WiPP: Workflow for Improved Peak Picking for Gas Chromatography-Mass Spectrometry (GC-MS) Data
Abstract
1. Introduction
2. Results
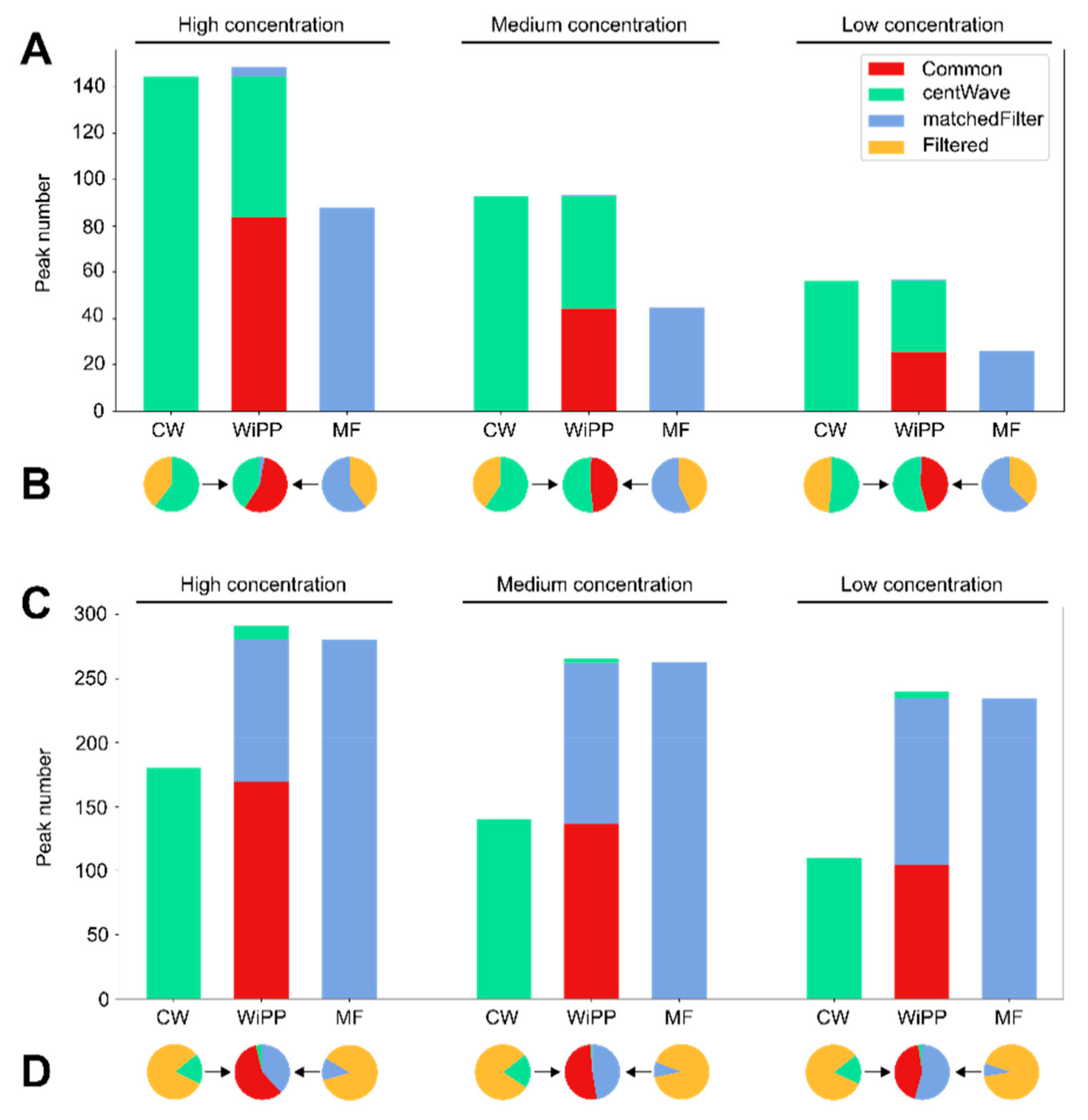
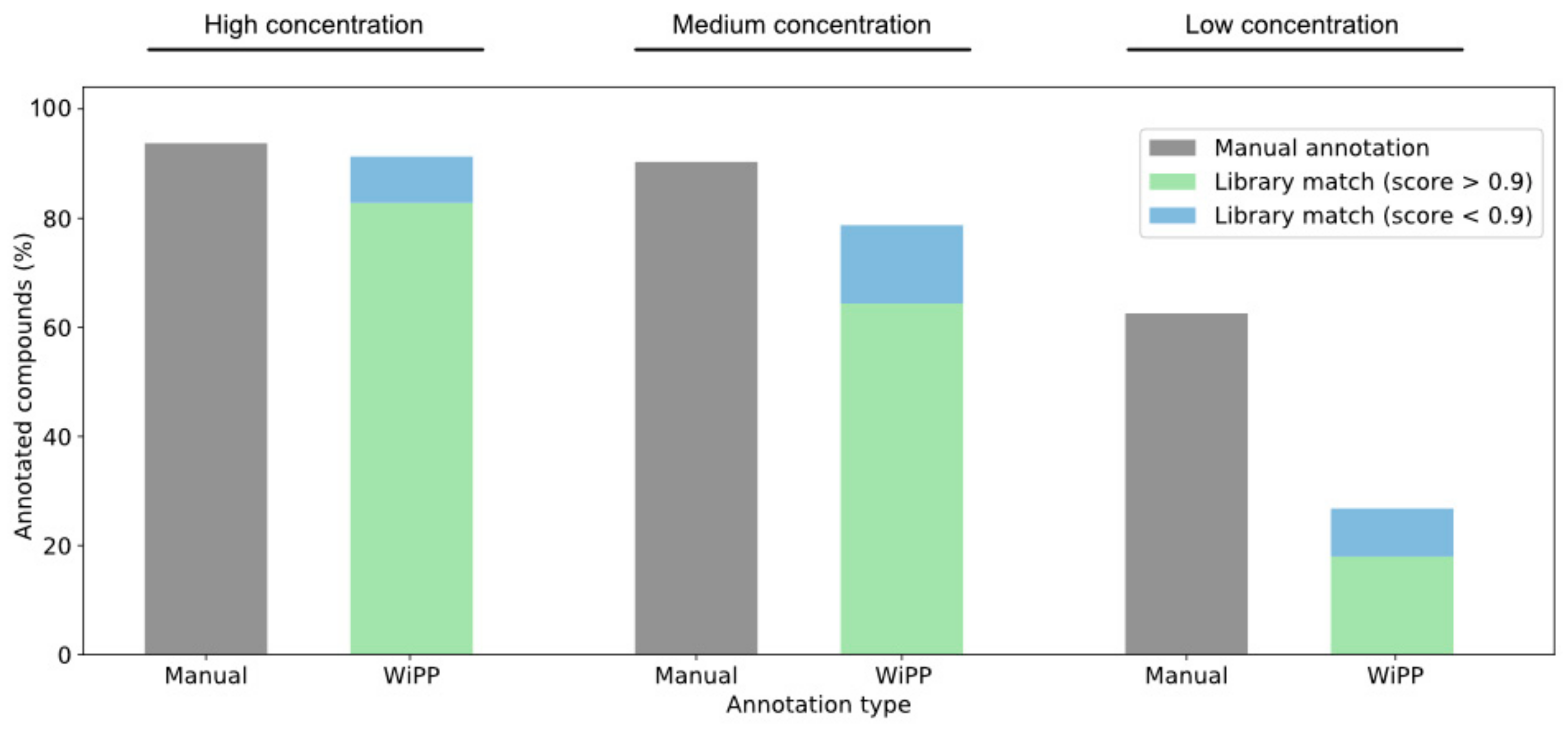
2.1. Validation and Benchmarking
2.2. Case Study
3. Discussion
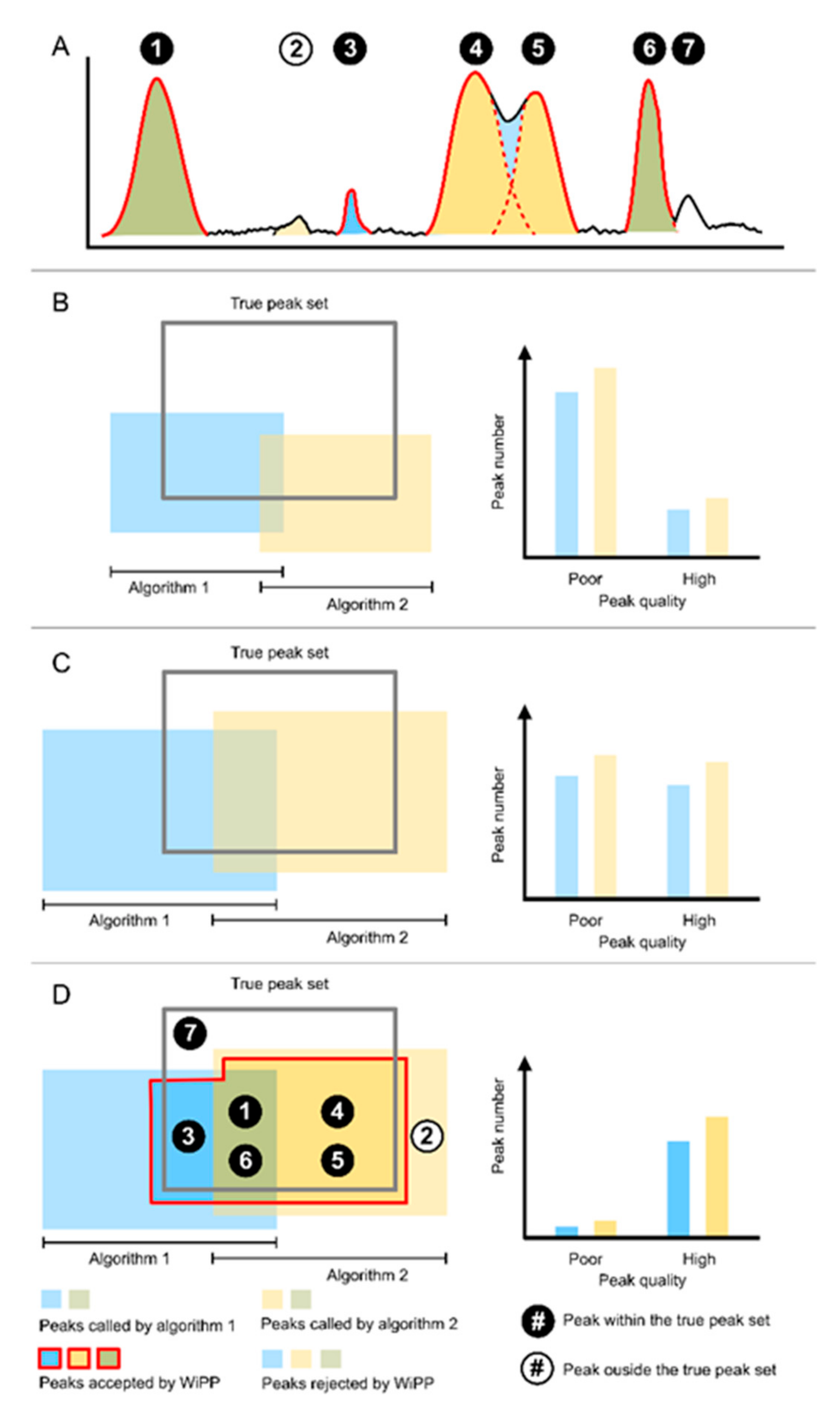
3.1. Automated Classification of Peak Picking Provides a Novel Way to Assess and Compare the Performance of Peak Picking Algorithms
3.2. Optimising Parameters for Peak Picking
3.3. Improving Overall Quality of the Final Picked Peak List
- Providing a novel way to classify peaks based on seven classes, and thus objectively assessing their quality.
- Enabling objective performance comparison of different peak picking algorithms.
- Enabling automated parameter optimisation for individual peak picking algorithms.
- Enabling a final, improved high quality peak list to be generated for further analyses.
- Reducing the operator time required by packaging WiPP within a fully automated workflow (once the initial training of data is completed).
4. Materials and Methods
4.1. Peak Detection
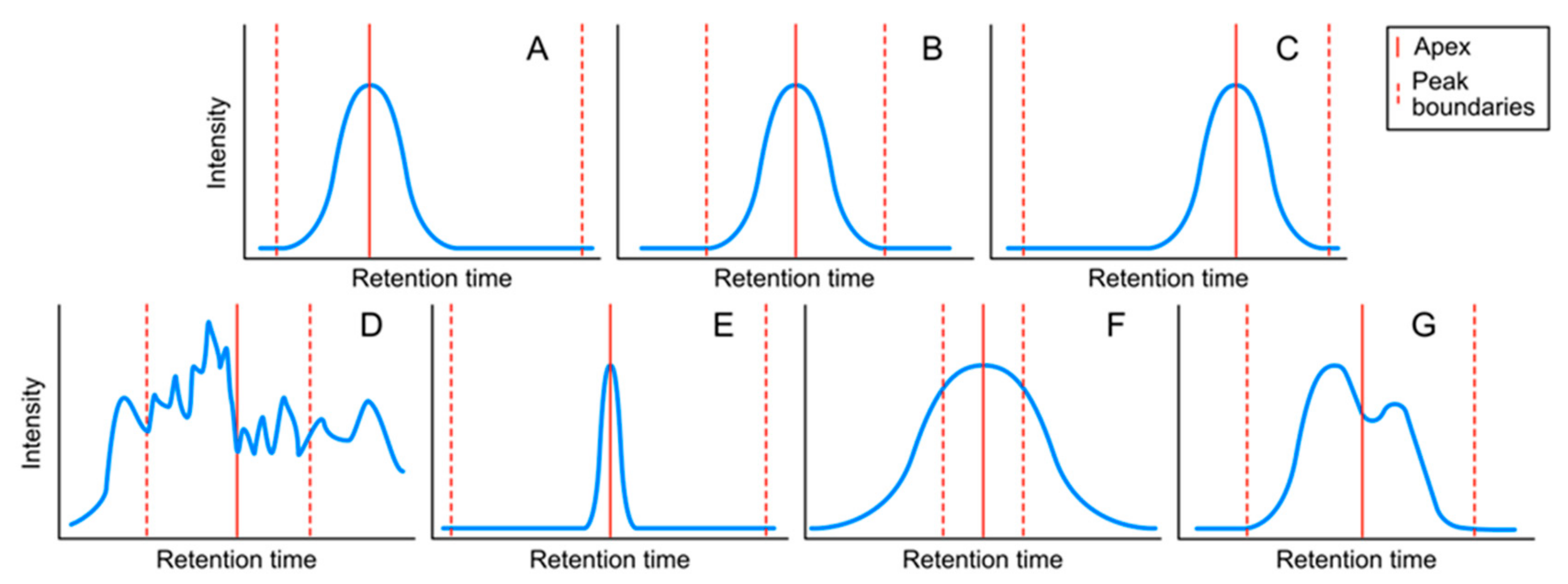
4.1.2. Peak Classes
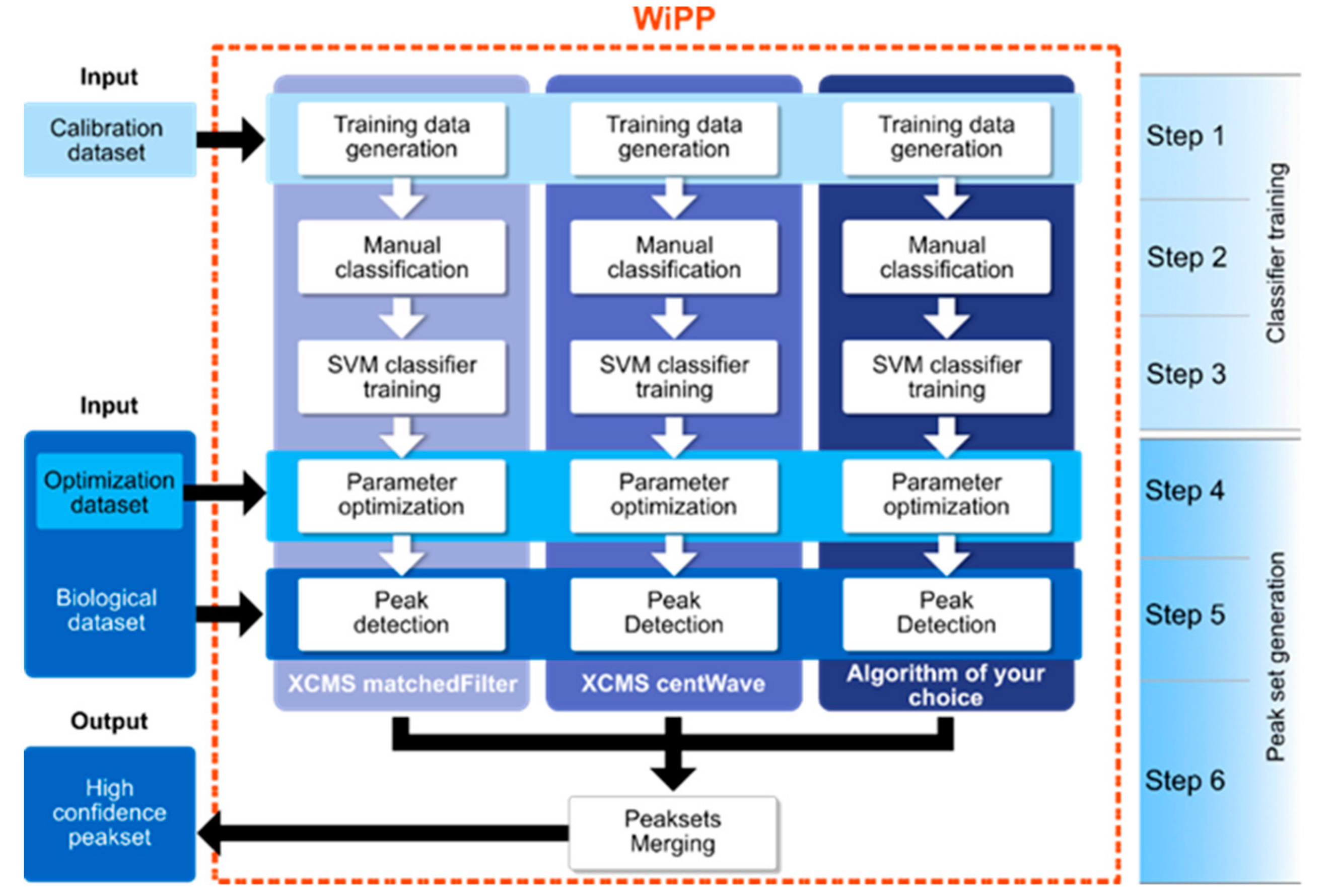
4.1.3. WiPP Workflow and Model
4.2. Implementation and Availability
4.3. Data
4.3.1. Datasets 1 and 2
4.3.2. Dataset 3
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Livera, A.M.; Dias, D.A.; De Souza, D.; Rupasinghe, T.; Pyke, J.; Tull, D.; Roessner, U.; McConville, M.; Speed, T.P. Normalizing and Integrating Metabolomics Data. Anal. Chem. 2012, 84, 10768–10776. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis−McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large−scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Haggarty, J.; Burgess, K.E. Recent advances in liquid and gas chromatography methodology for extending coverage of the metabolome. Curr. Opin. Biotechnol. 2017, 43, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; Maille, G.O.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Wehrens, R.; Weingart, G.; Mattivi, F. metaMS: An open−source pipeline for GC–MS−based untargeted metabolomics. J. Chromatogr. B 2014, 966, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Lommen, A. MetAlign: Interface−Driven, Versatile Metabolomics Tool for Hyphenated Full−Scan Mass Spectrometry Data Preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar−Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry−based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks. Anal. Chem. 2017, 89, 8696–8703. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Jia, W.; Walker, D.I.; Jones, D.P.; Du, X. ADAP−GC 3.2: Graphical Software Tool for Efficient Spectral Deconvolution of Gas Chromatography–High−Resolution Mass Spectrometry Metabolomics Data. J. Proteome Res. 2018, 17, 470–478. [Google Scholar] [CrossRef]
- O’Callaghan, S.; De Souza, D.P.; Isaac, A.; Wang, Q.; Hodkinson, L.; Olshansky, M.; Erwin, T.; Appelbe, B.; Tull, D.L.; Roessner, U.; et al. PyMS: A Python toolkit for processing of gas chromatography−mass spectrometry (GC−MS) data. Application and comparative study of selected tools. BMC Bioinform. 2012, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Domingo−Almenara, X.; Brezmes, J.; Vinaixa, M.; Samino, S.; Ramirez, N.; Ramon−Krauel, M.; Lerin, C.; Díaz, M.; Ibáñez, L.; Correig, X.; et al. eRah: A Computational Tool Integrating Spectral Deconvolution and Alignment with Quantification and Identification of Metabolites in GC/MS−Based Metabolomics. Anal. Chem. 2016, 88, 9821–9829. [Google Scholar] [CrossRef]
- Myers, O.D.; Sumner, S.J.; Li, S.; Barnes, S.; Du, X. Detailed Investigation and Comparison of the XCMS and MZmine 2 Chromatogram Construction and Chromatographic Peak Detection Methods for Preprocessing Mass Spectrometry Metabolomics Data. Anal. Chem. 2017, 89, 8689–8695. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning; Springer Series in Statistics; Springer: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Zhou, Z.; Tu, J.; Zhu, Z.J. Advancing the large−scale CCS database for metabolomics and lipidomics at the machine−learning era. Curr. Opin. Chem. Biol. 2018, 42, 34–41. [Google Scholar] [CrossRef]
- Zheng, H.; Zheng, P.; Zhao, L.; Jia, J.; Tang, S.; Xu, P.; Xie, P.; Gao, H. Predictive diagnosis of major depression using NMR−based metabolomics and least−squares support vector machine. Clin. Chim. Acta 2017, 464, 223–227. [Google Scholar] [CrossRef]
- Khitan, Z.; Shapiro, A.P.; Shah, P.T.; Sanabria, J.R.; Santhanam, P.; Sodhi, K.; Abraham, N.G.; Shapiro, J.I. Predicting Adverse Outcomes in Chronic Kidney Disease Using Machine Learning Methods: Data from the Modification of Diet in Renal Disease. Marshall J. Med. 2017, 3, 67. [Google Scholar] [CrossRef]
- Hsu, C.W.; Chang, C.C.; Lin, C.J. A Practical Guide to Support Vector Classification; Department of Computer Science, National Taiwan University: Taiwan, 2003.
- Libiseller, G.; Dvorzak, M.; Kleb, U.; Gander, E.; Eisenberg, T.; Madeo, F.; Neumann, S.; Trausinger, G.; Sinner, F.; Pieber, T.; et al. IPO: A tool for automated optimization of XCMS parameters. BMC Bioinform. 2015, 16, 118. [Google Scholar] [CrossRef] [PubMed]
- Kuich, P.H.J.L.; Hoffmann, N.; Kempa, S. Maui−VIA: A User−Friendly Software for Visual Identification, Alignment, Correction, and Quantification of Gas Chromatography–Mass Spectrometry Data. Front. Bioeng. Biotechnol. 2015, 2, 84. [Google Scholar] [CrossRef]
- Nezami Ranjbar, M.R.; Luo, Y.; Di Poto, C.; Varghese, R.S.; Ferrarini, A.; Zhang, C.; Sarhan, N.I.; Soliman, H.; Tadesse, M.G.; Ziada, D.H.; et al. GC-MS Based Plasma Metabolomics for Identification of Candidate Biomarkers for Hepatocellular Carcinoma in Egyptian Cohort. PLoS ONE 2015, 10, e0127299. [Google Scholar] [CrossRef] [PubMed]
- Coble, J.B.; Fraga, C.G. Comparative evaluation of preprocessing freeware on chromatography/mass spectrometry data for signature discovery. J. Chromatogr. A 2014, 1358, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of LC/MS data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef]
- Chicco, D. Ten quick tips for machine learning in computational biology. BioData Min. 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Pietzke, M.; Zasada, C.; Mudrich, S.; Kempa, S. Decoding the dynamics of cellular metabolism and the action of 3-bromopyruvate and 2-deoxyglucose using pulsed stable isotope−resolved metabolomics. Cancer Metab. 2014, 2, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca−Serra, P.; et al. MetaboLights—An open−access general−purpose repository for metabolomics studies and associated meta−data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef] [PubMed]
| ID | Identified (WiPP) | p-Value (Study) | Fold Change (Study) | p-Value (WiPP) | Fold Change (WiPP) |
|---|---|---|---|---|---|
| Glutamic Acid | + | 1.9 | 1.89 | ||
| α−tocopherol | + | 1.5 | 1.36 | ||
| Valine | + | 1.5 | 1.52 | ||
| Citric Acid | + | −1.3 | −1.20 | ||
| Sorbose | + | −2.4 | −1.66 | ||
| Cholesterol | + | 1.1 | 1.10 | ||
| Lactic Acid | + | −1.3 | X | X | |
| Leucine | + | 1.6 | X | X | |
| Isoleucine | + | 1.5 | X | X | |
| Hexose | + | – | – | −1.61 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borgsmüller, N.; Gloaguen, Y.; Opialla, T.; Blanc, E.; Sicard, E.; Royer, A.-L.; Le Bizec, B.; Durand, S.; Migné, C.; Pétéra, M.; et al. WiPP: Workflow for Improved Peak Picking for Gas Chromatography-Mass Spectrometry (GC-MS) Data. Metabolites 2019, 9, 171. https://doi.org/10.3390/metabo9090171
Borgsmüller N, Gloaguen Y, Opialla T, Blanc E, Sicard E, Royer A-L, Le Bizec B, Durand S, Migné C, Pétéra M, et al. WiPP: Workflow for Improved Peak Picking for Gas Chromatography-Mass Spectrometry (GC-MS) Data. Metabolites. 2019; 9(9):171. https://doi.org/10.3390/metabo9090171
Chicago/Turabian StyleBorgsmüller, Nico, Yoann Gloaguen, Tobias Opialla, Eric Blanc, Emilie Sicard, Anne-Lise Royer, Bruno Le Bizec, Stéphanie Durand, Carole Migné, Mélanie Pétéra, and et al. 2019. "WiPP: Workflow for Improved Peak Picking for Gas Chromatography-Mass Spectrometry (GC-MS) Data" Metabolites 9, no. 9: 171. https://doi.org/10.3390/metabo9090171
APA StyleBorgsmüller, N., Gloaguen, Y., Opialla, T., Blanc, E., Sicard, E., Royer, A.-L., Le Bizec, B., Durand, S., Migné, C., Pétéra, M., Pujos-Guillot, E., Giacomoni, F., Guitton, Y., Beule, D., & Kirwan, J. (2019). WiPP: Workflow for Improved Peak Picking for Gas Chromatography-Mass Spectrometry (GC-MS) Data. Metabolites, 9(9), 171. https://doi.org/10.3390/metabo9090171





