Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines
Abstract
1. Introduction
2. Results
2.1. Analysis of MiaPaCa-2 Cells
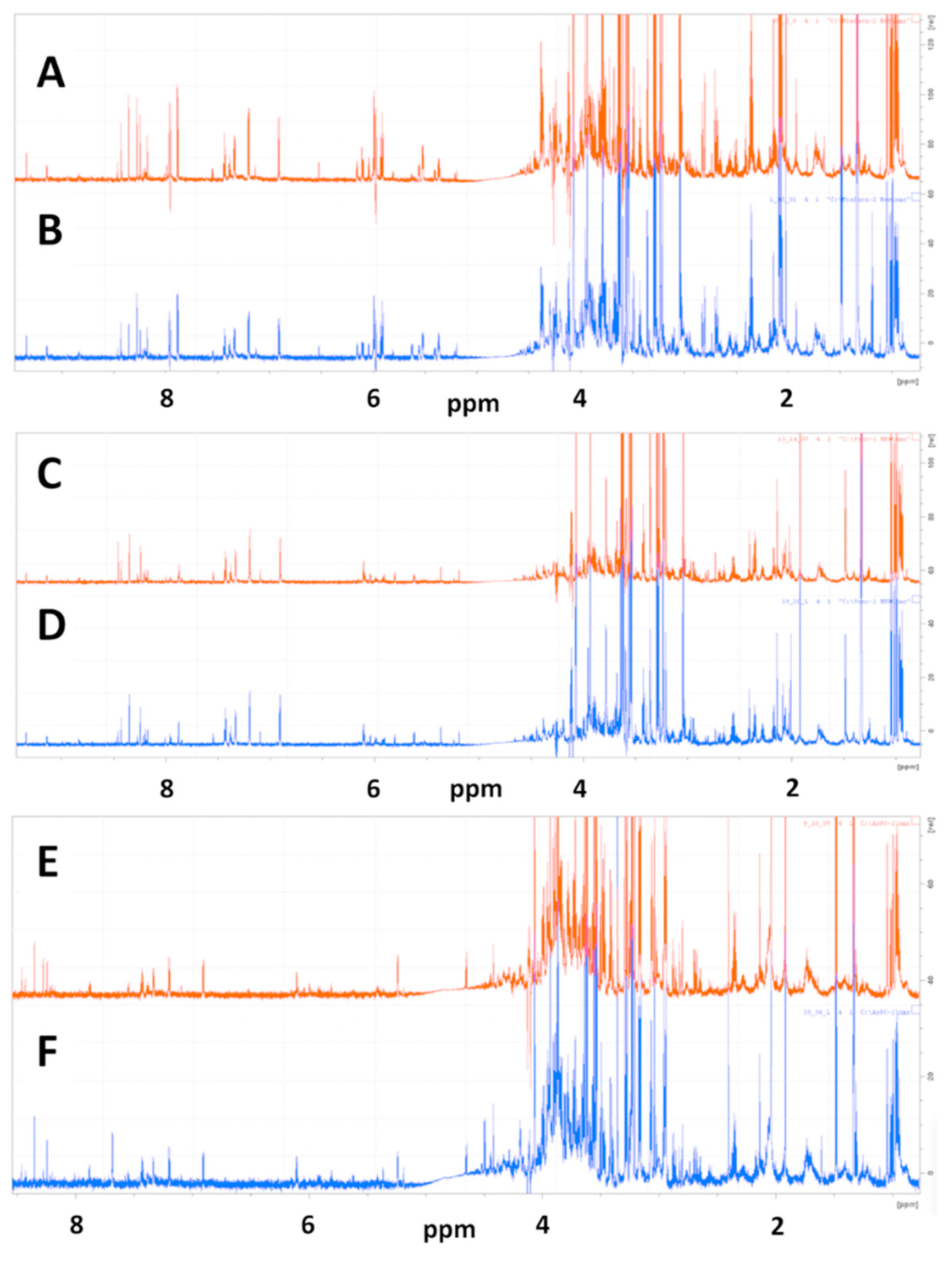
2.1.1. Representative NMR Data
2.1.2. Unsupervised Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) of MiaPaCa-2 Cells
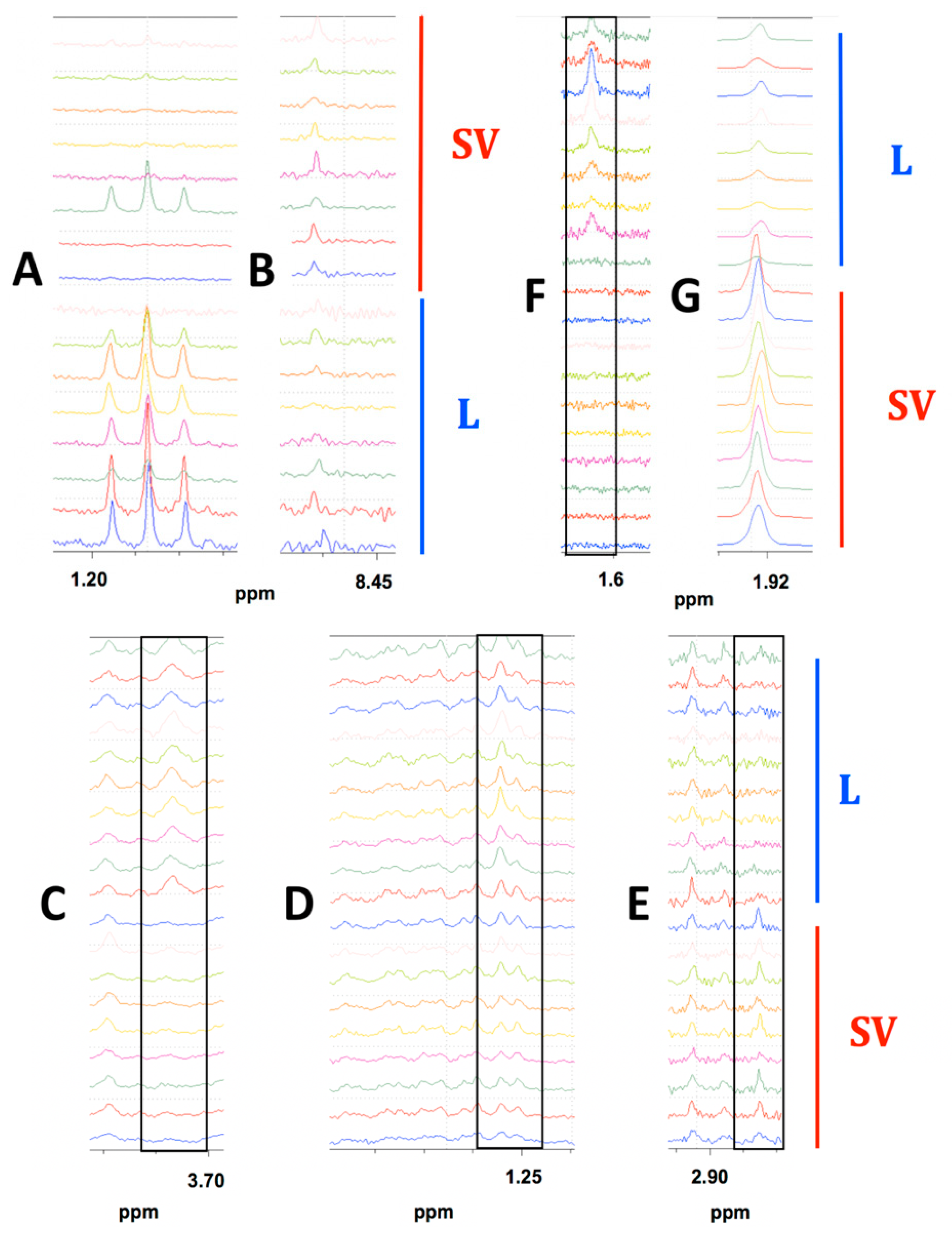
2.1.3. Prominent Differences in Metabolite Abundances Depending on Drying Method
2.2. Analysis of Panc-1 Cells
2.2.1. Representative NMR Data
2.2.2. Unsupervised Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) of Panc-1 Cells
2.2.3. Prominent Differences in Metabolite Abundances Depending on Drying Method
2.3. Analysis of AsPC-1 Cells
2.3.1. Representative NMR Data
2.3.2. Unsupervised Principal Component Analysis (PCA) and Partial Least Squares-Discriminant Analysis (PLS-DA) of AsPC-1 Cells
2.3.3. Prominent Differences in Metabolite Abundances Depending on Drying Method
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Preparation of Cell Extracts
4.2. Drying Methods
4.3. Preparation of Samples for NMR Data Collection
4.4. NMR Data Collection
4.5. Data Analysis
4.6. Metabolite Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. Metabonomics in pharmaceutical R & D. FEBS J. 2007, 274, 1140–1151. [Google Scholar] [PubMed]
- Robertson, D.G. Metabonomics in Toxicology: A Review. Toxicol. Sci. 2005, 85, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. BioMed Res. Int. 2015, 2015, 1–6. [Google Scholar] [CrossRef]
- Oldiges, M.; Lütz, S.; Pflug, S.; Schroer, K.; Stein, N.; Wiendahl, C. Metabolomics: Current state and evolving methodologies and tools. Appl. Microbiol. Biotechnol. 2007, 76, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; Van Der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef]
- Guasch-Ferre, M.; Bhupathiraju, S.N.; Hu, F.B. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin. Chem. 2018, 64, 82–98. [Google Scholar] [CrossRef]
- Brouwer-Brolsma, E.M.; Brennan, L.; Drevon, C.A.; Van Kranen, H.; Manach, C.; Dragsted, L.O.; Roche, H.M.; Andres-Lacueva, C.; Bakker, S.J.L.; Bouwman, J.; et al. Combining traditional dietary assessment methods with novel metabolomics techniques: Present efforts by the Food Biomarker Alliance. Proc. Nutr. Soc. 2017, 76, 619–627. [Google Scholar] [CrossRef]
- Gibbons, H.; Brennan, L. Metabolomics as a tool in the identification of dietary biomarkers. Proc. Nutr. Soc. 2017, 76, 42–53. [Google Scholar] [CrossRef]
- Bouhifd, M.; Hartung, T.; Hogberg, H.T.; Kleensang, A.; Zhao, L. Review: Toxicometabolomics. J. Appl. Toxicol. 2013, 33, 1365–1383. [Google Scholar] [CrossRef] [PubMed]
- Manchester, M.; Anand, A. Metabolomics: Strategies to Define the Role of Metabolism in Virus Infection and Pathogenesis. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 98, pp. 57–81. [Google Scholar]
- Romick-Rosendale, L.E.; Goodpaster, A.M.; Hanwright, P.J.; Patel, N.B.; Wheeler, E.T.; Chona, D.L.; Kennedy, M.A. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril). Magn. Reson. Chem. 2009, 47, S36–S46. [Google Scholar] [CrossRef] [PubMed]
- Romick-Rosendale, L.E.; Legomarcino, A.; Patel, N.B.; Morrow, A.L.; Kennedy, M.A. Prolonged antibiotic use induces intestinal injury in mice that is repaired after removing antibiotic pressure: Implications for empiric antibiotic therapy. Metabolomics 2014, 10, 8–20. [Google Scholar] [CrossRef]
- Rosendale, L.E.R.; Schibler, K.R.; Kennedy, M.A. A Potential Biomarker for Acute Kidney Injury in Preterm Infants from Metabolic Profiling. J. Mol. Biomark. Diagn. 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Chihanga, T.; Ma, Q.; Nicholson, J.D.; Ruby, H.N.; Edelmann, R.E.; Devarajan, P.; Kennedy, M.A. NMR spectroscopy and electron microscopy identification of metabolic and ultrastructural changes to the kidney following ischemia-reperfusion injury. Am. J. Physiol.-Ren. Physiol. 2018, 314, F154–F166. [Google Scholar] [CrossRef]
- Chihanga, T.; Ruby, H.N.; Ma, Q.; Bashir, S.; Devarajan, P.; Kennedy, M.A. NMR-based urine metabolic profiling and immunohistochemistry analysis of nephron changes in a mouse model of hypoxia-induced acute kidney injury. Am. J. Physiol. Physiol. 2018, 315, F1159–F1173. [Google Scholar] [CrossRef]
- Liu, X.J.; Locasale, J.W. Metabolomics: A Primer. Trends Biochem. Sci. 2017, 42, 274–284. [Google Scholar] [CrossRef]
- Watanabe, M.; Sheriff, S.; Ramelot, T.A.; Kadeer, N.; Cho, J.; Lewis, K.B.; Balasubramaniam, A.; Kennedy, M.A. NMR Based Metabonomics Study of DAG Treatment in a C2C12 Mouse Skeletal Muscle Cell Line Myotube Model of Burn-Injury. Int. J. Pept. Res. Ther. 2011, 17, 281–299. [Google Scholar] [CrossRef]
- Watanabe, M.; Sheriff, S.; Kadeer, N.; Cho, J.; Lewis, K.B.; Balasubramaniam, A.; Kennedy, M.A. NMR based metabonomics study of NPY Y5 receptor activation in BT-549, a human breast carcinoma cell line. Metabolomics 2012, 8, 854–868. [Google Scholar] [CrossRef]
- Watanabe, M.; Sheriff, S.; Lewis, K.B.; Cho, J.; Tinch, S.L.; Balasubramaniam, A.; Kennedy, M.A. Metabolic Profiling Comparison of Human Pancreatic Ductal Epithelial Cells and Three Pancreatic Cancer Cell Lines using NMR Based Metabonomics. J. Mol. Biomark. Diagn. 2012, 3, 1–17. [Google Scholar] [CrossRef]
- Nichols, A.C.; Black, M.; Yoo, J.; Pinto, N.; Fernandes, A.; Haibe-Kains, B.; Boutros, P.C.; Barrett, J.W. Exploiting high-throughput cell line drug screening studies to identify candidate therapeutic agents in head and neck cancer. BMC Pharmacol. Toxicol. 2014, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.L.; Bodmer, W. Cancer Cell Lines for Drug Discovery and Development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Burgess, K.; Rankin, N.; Weidt, S. Chapter 10-Metabolomics. In Handbook of Pharmacogenomics and Stratified Medicine; Padmanabhan, S., Ed.; Academic Press: San Diego, CA, USA, 2014; pp. 181–205. [Google Scholar]
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Lenz, E.M.; Wilson, I.D. Analytical Strategies in Metabonomics. J. Proteome Res. 2007, 6, 443–458. [Google Scholar] [CrossRef]
- Morgan, H.R. Latent viral infection of cells in tissue culture. I. Studies on latent infection of chick embryo tissues with psittacosis virus. J. Exp. Med. 1956, 103, 37–47. [Google Scholar] [CrossRef]
- Hayton, S.; Maker, G.L.; Mullaney, I.; Trengove, R.D. Experimental design and reporting standards for metabolomics studies of mammalian cell lines. Cell. Mol. Life Sci. 2017, 74, 4421–4441. [Google Scholar] [CrossRef]
- Halama, A. Metabolomics in cell culture—A strategy to study crucial metabolic pathways in cancer development and the response to treatment. Arch. Biochem. Biophys. 2014, 564, 100–109. [Google Scholar] [CrossRef]
- Kumar, A.; Misra, B.B. Challenges and Opportunities in Cancer Metabolomics. Proteomics 2019, e1900042. [Google Scholar] [CrossRef]
- Čuperlović-Culf, M.; Barnett, D.A.; Culf, A.S.; Chute, I. Cell culture metabolomics: Applications and future directions. Drug Discov. Today 2010, 15, 610–621. [Google Scholar] [CrossRef]
- Sato, G. Tissue culture: The unrealized potential. Cytotechnology 2008, 57, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Sano, D.; Pickering, C.R.; Jasser, S.A.; Henderson, Y.C.; Clayman, G.L.; Sturgis, E.M.; Ow, T.J.; Lotan, R.; Carey, T.E.; et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin. Cancer Res. 2011, 17, 7248–7264. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Leclercq, G. Relevance of Breast Cancer Cell Lines as Models for Breast Tumours: An Update. Breast Cancer Res. Treat. 2004, 83, 249–289. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Girard, L.; Lockwood, W.W.; Lam, W.L.; Minna, J.D. Lung Cancer Cell Lines as Tools for Biomedical Discovery and Research. J. Natl. Cancer Inst. 2010, 102, 1310–1321. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, A.-H.; Liu, S.-B.; Qiu, S.; Li, X.-N.; Zhang, T.-L.; Liu, L.; Wang, X.-J. Cell metabolomics identify regulatory pathways and targets of magnoline against prostate cancer. J. Chromatogr. 2018, 1102, 143–151. [Google Scholar] [CrossRef]
- Scheff, N.N.; Ye, Y.; Bhattacharya, A.; Macrae, J.; Hickman, D.N.; Sharma, A.K.; Dolan, J.C.; Schmidt, B.L.; Hickman, D.H. Tumor necrosis factor alpha secreted from oral squamous cell carcinoma contributes to cancer pain and associated inflammation. Pain 2017, 158, 2396–2409. [Google Scholar] [CrossRef]
- Han, S.E.; Park, K.-H.; Lee, G.; Huh, Y.-J.; Min, B.-M. Mutation and aberrant expression of Caveolin-1 in human oral squamous cell carcinomas and oral cancer cell lines. Int. J. Oncol. 2004, 24, 435–440. [Google Scholar] [CrossRef]
- Mohamadi, N.; Kazemi, S.M.; Mohammadian, M.; Toofani Milani, A.; Moradi, Y.; Yasemi, M. Toxicity of Cisplatin-Loaded Poly Butyl Cyanoacrylate Nanoparticles in a Brain Cancer Cell Line: Anionic Polymerization Results. Asian Pac. J. Cancer Prev. 2017, 18, 629–632. [Google Scholar]
- Fujita, M.; Imadome, K.; Imai, T. Metabolic characterization of invaded cells of the pancreatic cancer cell line, PANC-1. Cancer Sci. 2017, 108, 961–971. [Google Scholar] [CrossRef]
- Zhang, A.H.; Sun, H.; Xu, H.Y.; Qiu, S.; Wang, X.J. Cell Metabolomics. Omics 2013, 17, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kapoore, R.V.; Coyle, R.; Staton, C.A.; Brown, N.J.; Vaidyanathan, S. Cell line dependence of metabolite leakage in metabolome analyses of adherent normal and cancer cell lines. Metabolomics 2015, 11, 1743–1755. [Google Scholar] [CrossRef]
- Sapcariu, S.C.; Kanashova, T.; Weindl, D.; Ghelfi, J.; Dittmar, G.; Hiller, K. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 2014, 1, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Sellick, C.A.; Knight, D.; Croxford, A.S.; Maqsood, A.R.; Stephens, G.M.; Goodacre, R.; Dickson, A.J. Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: Matching extraction approaches to cell type and metabolite targets. Metabolomics 2010, 6, 427–438. [Google Scholar] [CrossRef]
- Sheikh, K.D.; Khanna, S.; Byers, S.W.; Fornace, A.J.; Cheema, A.K. Small molecule metabolite extraction strategy for improving LC/MS detection of cancer cell metabolome. J. Biomol. Tech. JBT 2011, 22, 1–4. [Google Scholar]
- Canavan, H.E.; Cheng, X.; Graham, D.J.; Ratner, B.D.; Castner, D.G. Cell sheet detachment affects the extracellular matrix: A surface science study comparing thermal liftoff, enzymatic, and mechanical methods. J. Biomed. Mater. Res. Part A 2005, 75, 1–13. [Google Scholar] [CrossRef]
- Masson, P.; Alves, A.C.; Ebbels, T.M.D.; Nicholson, J.K.; Want, E.J.; Alves, A. Optimization and Evaluation of Metabolite Extraction Protocols for Untargeted Metabolic Profiling of Liver Samples by UPLC-MS. Anal. Chem. 2010, 82, 7779–7786. [Google Scholar] [CrossRef]
- Tambellini, N.P.; Zaremberg, V.; Turner, R.J.; Weljie, A.M. Evaluation of Extraction Protocols for Simultaneous Polar and Non-Polar Yeast Metabolite Analysis Using Multivariate Projection Methods. Metabolites 2013, 3, 592–605. [Google Scholar] [CrossRef]
- Chihanga, T.; Hausmann, S.M.; Ni, S.; Kennedy, M.A. Influence of media selection on NMR based metabolic profiling of human cell lines. Metabolomics 2018, 14, 28. [Google Scholar] [CrossRef]
- Villas-Bôas, S.G.; Koulman, A.; Lane, G.A. Analytical methods from the perspective of method standardization. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin, Germany, 2007; Volume 18, pp. 11–52. [Google Scholar]
- The Standard Metabolic Reporting Structures working group; Lindon, J.C.; Nicholson, J.K.; Holmes, E.; Keun, H.C.; Craig, A.; Pearce, J.T.M.; Bruce, S.J.; Hardy, N.; Sansone, S.-A.; et al. Summary recommendations for standardization and reporting of metabolic analyses. Nat. Biotechnol. 2005, 23, 833–838. [Google Scholar]
- van der Werf, M.J.; Takors, R.; Smedsgaard, J.; Nielsen, J.; Ferenci, T.; Portais, J.C.; Wittmann, C.; Hooks, M.; Tomassini, A.; Oldiges, M.; et al. Standard reporting requirements for biological samples in metabolomics experiments: Microbial and in vitro biology experiments. Metabolomics 2007, 3, 189–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alvarez-Sanchez, B.; Priego-Capote, F.; de Castro, M.D.L. Metabolomics analysis II. Preparation of biological samples prior to detection. TrAC Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- León, Z.; García-Cañaveras, J.C.; Donato, M.T.; Lahoz, A. Mammalian cell metabolomics: Experimental design and sample preparation. Electrophoresis 2013, 34, 2762–2775. [Google Scholar] [CrossRef]
- Åkesson, M.; Smedsgaard, J.; Nielsen, J.; Villas-Bôas, S.G.; Højer-Pedersen, J. Global metabolite analysis of yeast: Evaluation of sample preparation methods. Yeast 2005, 22, 1155–1169. [Google Scholar]
- Schmahl, M.J.; Regan, D.P.; Rivers, A.C.; Joesten, W.C.; Kennedy, M.A. NMR-based metabolic profiling of urine, serum, fecal, and pancreatic tissue samples from the Ptf1a-Cre; LSL-KrasG12D transgenic mouse model of pancreatic cancer. PLoS ONE 2018, 13, e0200658. [Google Scholar] [CrossRef]
- Wang, B.; Sheriff, S.; Balasubramaniam, A.; Kennedy, M.A. NMR based metabolomics study of Y2 receptor activation by neuropeptide Y in the SK-N-BE2 human neuroblastoma cell line. Metabolomics 2015, 11, 1243–1252. [Google Scholar] [CrossRef]
- Goodpaster, A.M.; Romick-Rosendale, L.E.; Kennedy, M.A. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal. Biochem. 2010, 401, 134–143. [Google Scholar] [CrossRef]
- Goodpaster, A.M.; Kennedy, M.A. Quantification and statistical significance analysis of group separation in NMR-based metabonomics studies. Chemom. Intell. Lab. Syst. 2011, 109, 162–170. [Google Scholar] [CrossRef]
- Hyberts, S.G.; Arthanari, H.; Wagner, G. Applications of non-uniform sampling and processing. Top. Curr. Chem. 2012, 316, 125–148. [Google Scholar]
- Delaglio, F.; Grzesiek, S.; Zhu, G.; Pfeifer, J.; Bax, A.; Vuister, G.; Vuister, G.W. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Ulrich, E.L.; Akutsu, H.; Doreleijers, J.F.; Harano, Y.; Ioannidis, Y.E.; Lin, J.; Livny, M.; Mading, S.; Maziuk, D.; Miller, Z.; et al. BioMagResBank. Nucleic Acids Res. 2008, 36, D402–D408. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Watson, M.S.; Boyko, R.F.; Sykes, B.D. Automated H-1 and C-13 chemical shift prediction using the BioMagResBank. J. Biomol. NMR 1997, 10, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Bingol, K.; Li, D.-W.; Zhang, B.; Brüschweiler, R. Comprehensive Metabolite Identification Strategy Using Multiple Two-Dimensional NMR Spectra of a Complex Mixture Implemented in the COLMARm Web Server. Anal. Chem. 2016, 88, 12411–12418. [Google Scholar] [CrossRef]
- Bingol, K.; Bruschweiler-Li, L.; Li, D.-W.; Brüschweiler, R. Customized Metabolomics Database for the Analysis of NMR 1H–1H TOCSY and 13C–1H HSQC-TOCSY Spectra of Complex Mixtures. Anal. Chem. 2014, 86, 5494–5501. [Google Scholar] [CrossRef]
- Joesten, W.C.; Kennedy, M.A. RANCM: A new ranking scheme for assigning confidence levels to metabolite assignments in NMR-based metabolomics studies. Metabolomics 2019, 15, 5. [Google Scholar] [CrossRef]

| Only in Lyophilized | More Abundant in Lyophilized Cell Extracts | More Abundant in Speed-Vacuum Dried Extracts |
|---|---|---|
| 3-Hydroxyisovaleric acid 3 | 1,3,7-trimethyluric acid13 | Acetate 2,3 |
| N-methyl-α-aminoisobutyric acid 1,2,3 | Creatinine13 | 1,9-Dimethyluric acid 1,3 |
| N-Methylnicotinamide 3 | Methanol13 | Cytidine Triphosphate 3 |
| Sarcosine 3 | 3-Cresotinic acid 3 | Glucose-6-phosphate 3 |
| Leucine 2 | Threonine 3 | Glucose 3 |
| Ethanol 1 | 1,3-Dihydroxyacetone 3 | 1-methyladenosine 1,3 |
| Only in Speed-Vacuum | l-Aspartic acid 1,3 | Carnosine 3 |
| Trimethylamine 2 | 2-phosphoglycerate 3 | l-Threonine 3 |
| l-Methionine 1,3 | Tyramine 3 | |
| Creatine 2,3 | Mannose-6-phosphate 3 | |
| Glycerophosphocholine 1,3 | Uracil 3 | |
| Butanone 3 | D-Alanine13 | |
| Methylcysteine 3 | Glycine 3 | |
| Leucine 1,3 | l-Lactic acid 3 | |
| Capric acid 3 | l-Valine 3 | |
| Galactaric acid 3 | Isoleucine 3 | |
| Phosphocreatine 3 | l-Phenylalanine 3 | |
| Pyruvate 3 | Glycerate 3 | |
| Uridine diphosphategalactose 1,2 | N-Carbamoyl-β-alanine 3 | |
| Glucaric acid 1 | Glycerol 3 | |
| Threonic acid 1 | Trimethylamine 1 | |
| Uracil 1 | Fumaric acid 1 | |
| Acetylglycine 1 | Formic acid 1,2 | |
| l-Phenylalanine 1,2 | 1-Butanol 1 | |
| creatine phosphate 1,2 | 2-hydroxybutyrate 1 | |
| 2,3-Diphosphoglyceric acid 1 | Taurine 1 | |
| l-Tyrosine 1,2 | 1-Methyluric acid 1 | |
| l-Isoleucine 1,2 | NAD+1 | |
| Uridine 1 | l-Malic acid 1 | |
| Inosine 1,2 | Methanol 2 | |
| S-Adenosylhomocysteine 1 | l-Aspartic acid 2 | |
| 1-Butanol 2 | Uracil 2 | |
| S-Adenosylhomocysteine 1,2 | Acetylcholine 2 | |
| Histidine 2 | l-Methionine 2 | |
| Glutathione 1,2 | Glycerophosphocholine 2 | |
| Methionine sulfoxide 1 | Phosphoenolpyruvic acid 2 | |
| Phosphorylcholine 1 | 1-methylguanosine 2 | |
| Cellobiose 1 | 3-Methyladipic acid 2 | |
| 3-Hydroxyisovaleric acid 1 | Oxypurinol 2 | |
| l-Valine 1,2 | Adenine 2 | |
| Pyroglutamate 1 | Guanosine 2 | |
| Acetylphosphate 1 | ||
| Glucose 1,2 | ||
| Glucose-6-phosphate 1 | ||
| Myoinositol 1,2 | ||
| 1-Methylguanine 1 | ||
| 1,1-Dimethylbiguanide 1 | ||
| Pyruvatoxime 1,2 | ||
| l-lactic acid 1,2 | ||
| 3,7-Dimethyluric acid 2 | ||
| Succinic acid 2 | ||
| Adenosine phosphosulfate 2 | ||
| Mannose 6-phosphate 2 | ||
| 3-Phosphoglyceric acid 2 | ||
| NAD+2 | ||
| N-acetylneuraminic acid 2 | ||
| Lysine 2 | ||
| 5-aminolevulinate 2 | ||
| Trimethylamine N-oxide 2 | ||
| Taurine 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrova, I.; Xu, S.; Joesten, W.C.; Ni, S.; Kennedy, M.A. Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines. Metabolites 2019, 9, 256. https://doi.org/10.3390/metabo9110256
Petrova I, Xu S, Joesten WC, Ni S, Kennedy MA. Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines. Metabolites. 2019; 9(11):256. https://doi.org/10.3390/metabo9110256
Chicago/Turabian StylePetrova, Irina, Shenyuan Xu, William C. Joesten, Shuisong Ni, and Michael A. Kennedy. 2019. "Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines" Metabolites 9, no. 11: 256. https://doi.org/10.3390/metabo9110256
APA StylePetrova, I., Xu, S., Joesten, W. C., Ni, S., & Kennedy, M. A. (2019). Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines. Metabolites, 9(11), 256. https://doi.org/10.3390/metabo9110256






