry.
- text: a text representation of the reaction in a canonical form conforming to the standards of and parseable by the PyValem library [8].
- html, latex: HTML and LaTeX representations of the reaction, for display in the browser and export.
- comment: a free-text comment field providing further information about the reaction.
- ordered_text: a text representation of the reaction in which the reactants and products are ordered in an arbitrary but consistent way to facilitate indexing, searching and comparison of reactions.
- rxn_reaction_reactants and rxn_reaction_products: these tables provide a many-to-many relationship between each reaction and its individual reactant and product species (including their quantum states, where relevant), which are held in the rp_rp table.
- rxn_reaction_process_types: this table provides a many-to-many relationship between a reaction and its classifying process codes (e.g., EIN = electron-impact ionization); a complete list of these codes is given in Ref. [9].
Reactants and products are stored in the rp_rp table in plain-text format conforming to the standardized syntax implemented by the PyValem library. They can be “stateful” in the sense that, in addition to identifying the chemical species, they can contain information about any relevant quantum numbers, labels and symmetries. Species themselves can be atoms, molecules, ions or isotopes, including the electron (e-) and the photon (h).
To aid with the search functionality, these stateful species can have one or more aliases (stored in the table rp_speciesalias). For example, the isotopologue of molecular hydrogen usually written as HD has aliases DH, H(2H), (2H)H, (1H)(2H) and (2H)(1H). Aliases can also be other chemical identifiers, for example, the InChIKey, and quantum states can be specified as atomic or molecular electronic configurations, term symbols and individual quantum numbers in key–value pairs. Some examples are given in Table 1.
CollisionDB also supports analytic fits to collisional data where available. Metadata for fitting functions can be stored in the cds_fitfunction table, including the arguments of the fitting function, their descriptions, types and units in the json_args attribute. CollisionDB also provides implementations (source code) of the fitting functions themselves in the Python programming language.
A description of the metadata formats and standards used to populate this database is given in the following subsections.
3.2. Data Transfer Format for Download: JSON
Each dataset in CollisionDB is described by metadata which are stored in the database and also presented as a header, in JSON format, to each data file. The meaning of the JSON metadata objects is given in Table 2, and an example metadata object is given in Listing 1. The data are separated from the metadata header by a single line of at least five hyphens.
Uncertainties in the numerical data may be given as a global estimate applying to all data points (using the unc_perc key) or per-data point; in the latter case, the uncertainty is separated from the data value by a colon and may be either a symmetric range v:u implying or asymmetric v:l:u implying .
3.3. Data Transfer Format for Upload
3.3.1. Plain Text
For the convenience of data providers, data can be submitted to the CollisionDB database in UTF-8 encoded, plain-text format; an example template file is given in Listing 10. The file includes metadata specified as key=value pairs, followed by the numerical data in space-delimited columns. The metadata keys are described in Table 2 and online at https://amdis.iaea.org/db/collisiondb/submitting-data/ (accessed on 23 March 2024). Their values may be numbers, strings (inside double quotes, "...") or lists (a comma-separated sequence of values, enclosed in square brackets, [...], depending on the key. Blank lines and any content following the “#” symbol are ignored; missing data values are given by asterisks.
| Listing 10. An example template file for submission. |
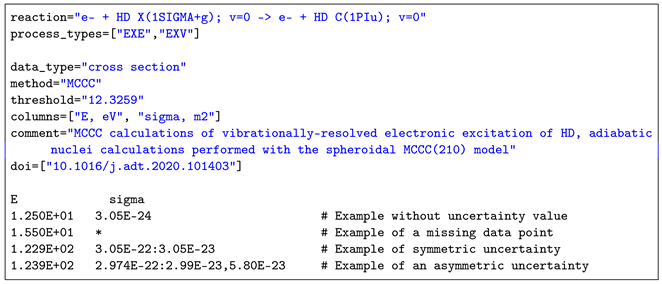 |
3.3.2. JSON
Data are uploaded to the CollisionDB database from a JSON object, and data providers can also choose to submit their data directly in JSON format, as shown in Listing 11. This example represents the JSON object containing metadata and numerical data of a collisional dataset, as specified in Listing 10 in the plain-text format. Numerical data undergo standardized unit conversions using the the PyQn Library [10] as well as numerical checks. These verify, for example, that the grid of energies or temperatures is monotonically increasing.
This JSON object is processed into a well-defined format conforming to the standardized schemas established by the IAEA’s AMD Unit and other experts and validated prior to uploading it to the database. This involves the use of the PyValem Library [8] to canonicalize reactions and validate charge balance and stoichiometry conservation, etc. Additionally, the DOIs (Digital Object Identifiers) are resolved into the citable format using the Python package django-pyref [17]. In order to prevent duplication, the import script checks for pre-existing metadata in CollisionDB.
Although the metadata are uploaded as cdataset instances into the relational database, numerical data (including the JSON metadata header) are saved as static files.
| Listing 11. An example JSON dataset file for submission. |
 |
4. Conclusions
In providing easy access to peer-reviewed published data, conforming to FAIR princples and being facilitated by structured metadata and API integration, it is hoped that CollisionDB can provide a useful resource to the plasma collisional physics community and open up new possibilities for machine-learning-driven advances in fusion and other areas of plasma research.
Author Contributions
C.H. conceived the idea and the initial schema of the CollisionDB database. D. and C.H. wrote the manuscript and managed data uploading and database maintenance. C.H., D. and M.H. contributed to software development. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The first release version of all datasets, v2023.1, is available on the CollisionDB website at https://amdis.iaea.org/db/collisiondb/ (accessed on 23 March 2024) under the terms of the CC BY 4.0 license. The PyCollisionDB Python library for interacting with the database is available at https://github.com/xnx/pycollisiondb/ (accessed on 23 March 2024) and is released under the terms of Apache license.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Pitchford, L.C.; Alves, L.L.; Bartschat, K.; Biagi, S.F.; Bordage, M.-C.; Bray, I.; Brion, C.E.; Brunger, M.J.; Campbell, L.; Chachereau, A.; et al. LXCat: An Open-Access, Web-Based Platform for Data Needed for modeling low temperature plasmas. Plasma Process. Polym. 2017, 14, 1600098. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, H.; Chang, W.S.; Chung, S.Y.; Kwon, D.C.; Song, M.Y.; Yoon, J.S. A New Version of the Plasma Database for Plasma Physics in the Data Center for Plasma Properties. Appl. Sci. Converg. Technol. 2020, 29, 5–9. [Google Scholar] [CrossRef]
- Celiberto, R.; Armenise, I.; Cacciatore, M.; Capitelli, M.; Esposito, F.; Gamallo, P.; Janev, R.K.; Laganà, A.; Laporta, V.; Laricchiuta, A.; et al. Atomic and molecular data for spacecraft re-entry plasmas. Plasma Sources Sci. Technol. 2016, 25, 033004. [Google Scholar] [CrossRef]
- Tennyson, J.; Mohr, S.; Hanicinec, M.; Dzarasova, A.; Smith, C.; Waddington, S.; Liu, B.; Alves, L.L.; Bartschat, K.; Bogaerts, A.; et al. The 2021 release of the Quantemol database (QDB) of plasma chemistries and reactions. Plasma Sources Sci. Technol. 2022, 31, 095020. [Google Scholar] [CrossRef]
- Summers, H.P. The ADAS User Manual, Version 2.6.2004. Available online: http://www.adas.ac.uk (accessed on 23 March 2024).
- Hulse, R.A. The ALADDIN atomic physics database system. AIP Conf. Proc. 1990, 206, 63–72. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Hill, C. PyValem, GitHub Repository. 2022. Available online: https://github.com/xnx/pyvalem (accessed on 23 March 2024).
- Hill, C.; Dubernet, M.L.; Endres, C.; Karwasz, G.; Marinković, B.; Marquart, T.; Heinola, K.; Zwölf, C.M.; Moreau, N.; Dipti; et al. “Classification of Processes in Plasma Physics” Version 2.4. 2022. Available online: https://amdis.iaea.org/media/miscellaneous-publications/plasma-processes-classification-v2.4.pdf (accessed on 23 March 2024).
- Hill, C. PyQn, GitHub Repository. 2022. Available online: https://github.com/xnx/pyqn (accessed on 23 March 2024).
- Hill, C. PyCollisionDB, GitHub Repository. 2022. Available online: https://github.com/xnx/pycollisiondb (accessed on 23 March 2024).
- Shah, M.B.; Gilbody, H.B. Experimental study of the ionisation of atomic hydrogen by fast H+ and He2+ ions. J. Phys. B At. Mol. Opt. Phys. 1981, 14, 2361. [Google Scholar] [CrossRef]
- Shah, M.B.; Elliott, D.S.; Gilbody, H.B. Ionisation of atomic hydrogen by 9–75 keV protons. J. Phys. B At. Mol. Opt. Phys. 1987, 20, 2481. [Google Scholar] [CrossRef]
- Agueny, H.; Hansen, J.P.; Dubois, A.; Makhoute, A.; Taoutioui, A.; Sisourat, N. Electron capture, ionization and excitation cross sections for keV collisions between fully stripped ions and atomic hydrogen in ground and excited states. At. Data Nucl. Data Tables 2019, 129–130, 101281. [Google Scholar] [CrossRef]
- Leung, A.C.K.; Kirchner, T. Proton impact on ground and excited states of atomic hydrogen. Eur. Phys. J. D 2019, 73, 246. [Google Scholar] [CrossRef]
- Hill, C.; Dipti; Heinola, K.; Dubois, A.; Sisourat, N.; Taoutioui, A.; Agueny, H.; Tőkési, K.; Ziaeian, I.; Illescas, C.; et al. Atomic collisional data for neutral beam modeling in fusion plasmas. Nucl. Fusion 2023, 63, 125001. [Google Scholar] [CrossRef]
- Hill, C. django-pyref, GitHub Repository. 2022. Available online: https://github.com/xnx/django-pyref (accessed on 23 March 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).