Drying Enhances Signal Intensities for Global GC–MS Metabolomics
Abstract
1. Introduction
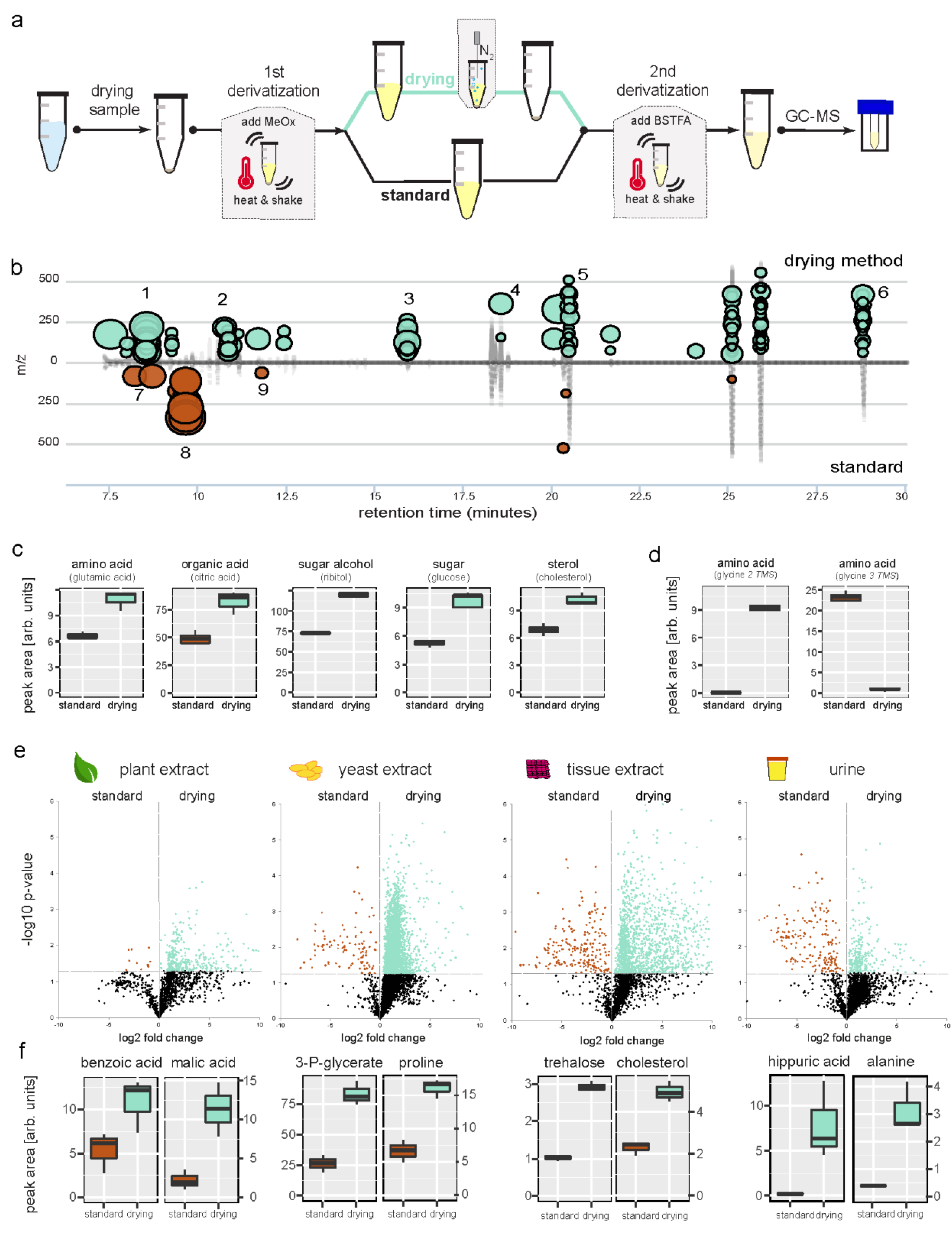
2. Results
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Metabolite Standard Mixture
4.3. Metabolite Extraction
4.4. Derivatization Procedures
4.5. GC–MS Method
4.6. GC–MS Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Anwar, M.A.; Vorkas, P.A.; Li, J.V.; Shalhoub, J.; Want, E.J.; Davies, A.H.; Holmes, E. Optimization of metabolite extraction of human vein tissue for ultra performance liquid chromatography-mass spectrometry and nuclear magnetic resonance-based untargeted metabolic profiling. Analyst 2015, 140, 7586–7597. [Google Scholar] [CrossRef] [PubMed]
- Geier, F.M.; Want, E.J.; Leroi, A.M.; Bundy, J.G. Cross-platform comparison of Caenorhabditis elegans tissue extraction strategies for comprehensive metabolome coverage. Anal. Chem. 2011, 83, 3730–3736. [Google Scholar] [CrossRef] [PubMed]
- Kuehnbaum, N.L.; Britz-McKibbin, P. New advances in separation science for metabolomics: resolving chemical diversity in a post-genomic era. Chem. Rev. 2013, 113, 2437–2468. [Google Scholar] [CrossRef] [PubMed]
- Zaikin, V.G.; Halket, J.M. Review: derivatization in mass spectrometry-6. Formation of mixed derivatives of polyfunctional compounds. Eur. J. Mass Spectrom. (Chichester) 2005, 11, 611–636. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.; Steinhauser, D.; Strelkov, S.; Schomburg, D.; Allison, G.; Moritz, T.; Lundgren, K.; Roessner-Tunali, U.; Forbes, M.G.; Willmitzer, L.; et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005, 579, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: the golm metabolome database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Bassey, E.; Bishop, A.C.; Kusel, D.T.; Cox, L.A.; Olivier, M. High Resolution GC/MS Metabolomics of Non-Human Primate Serum. Rapid Commun. Mass Spectrom. 2018, 32, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Koek, M.M.; van der Kloet, F.M.; Kleemann, R.; Kooistra, T.; Verheij, E.R.; Hankemeier, T. Semi-automated non-target processing in GC × GC-MS metabolomics analysis: applicability for biomedical studies. Metabolomics 2011, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Sobolevsky, T.; Revelsky, A.I.; Miller, B.; Oriedo, V.; Chernetsova, E.S.; Revelsky, I.A. Comparison of silylation and esterification/acylation procedures in GC-MS analysis of amino acids. J. Sep. Sci. 2003, 26, 1474–1478. [Google Scholar] [CrossRef]
- Ruiz-Matute, A.I.; Hernández-Hernández, O.; Rodríguez-Sánchez, S.; Sanz, M.L.; Martínez-Castro, I. Derivatization of carbohydrates for GC and GC-MS analyses. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 1226–1240. [Google Scholar] [CrossRef] [PubMed]
- Little, J.L. Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. J. Chromatogr. A 1999, 844, 1–22. [Google Scholar] [CrossRef]
- Drozd, J. Chemical Derivatization in Gas Chromatography, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1986. [Google Scholar]
- Lees, H.J.; Swann, J.R.; Wilson, I.D.; Nicholson, J.K.; Holmes, E. Hippurate: the natural history of a mammalian-microbial cometabolite. J. Proteome Res. 2013, 12, 1527–1546. [Google Scholar] [CrossRef]
- Tian, J.; Chen, G.; He, Z. Overcoming matrix effects: GC method development for the determination of triethylamine and dimethyl sulfoxide in a drug substance. J. Chromatogr. Sci. 2014, 52, 36–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chan, E.C.; Pasikanti, K.K.; Nicholson, J.K. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat. Protoc. 2011, 6, 1483–1499. [Google Scholar] [CrossRef] [PubMed]
- Liebeke, M.; Strittmatter, N.; Fearn, S.; Morgan, A.J.; Kille, P.; Fuchser, J.; Wallis, D.; Palchykov, V.; Robertson, J. Unique metabolites protect earthworms against plant polyphenols. Nat. Commun. 2015, 6, 7869. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Salek, R.M.; Conesa, P.; Hastings, J.; de Matos, P.; Rijnbeek, M.; Mahendraker, T.; Williams, M.; Neumann, S.; Rocca-Serra, P. MetaboLights―an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013, 41, D781–D786. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liebeke, M.; Puskás, E. Drying Enhances Signal Intensities for Global GC–MS Metabolomics. Metabolites 2019, 9, 68. https://doi.org/10.3390/metabo9040068
Liebeke M, Puskás E. Drying Enhances Signal Intensities for Global GC–MS Metabolomics. Metabolites. 2019; 9(4):68. https://doi.org/10.3390/metabo9040068
Chicago/Turabian StyleLiebeke, Manuel, and Erik Puskás. 2019. "Drying Enhances Signal Intensities for Global GC–MS Metabolomics" Metabolites 9, no. 4: 68. https://doi.org/10.3390/metabo9040068
APA StyleLiebeke, M., & Puskás, E. (2019). Drying Enhances Signal Intensities for Global GC–MS Metabolomics. Metabolites, 9(4), 68. https://doi.org/10.3390/metabo9040068





