Untargeted Metabolomics Approach Reveals Diverse Responses of Pastinaca Sativa to Ozone and Wounding Stresses
Abstract
1. Introduction
2. Material and Methods
2.1. Stress Application Settings
2.2. Metabolite Extraction
2.3. UHPLC-MS
2.4. High-Resolution Mass Spectrometry
2.5. ROS Analysis
2.6. Metabolic Data Analysis
3. Results
3.1. Impact of Treatment on the Oxidative Status of Parsnip Leaves
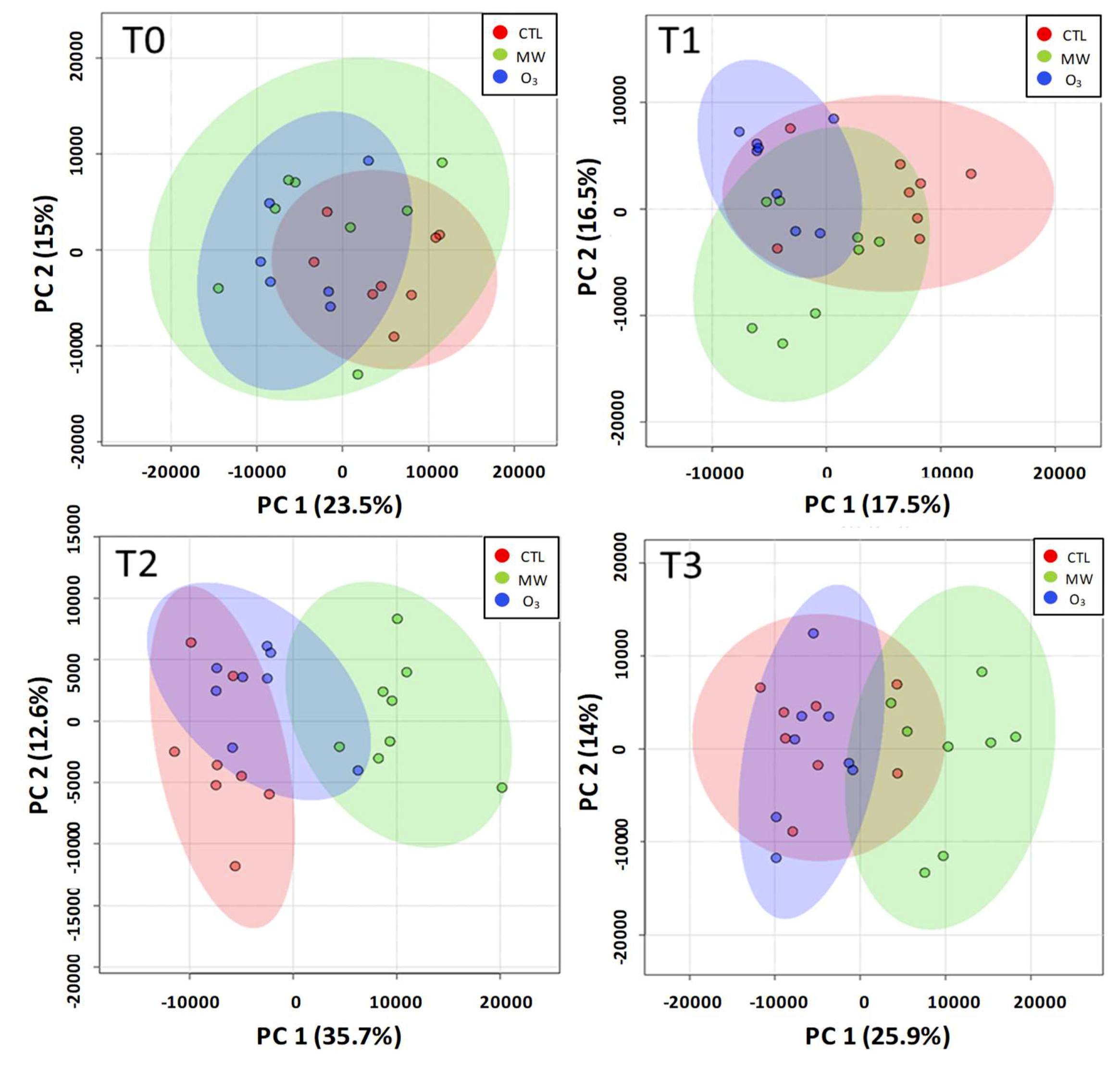
3.2. Global Overview of the Metabolomic Profile.
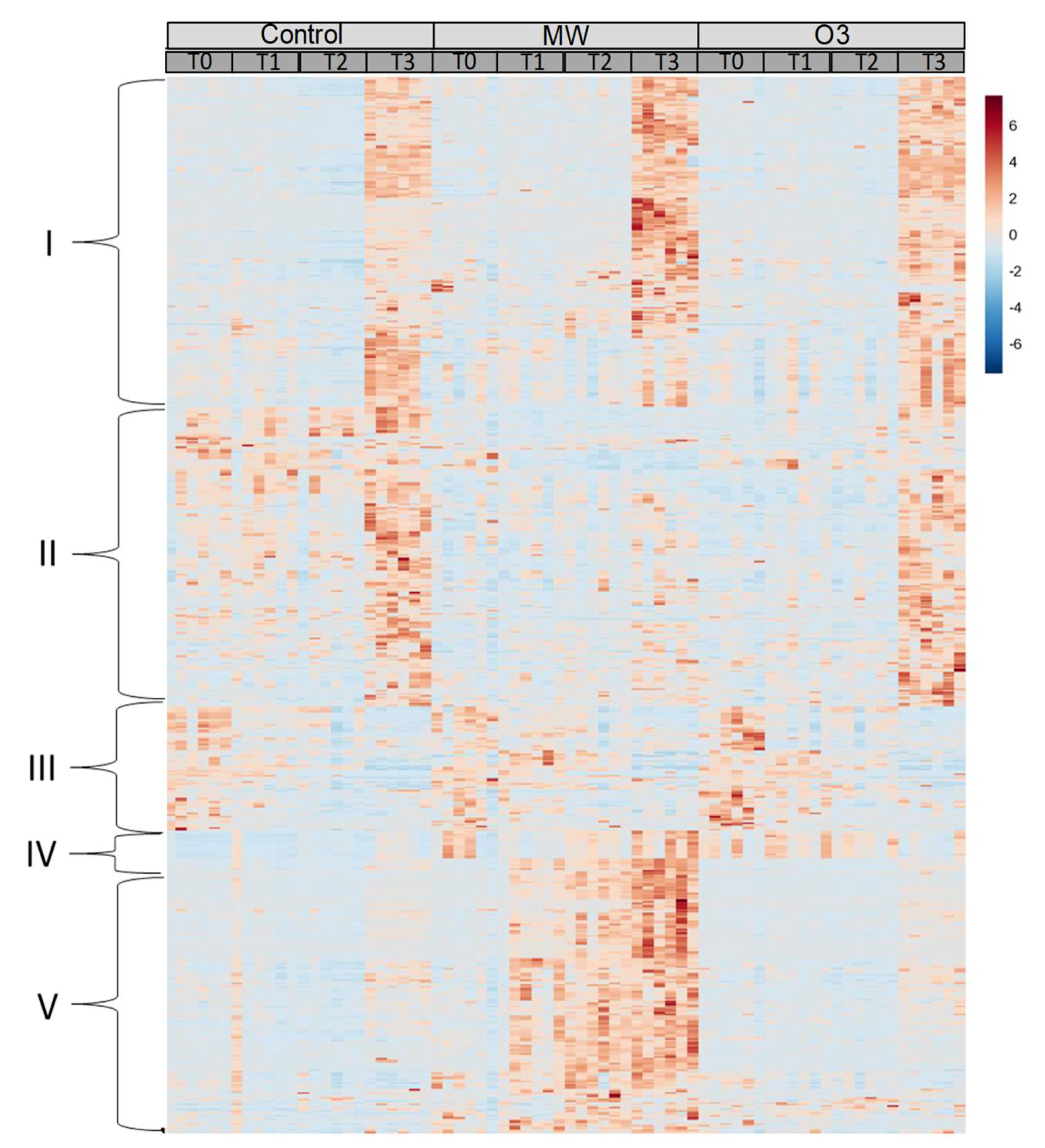
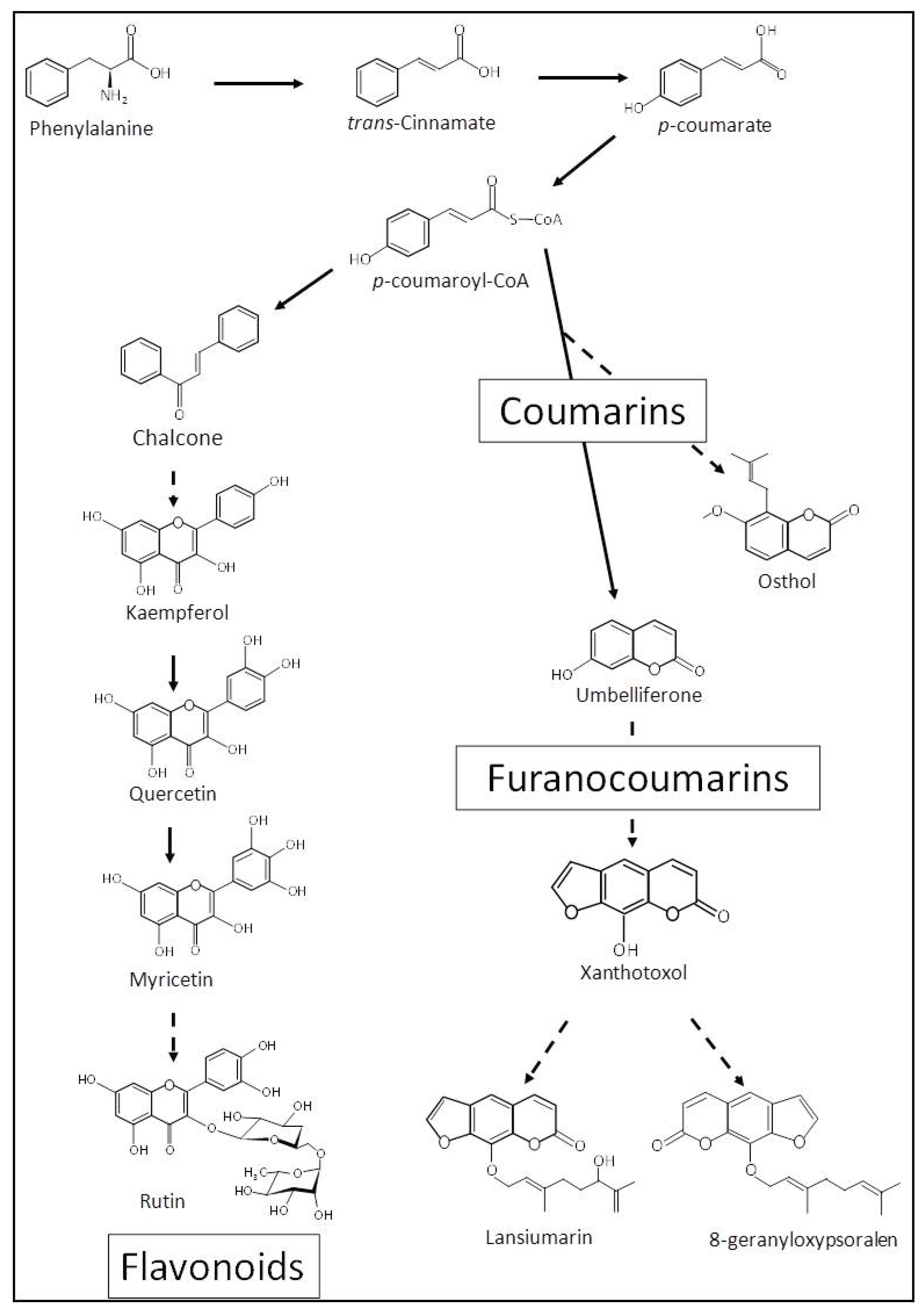
3.3. Metabolite Identification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Graham, L.E.; Kodner, R.B.; Fisher, M.M.; Graham, J.M.; Wilcox, L.W.; Hackney, J.M.; Obst, J.; Bilkey, P.C.; Hanson, D.T.; Cook, M.E. 9—Early Land Plant Adaptations to Terrestrial Stress: A Focus on Phenolics. In The Evolution of Plant Physiology; Hemsley, A.R., Poole, I., Eds.; Academic Press: Oxford, UK, 2004; pp. 155–169. [Google Scholar]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat Tolerance in Plants: An Overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Walley, J.W.; Dehesh, K. Molecular Mechanisms Regulating Rapid Stress Signaling Networks in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor Signal Transduction Leading to Production of Plant Secondary Metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Klessig, D.F. MAPK Cascades in Plant Defense Signaling. Trends Plant Sci. 2001, 6, 520–527. [Google Scholar] [CrossRef]
- Ludwig, A.A.; Romeis, T.; Jones, J.D.G. CDPK-mediated Signalling Pathways: Specificity and Cross-talk. J. Exp. Bot. 2004, 55, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Vandelle, E.; Vannozzi, A.; Wong, D.; Danzi, D.; Digby, A.-M.; Dal Santo, S.; Astegno, A. Identification, Characterization, and Expression Analysis of Calmodulin and Calmodulin-like Genes in Grapevine (Vitis Vinifera) Reveal Likely Roles in Stress Responses. Plant Physiol. Biochem. 2018, 129, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Per, T.S.; Khan, M.I.R.; Anjum, N.A.; Masood, A.; Hussain, S.J.; Khan, N.A. Jasmonates in Plants under Abiotic Stresses: Crosstalk with Other Phytohormones Matters. Environ. Exp. Bot. 2018, 145, 104–120. [Google Scholar] [CrossRef]
- Crutzen, P.J. The “Anthropocene.” In Earth System Science in the Anthropocene; Ehlers, E., Krafft, T., Eds.; Springer: Heidelberg/Berlin, Germany, 2006; pp. 13–18. [Google Scholar]
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland, 2014; 151p. [Google Scholar]
- Ainsworth, E.A. Understanding and Improving Global Crop Response to Ozone Pollution. Plant J. 2017, 90, 886–897. [Google Scholar] [CrossRef]
- Baier, M.; Kandlbinder, A.; Golldack, D.; Dietz, K.J. Oxidative Stress and Ozone: Perception, Signalling and Response. Plant Cell Environ. 2005, 28, 1012–1020. [Google Scholar] [CrossRef]
- Castagna, A.; Ranieri, A. Detoxification and Repair Process of Ozone Injury: From O3 Uptake to Gene Expression Adjustment. Environ. Pollut. 2009, 157, 1461–1469. [Google Scholar] [CrossRef]
- Delcour, I.; Spanoghe, P.; Uyttendaele, M. Literature Review: Impact of Climate Change on Pesticide Use. Food Res. Int. 2015, 68, 7–15. [Google Scholar] [CrossRef]
- Alamgir, K.M.; Hojo, Y.; Christeller, J.T.; Fukumoto, K.; Isshiki, R.; Shinya, T.; Baldwin, I.T.; Galis, I. Systematic Analysis of Rice (Oryza Sativa) Metabolic Responses to Herbivory. Plant Cell Environ. 2016, 39, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Mishra, S.; Vadassery, J. Spodoptera Litura-Mediated Chemical Defense Is Differentially Modulated in Older and Younger Systemic Leaves of Solanum Lycopersicum. Planta 2018. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Saito, K. Integrated Metabolomics for Abiotic Stress Responses in Plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for Plant Stress Response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef]
- Berenbaum, M.; Feeny, P. Toxicity of Angular Furanocoumarins to Swallowtail Butterflies: Escalation in a Coevolutionary Arms Race? Science 1981, 212, 927–929. [Google Scholar] [CrossRef]
- Bourgaud, F.; Hehn, A.; Larbat, R.; Doerper, S.; Gontier, E.; Kellner, S.; Matern, U. Biosynthesis of Coumarins in Plants: A Major Pathway Still to Be Unravelled for Cytochrome P450 Enzymes. Phytochem. Rev. 2006, 5, 293–308. [Google Scholar] [CrossRef]
- Smith, S.I.; Brodbelt, J.S. Rapid Characterization of Cross-Links, Mono-Adducts, and Non-Covalent Binding of Psoralens to Deoxyoligonucleotides by LC-UV/ESI-MS and IRMPD Mass Spectrometry. Analyst 2010, 135, 943–952. [Google Scholar] [CrossRef]
- Larbat, R.; Kellner, S.; Specker, S.; Hehn, A.; Gontier, E.; Hans, J.; Bourgaud, F.; Matern, U. Molecular Cloning and Functional Characterization of Psoralen Synthase, the First Committed Monooxygenase of Furanocoumarin Biosynthesis. J. Biol. Chem. 2007, 282, 542–554. [Google Scholar] [CrossRef]
- Larbat, R.; Hehn, A.; Hans, J.; Schneider, S.; Jugdé, H.; Schneider, B.; Matern, U.; Bourgaud, F. Isolation and Functional Characterization of CYP71AJ4 Encoding for the First P450 Monooxygenase of Angular Furanocoumarin Biosynthesis. J. Biol. Chem. 2009, 284, 4776–4785. [Google Scholar] [CrossRef]
- Roselli, S.; Olry, A.; Vautrin, S.; Coriton, O.; Ritchie, D.; Galati, G.; Navrot, N.; Krieger, C.; Vialart, G.; Bergès, H.; et al. A Bacterial Artificial Chromosome (BAC) Genomic Approach Reveals Partial Clustering of the Furanocoumarin Pathway Genes in Parsnip. Plant J. 2017, 89, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Krieger, C.; Roselli, S.; Kellner-Thielmann, S.; Galati, G.; Schneider, B.; Grosjean, J.; Olry, A.; Ritchie, D.; Matern, U.; Bourgaud, F.; et al. The CYP71AZ P450 Subfamily: A Driving Factor for the Diversification of Coumarin Biosynthesis in Apiaceous Plants. Front. Plant Sci. 2018, 9, 820. [Google Scholar] [CrossRef] [PubMed]
- Munakata, R.; Olry, A.; Karamat, F.; Courdavault, V.; Sugiyama, A.; Date, Y.; Krieger, C.; Silie, P.; Foureau, E.; Papon, N.; et al. Molecular Evolution of Parsnip (Pastinaca Sativa) Membrane-Bound Prenyltransferases for Linear and/or Angular Furanocoumarin Biosynthesis. New Phytol. 2016, 211, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Junglee, S.; Urban, L.; Sallanon, H.; Lopez-Lauri, F. Optimized Assay for Hydrogen Peroxide Determination in Plant Tissue Using Potassium Iodide. Am. J. Anal. Chem. 2014, 5, 730–736. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Web-Based Inference of Biological Patterns, Functions and Pathways from Metabolomic Data Using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Djoumbou Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef]
- Kräutler, B. Chlorophyll Breakdown and Chlorophyll Catabolites in Leaves and Fruit. Photochem. Photobiol. Sci. 2008, 7, 1114–1120. [Google Scholar] [CrossRef]
- Müller, T.; Ulrich, M.; Ongania, K.-H.; Kräutler, B. Colorless Tetrapyrrolic Chlorophyll Catabolites Found in Ripening Fruit Are Effective Antioxidants. Angew. Chem. Int. Ed. 2007, 46, 8699–8702. [Google Scholar] [CrossRef]
- Oltmans, S.J.; Lefohn, A.S.; Shadwick, D.; Harris, J.M.; Scheel, H.E.; Galbally, I.; Tarasick, D.W.; Johnson, B.J.; Brunke, E.-G.; Claude, H.; et al. Recent Tropospheric Ozone Changes—A Pattern Dominated by Slow or No Growth. Atmos. Environ. 2013, 67, 331–351. [Google Scholar] [CrossRef]
- Paudel, R.; Grantz, D.A.; Vu, H.-B.; Shrestha, A. Tolerance of Elevated Ozone and Water Stress in a California Population of Palmer Amaranth (Amaranthus Palmeri). Weed Sci. 2016, 64, 276–284. [Google Scholar] [CrossRef]
- Gandin, A.; Davrinche, A.; Jolivet, Y. Deciphering the Main Determinants of O3 Tolerance in Euramerican Poplar Genotypes. Sci. Total Environ. 2019, 656, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Khaling, E.; Papazian, S.; Poelman, E.H.; Holopainen, J.K.; Albrectsen, B.R.; Blande, J.D. Ozone Affects Growth and Development of Pieris Brassicae on the Wild Host Plant Brassica nigra. Environ. Pollut. 2015, 199, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Takahama, U.; Oniki, T. A Peroxidase/Phenolics/Ascorbate System Can Scavenge Hydrogen Peroxide in Plant Cells. Physiol. Plant. 1997, 101, 845–852. [Google Scholar] [CrossRef]
- Reich, P.B. Effects of Low Concentrations of O3 on Net Photosynthesis, Dark Respiration, and Chlorophyll Contents in Aging Hybrid Poplar Leaves. Plant Physiol. 1983, 73, 291–296. [Google Scholar] [CrossRef]
- Pell, E.J.; Schlagnhaufer, C.D.; Arteca, R.N. Ozone-Induced Oxidative Stress: Mechanisms of Action and Reaction. Physiol. Plant. 1997, 100, 264–273. [Google Scholar] [CrossRef]
- Jolivet, Y.; Bagard, M.; Cabané, M.; Vaultier, M.-N.; Gandin, A.; Afif, D.; Dizengremel, P.; Le Thiec, D. Deciphering the Ozone-Induced Changes in Cellular Processes: A Prerequisite for Ozone Risk Assessment at the Tree and Forest Levels. Ann. For. Sci. 2016, 73, 923–943. [Google Scholar] [CrossRef]
- Escarré, J.; Lepart, J.; Sentuc, J.J. Effects of Simulated Herbivory in Three Old Field Compositae with Different Inflorescence Architectures. Oecologia 1996, 105, 501–508. [Google Scholar] [CrossRef]
- Mithofer, A.; Wanner, G.; Boland, W. Effects of Feeding Spodoptera Littoralis on Lima Bean Leaves. II. Continuous Mechanical Wounding Resembling Insect Feeding Is Sufficient to Elicit Herbivory-Related Volatile Emission. Plant Physiol. 2005, 137, 1160–1168. [Google Scholar] [CrossRef] [PubMed]
- Constabel, C.P.; Yip, L.; Patton, J.J.; Christopher, M.E. Polyphenol Oxidase from Hybrid Poplar. Cloning and Expression in Response to Wounding and Herbivory. Plant Physiol. 2000, 124, 285–295. [Google Scholar] [CrossRef]
- Li, G.; Bartram, S.; Guo, H.; Mithöfer, A.; Kunert, M.; Boland, W. SpitWorm, an Herbivorous Robot: Mechanical Leaf Wounding with Simultaneous Application of Salivary Components. bioRxiv 2018. [Google Scholar] [CrossRef]
- Zangerl, A.R. Furanocoumarin Induction in Wild Parsnip: Evidence for an Induced Defense against Herbivores. Ecology 1990, 71, 1926–1932. [Google Scholar] [CrossRef]
- Berenbaum, M.R.; Zangerl, A.R. Chemical Phenotype Matching between a Plant and Its Insect Herbivore. Proc. Natl. Acad. Sci. USA 1998, 95, 13743–13748. [Google Scholar] [CrossRef] [PubMed]
- Likic, S.; Rusak, G. Changes in Phenolic Compounds in Nicotiana Species as a Response to Wounding and Viral Infection. J. Plant Pathol. 2014, 96, 569–575. [Google Scholar]
- Tzin, V.; Malitsky, S.; Aharoni, A.; Galili, G. Expression of a Bacterial Bi-Functional Chorismate Mutase/Prephenate Dehydratase Modulates Primary and Secondary Metabolism Associated with Aromatic Amino Acids in Arabidopsis. Plant J. 2009, 60, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.-X.; Li, J.-X.; Fang, X.; Wang, L.-J.; Hu, W.-L.; Chen, X.-Y.; Yang, C.-Q. Isolation and Characterization of Three New Monoterpene Synthases from Artemisia Annua. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lincoln, D.E. Effects of Light Intensity and Artificial Wounding on Monoterpene Production in Myrica Cerifera from Two Different Ecological Habitats. Can. J. Bot. 2004, 82, 1501–1508. [Google Scholar] [CrossRef]
- Lee, S.; Suh, S.; Kim, S.; Crain, R.C.; Kwak, J.M.; Nam, H.-G.; Lee, Y. Systemic Elevation of Phosphatidic Acid and Lysophospholipid Levels in Wounded Plants. Plant J. 1997, 12, 547–556. [Google Scholar] [CrossRef]
- Ryu, S.B.; Wang, X. Activation of Phospholipase D and the Possible Mechanism of Activation in Wound-Induced Lipid Hydrolysis in Castor Bean Leaves. Biochim. Biophys. Acta (BBA)—Lipids Lipid Metab. 1996, 1303, 243–250. [Google Scholar] [CrossRef]
- Zhang, M.; Demeshko, Y.; Dumbur, R.; Iven, T.; Feussner, I.; Lebedov, G.; Ganim, M.; Barg, R.; Ben-Hayyim, G. Elevated α-Linolenic Acid Content in Extra-Plastidial Membranes of Tomato Accelerates Wound-Induced Jasmonate Generation and Improves Tolerance to the Herbivorous Insects Heliothis peltigera and Spodoptera littoralis. J. Plant Growth Regul. 2019, 38, 723–738. [Google Scholar] [CrossRef]
- Marchica, A.; Lorenzini, G.; Papini, R.; Bernardi, R.; Nali, C.; Pellegrini, E. Signalling Molecules Responsive to Ozone-Induced Oxidative Stress in Salvia officinalis. Sci. Total Environ. 2019, 657, 568–576. [Google Scholar] [CrossRef]
- Savatin, D.V.; Gramegna, G.; Modesti, V.; Cervone, F. Wounding in the Plant Tissue: The Defense of a Dangerous Passage. Front. Plant Sci. 2014, 5, 470. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.S.; Shiva, S.; Roth, M.R.; Tamura, P.; Zheng, L.; Li, M.; Sarowar, S.; Honey, S.; McEllhiney, D.; Hinkes, P.; et al. Lipid Changes after Leaf Wounding in Arabidopsis thaliana: Expanded Lipidomic Data Form the Basis for Lipid Co-Occurrence Analysis. Plant J. 2014, 80, 728–743. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.S.; Roston, R.; Shiva, S.; Hur, M.; Wurtele, E.S.; Wang, X.; Shah, J.; Welti, R. Modifications of Membrane Lipids in Response to Wounding of Arabidopsis thaliana Leaves. Plant Signal. Behav. 2015, 10, e1056422. [Google Scholar] [CrossRef] [PubMed]
- Zangerl, A.R.; Berenbaum, M.R. Damage-Inducibility of Primary and Secondary Metabolites in the Wild Parsnip (Pastinaca sativa). Chemoecology 1998, 8, 187–193. [Google Scholar] [CrossRef]
| O3/Control | MW/Control | |||
|---|---|---|---|---|
| Down | Up | Down | Up | |
| T0 | 1 | 1 | 1 | 0 |
| T1 | 19 | 42 | 17 | 111 |
| T2 | 19 | 6 | 41 | 304 |
| T3 | 10 | 16 | 82 | 185 |
| Group | Feature Name | FC O3/Control | FC MW/Control | Ion Adduct | RT (min) | [M + H] + m/z | Molecular Weight | Molecular Formula | Assigned Compound | Classyfire Class | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T1 | T2 | T3 | |||||||||
| A | M293T11 | _ | _ | _ | _ | _ | 0.4 | [M + Na]+ | 10.56 | 293.078 | 270.28 | C16H14O4 | Imperatorin | Coumarins and derivatives |
| A | M595T8 | _ | _ | _ | _ | _ | 0.7 | [M + H]+ | 7.55 | 595.165 | 594.52 | C27H30O15 | Kaempferol 3-rhamnoside-7-glucoside | Flavonoids |
| A | M611T6 | _ | _ | _ | _ | _ | 0.5 | [M + H]+ | 6.43 | 611.160 | 610.52 | C27H30O16 | Rutin | Flavonoids |
| A | M627T5 | _ | _ | _ | _ | _ | 0.6 | [M + H]+ | 5.09 | 627.154 | 626.52 | C27H30O17 | Quercetin 3, 4-diglucoside | Flavonoids |
| A | M625T8 | 0.5 | 0.6 | _ | _ | _ | 0.5 | [M + H]+ | 7.75 | 625.178 | 624.54 | C28H32O16 | Myricetin 7-methyl ether 3,4’-di-O-alpha-L-rhamnopyranoside | Flavonoids |
| A | M757T6 | _ | 0.5 | _ | _ | 0.7 | 0.5 | [M + H]+ | 6.46 | 757.218 | 756.66 | C33H40O20 | Quercetin-3-O-alpha-L-rhamnopyranosyl(1-2)-beta-D-glucopyranoside-7-O-alpha-L-rhamnopyranoside | Flavonoids |
| A | M751T13 | _ | _ | _ | _ | 0.4 | 0.3 | [M + Na]+ | 13.37 | 751.422 | 728.99 | C43H68O9 | CID 11104554 | Fatty Acyls |
| A | M585T16 | _ | 0.7 | _ | _ | 0.2 | 0.4 | [M + H]+ | 16.01 | 585.447 | 584.87 | C37H60O5 | DG(20:5(5Z,8Z,11Z,14Z,17Z)/14:1(9Z)/0:0) | Glycerolipids |
| A | M583T15 | _ | _ | _ | _ | _ | 0.5 | [M + Na]+ | 14.58 | 583.435 | 560.84 | C35H60O5 | DG(14:1(9Z)/18:3(9Z,12Z,15Z)/0:0) | Glycerolipids |
| A | M769T16 | _ | _ | _ | _ | 0.3 | 0.4 | [M + Na]+ | 16.00 | 769.482 | 747.01 | C43H70O10 | 18:3/16:3-MGD | Glycerolipids |
| A | M783T15 | _ | _ | _ | _ | 0.5 | 0.4 | [M + Na]+ | 14.71 | 783.460 | 760.99 | C43H68O11 | oxy phytodienoic acid/16:3-MGD | Glycerolipids |
| B | M677T8 | 3.0 | 2.2 | 0.4 | _ | 2.1 | 1.6 | [M + H]+ | 8.43 | 677.263 | 676.72 | C38H36N4O8 | (2S,2(1)R)-2(1),2(2)-dicarboxy-8-ethenyl-2,7,12,18-tetramethyl-2,2(1)-dihydrobenzo[b]porphyrin-13,17-dipropanoic acid | Tetrapyrroles and derivatives |
| B | M661T9 | 3.9 | 2.6 | _ | _ | 2.4 | _ | [M + H]+ | 8.67 | 661.283 | 660.76 | C42H36N4O4 | Putative chlorophyll catabolite | Tetrapyrroles and derivatives |
| B | M707T9 | 3.2 | 2.9 | 0.4 | _ | 2.7 | 2.2 | [M + H]+ | 9.45 | 707.293 | 706.74 | C36H42N4O11 | Putative chlorophyll catabolite | Tetrapyrroles and derivatives |
| B | M693T9 | 3.0 | 2.4 | _ | _ | 2.1 | _ | [M + H]+ | 9.14 | 693.277 | 692.71 | C35H40N4O11 | Putative chlorophyll catabolite | Tetrapyrroles and derivatives |
| B | M691T10 | 2.9 | 2.5 | 0.6 | _ | 3.1 | 2.4 | [M + H]+ | 9.52 | 691.261 | 690.70 | C35H38N4O11 | Putative chlorophyll catabolite | Tetrapyrroles and derivatives |
| C | M135T11 | _ | _ | _ | 3.2 | 6.9 | 4.2 | [M + H]+ | 10.64 | 135.116 | 134.22 | C10H14 | p-cymene | Prenol lipids |
| C | M151T11 | _ | _ | _ | 2.6 | 3.9 | 2.0 | [M + H]+ | 10.74 | 151.111 | 150.22 | C10H14O | Thymol | Prenol lipids |
| C | M387T6 | _ | _ | _ | 1.7 | 2.2 | 1.9 | [M + Na]+ | 5.60 | 387.066 | 364.30 | C17H16O9 | Xanthotoxol glucoside | Coumarins and derivatives |
| C | M205T4 | _ | _ | _ | _ | 1.8 | _ | [M + H]+ | 4.20 | 205.096 | 204.23 | C11H12N2O2 | Tryptophan | Indoles and derivatives |
| C | M197T7 | _ | _ | _ | 1.8 | [M + H]+ | 6.86 | 197.116 | 196.24 | C11H16O3 | 4-(3-hydroxybutyl)-2-methoxyphenol | Phenols | ||
| C | M267T11 | _ | _ | _ | _ | 3.2 | 3.1 | [M + Na]+ | 10.73 | 245.116 | 244.28 | C15H16O3 | Osthole | Coumarins and derivatives |
| C | M335T10 | _ | _ | _ | _ | 8.4 | 6.2 | [M + H]+ | 9.57 | 335.127 | 334.36 | C21H18O4 | Anhydronotoptol derivative 1 | Coumarins and derivatives |
| C | M335T12 | _ | _ | _ | 2.2 | 10.2 | 2.5 | [M + H]+ | 12.01 | 335.127 | 334.36 | C21H18O4 | Anhydronotoptol derivative 2 | Coumarins and derivatives |
| C | M337T11 | _ | _ | _ | 2.9 | 6.3 | 3.0 | [M + H]+ | 10.62 | 337.143 | 336.38 | C21H20O4 | Anhydronotoptol | Coumarins and derivatives |
| C | M353T10 | _ | _ | _ | 3.4 | 17.4 | 11.0 | [M + H]+ | 9.63 | 353.136 | 352.38 | C21H20O5 | Lansiumarin A derivative 1 | Coumarins and derivatives |
| C | M353T11 | _ | _ | _ | _ | 7.9 | 2.3 | [M + H]+ | 10.72 | 353.138 | 352.38 | C21H20O5 | Lansiumarin A derivative 2 | Coumarins and derivatives |
| C | M339T13 | _ | _ | _ | _ | 1.7 | 2.9 | [M + H]+ | 12.68 | 339.159 | 338.39 | C21H22O4 | 8-geranyloxy psoralen | Coumarins and derivatives |
| C | M613T15 | _ | _ | _ | _ | 2.7 | 6.2 | [M + H]+ | 14.66 | 613.480 | 612.92 | C39H64O5 | DG(18:3(9Z,12Z,15Z)/18:3(9Z,12Z,15Z)/0:0) | Glycerolipids |
| C | M277T12 | _ | _ | _ | 2.5 | 4.3 | 3.3 | [M + H]+ | 12.33 | 277.213 | 276.41 | C18H28O2 | Stearidonic acid | Fatty Acyls |
| C | M279T12 | _ | _ | _ | _ | 6.0 | 5.7 | [M + H]+ | 12.18 | 279.231 | 278.43 | C18H30O2 | Linolenic acid | Fatty Acyls |
| C | M279T13 | _ | _ | _ | _ | 5.4 | _ | [M + H]+ | 12.80 | 279.231 | 278.43 | C18H30O2 | Linolenic acid | Fatty Acyls |
| C | M283T14 | _ | _ | _ | 3.6 | 14.3 | 15.0 | [M + H]+ | 13.22 | 283.262 | 282.46 | C18H34O2 | Oleic acid | Fatty Acyls |
| C | M295T12 | _ | _ | _ | 3.0 | 17.5 | 21.4 | [M + H]+ | 11.81 | 295.226 | 294.42 | C18H30O3 | 13-HOTE | Fatty Acyls |
| C | M496T14 | _ | _ | _ | 3.0 | 4.8 | 2.7 | [M + H]+ | 14.08 | 496.338 | 495.63 | C24H50NO7P | LysoPC(16:0) | Glycerophospholipids |
| C | M518T12 | _ | _ | _ | _ | _ | 4.9 | [M + H]+ | 11.68 | 518.320 | 517.63 | C26H48NO7P | LysoPC(18:3) | Glycerophospholipids |
| C | M534T12 | _ | _ | _ | 3.3 | 7.2 | 5.2 | [M + H]+ | 11.66 | 534.318 | 533.67 | C27H52NO7P | LysoPC(18:2/0:0) | Glycerophospholipids |
| C | M536T12 | _ | _ | _ | 4.0 | 11.1 | 6.5 | [M + H]+ | 11.66 | 536.334 | 535.65 | C26H50NO8P | PC(16:1(9Z)/2:0) | Glycerophospholipids |
| C | M791T16 | _ | _ | _ | 2.0 | 3.8 | 2.9 | [M + H]+ | 15.53 | 790.558 | 790.06 | C42H80NO10P | PS(18:0/18:1(9Z)) | Glycerophospholipids |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galati, G.; Gandin, A.; Jolivet, Y.; Larbat, R.; Hehn, A. Untargeted Metabolomics Approach Reveals Diverse Responses of Pastinaca Sativa to Ozone and Wounding Stresses. Metabolites 2019, 9, 153. https://doi.org/10.3390/metabo9070153
Galati G, Gandin A, Jolivet Y, Larbat R, Hehn A. Untargeted Metabolomics Approach Reveals Diverse Responses of Pastinaca Sativa to Ozone and Wounding Stresses. Metabolites. 2019; 9(7):153. https://doi.org/10.3390/metabo9070153
Chicago/Turabian StyleGalati, Gianni, Anthony Gandin, Yves Jolivet, Romain Larbat, and Alain Hehn. 2019. "Untargeted Metabolomics Approach Reveals Diverse Responses of Pastinaca Sativa to Ozone and Wounding Stresses" Metabolites 9, no. 7: 153. https://doi.org/10.3390/metabo9070153
APA StyleGalati, G., Gandin, A., Jolivet, Y., Larbat, R., & Hehn, A. (2019). Untargeted Metabolomics Approach Reveals Diverse Responses of Pastinaca Sativa to Ozone and Wounding Stresses. Metabolites, 9(7), 153. https://doi.org/10.3390/metabo9070153





